Proteomic Analysis of the Effect of Salmonella Challenge on Broiler Chicken
Abstract
1. Introduction
2. Results
2.1. Performance Parameters and Concentration of SE in Ceca of Broiler-Chicken
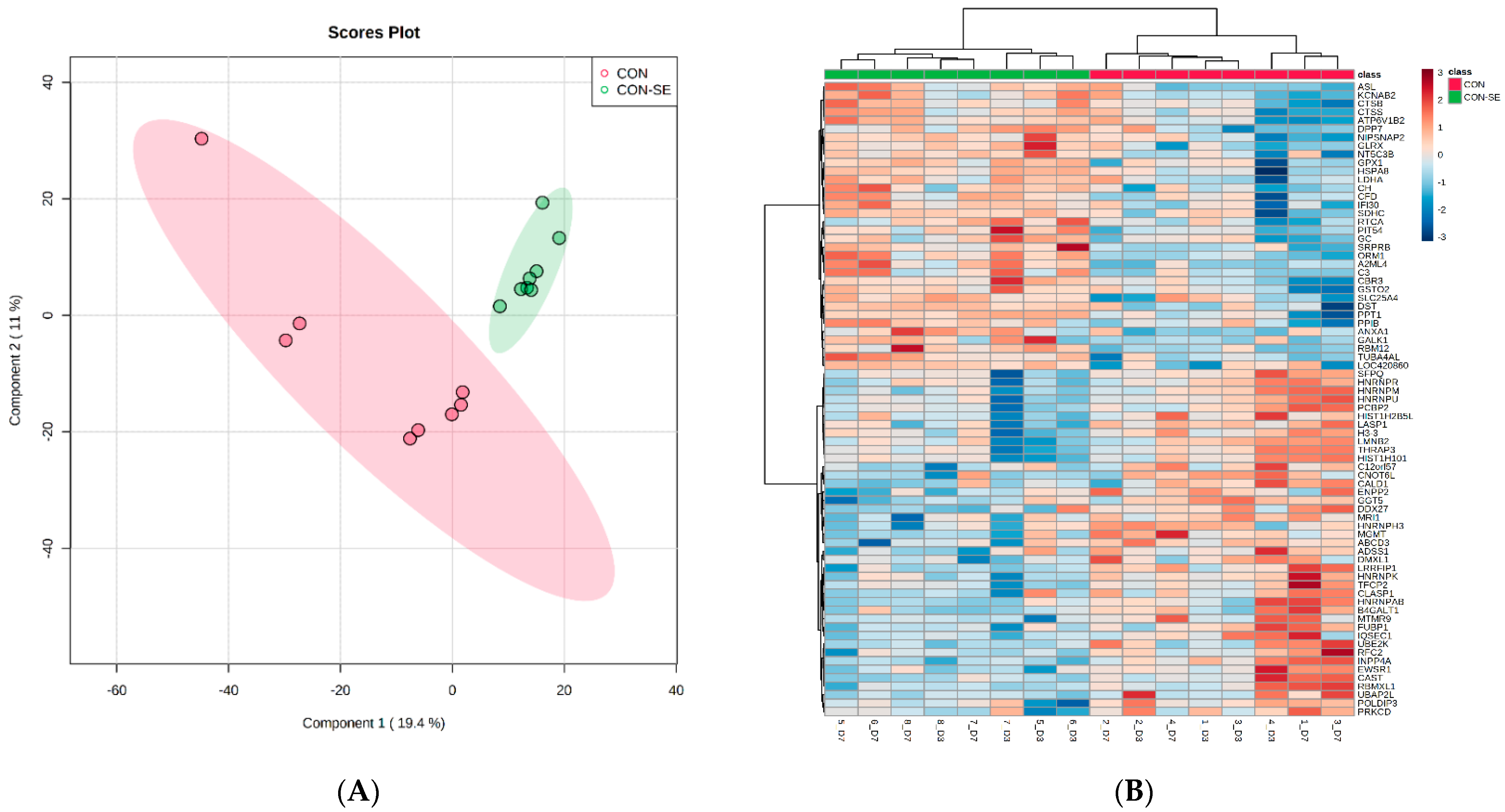
2.2. Correlation Coefficients and Number of Differentially Abundant Proteins between Time Points and Groups
2.3. Differences of Protein Abundances between Time Points
2.4. Effect of Salmonella Challenge on Protein Abundance
2.5. Differences in Proteins Abundance due to Salmonella Challenge within Day 3 and Day 7
2.6. Effect of Salmonella Challenge on Protein Abundance
2.7. Differences in Proteins Abundance Due to Salmonella Challenge within Day 3 and Day 7
3. Discussion
4. Materials and Methods
4.1. Experiment Design, and Salmonella Strain used for the Experiment
4.2. Growth Performance Evaluation and Spleen Sample Collection
4.3. Protein Extraction and Proteomic Analysis
4.4. LC-MS/MS Analysis
4.5. Data Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Denagamage, T.N.; Jayarao, B.M.; Wallner-Pendleton, E.; Patterson, P.H.; Kariyawasam, S. A Retrospective Study of Salmonella Enteritidis Isolated from Commercial Layer Flocks. Avian Dis. 2017, 61, 330–334. [Google Scholar] [CrossRef] [PubMed]
- Ehuwa, O.; Jaiswal, A.K.; Jaiswal, S. Salmonella, Food Safety and Food Handling Practices. Foods 2021, 10, 907. [Google Scholar] [CrossRef] [PubMed]
- Heredia, N.; García, S. Animals as sources of food-borne pathogens: A review. Anim. Nutr. 2018, 4, 250–255. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Wang, H.; Li, T.; Liu, F.; Cheng, Y.; Guo, X.; Wen, G.; Luo, Q.; Shao, H.; Pan, Z. Characterization of Salmonella spp. isolated from chickens in Central China. BMC Vet. Res. 2020, 16, 299. [Google Scholar] [CrossRef] [PubMed]
- Shah, D.H.; Casavant, C.; Hawley, Q.; Addwebi, T.; Call, D.R.; Guard, J. Salmonella Enteritidis Strains from Poultry Exhibit Differential Responses to Acid Stress, Oxidative Stress, and Survival in the Egg Albumen. Foodborne Pathog. Dis. 2012, 9, 258–264. [Google Scholar] [CrossRef] [PubMed]
- Gast, R.K.; Jones, D.R.; Guraya, R.; Anderson, K.E.; Karcher, D.M. Research Note: Contamination of eggs by Salmonella Enteritidis and Salmonella Typhimurium in experimentally infected laying hens in indoor cage-free housing. Poult. Sci. 2021, 100, 101438. [Google Scholar] [CrossRef]
- Jeurissen, S.H.M. Structure and function of the chicken spleen. Res. Immunol. 1991, 142, 352–355. [Google Scholar] [CrossRef]
- Elsharkawy, M.S.; Wang, H.; Ding, J.; Madkour, M.; Wang, Q.; Zhang, Q.; Zhang, N.; Li, Q.; Zhao, G.; Wen, J. Transcriptomic Analysis of the Spleen of Different Chicken Breeds Revealed the Differential Resistance of Salmonella Typhimurium. Genes 2022, 13, 811. [Google Scholar] [CrossRef]
- Barrow, P.A.; Bumstead, N.; Martson, K.; Lovell, M.A.; Wigley, P. Faecal shedding and intestinal colonization of Salmonella enterica in in-bred chickens: The effect of host-genetic background. Epidemiol. Infect. 2004, 132, 117–126. [Google Scholar] [CrossRef]
- Fasina, Y.O.; Obanla, T.O.; Ferket, P.R.; Shah, D.H. Comparative efficacy of spray-dried plasma and bacitracin methylene disalicylate in reducing cecal colonization by Salmonella Enteritidis in broiler chickens. Poult. Sci. 2021, 100, 101134. [Google Scholar] [CrossRef]
- Yan, G.L.; Guo, Y.M.; Yuan, J.M.; Liu, D.; Zhang, B.K. Sodium alginate oligosaccharides from brown algae inhibit Salmonella Enteritidis colonization in broiler chickens. Poult. Sci. 2011, 90, 1441–1448. [Google Scholar] [CrossRef] [PubMed]
- Neyt, K.; Perros, F.; GeurtsvanKessel, C.H.; Hammad, H.; Lambrecht, B.N. Tertiary lymphoid organs in infection and autoimmunity. Trends Immunol. 2012, 33, 297–305. [Google Scholar] [CrossRef]
- Dally, S.; Bredoux, R.; Corvazier, E.; Andersen, J.P.; Clausen, J.D.; Dode, L.; Fanchaouy, M.; Gelebart, P.; Monceau, V.; Del Monte, F. Ca2+-ATPases in non-failing and failing heart: Evidence for a novel cardiac sarco/endoplasmic reticulum Ca2+-ATPase 2 isoform (SERCA2c). Biochem. J. 2006, 395, 249–258. [Google Scholar] [CrossRef] [PubMed]
- He, G.Z.; Tian, W.Y.; Qian, N.; Cheng, A.C.; Deng, S.X. Quantitative studies of the distribution pattern for Salmonella Enteritidis in the internal organs of chicken after oral challenge by a real-time PCR. Vet. Res. Commun. 2010, 34, 669–676. [Google Scholar] [CrossRef] [PubMed]
- Brostrom, C.O.; Brostrom, M.A. Calcium-Dependent Regulation of Protein Synthesis in Intact Mammalian Cells. Annu. Rev. Physiol. 1990, 52, 577–590. [Google Scholar] [CrossRef] [PubMed]
- Shigene, K.; Hiasa, Y.; Otake, Y.; Soufi, M.; Janewanthanakul, S.; Nishimura, T.; Sato, Y.; Suetsugu, S. Translation of Cellular Protein Localization Using Convolutional Networks. Front. Cell Dev. Biol. 2021, 9, 635231. [Google Scholar] [CrossRef]
- Labib, K.; Kearsey, S.E.; Diffley, J.F.X. MCM2–7 Proteins Are Essential Components of Prereplicative Complexes that Accumulate Cooperatively in the Nucleus during G1-phase and Are Required to Establish, But Not Maintain, the S-phase Checkpoint. Mol. Biol. Cell 2001, 12, 3658–3667. [Google Scholar] [CrossRef]
- Mori, Y.; Inoue, Y.; Taniyama, Y.; Tanaka, S.; Terada, Y. Phosphorylation of the centrosomal protein, Cep169, by Cdk1 promotes its dissociation from centrosomes in mitosis. Biochem. Biophys. Res. Commun. 2015, 468, 642–646. [Google Scholar] [CrossRef]
- Ye, W.; Zhang, L. Heme controls the expression of cell cycle regulators and cell growth in HeLa cells. Biochem. Biophys. Res. Commun. 2004, 315, 546–554. [Google Scholar] [CrossRef]
- Azenabor, A.A.; Mahony, J.B. Generation of reactive oxygen species and formation of membrane lipid peroxides in cells infected with Chlamydia trachomatis. Int. J. Infect. Dis. 2000, 4, 46–50. [Google Scholar] [CrossRef]
- Brigelius-Flohé, R. Glutathione peroxidases and redox-regulated transcription factors. Biol. Chem. 2006, 387, 1329–1335. [Google Scholar] [CrossRef] [PubMed]
- Kato, T.; Ono, H.; Fujii, M.; Akahoshi, K.; Ogura, T.; Ogawa, K.; Ban, D.; Kudo, A.; Tanaka, S.; Tanabe, M. Cytoplasmic RRM1 activation as an acute response to gemcitabine treatment is involved in drug resistance of pancreatic cancer cells. PLoS ONE 2021, 16, e0252917. [Google Scholar] [CrossRef] [PubMed]
- Kang, S.W.; Baines, I.C.; Rhee, S.G. Characterization of a Mammalian Peroxiredoxin That Contains One Conserved Cysteine. J. Biol. Chem. 1998, 273, 6303–6311. [Google Scholar] [CrossRef] [PubMed]
- Rojkind, M.; Domínguez-Rosales, J.-A.; Nieto, N.; Greenwel, P. Role of hydrogen peroxide and oxidative stress in healing responses. CMLS Cell. Mol. Life Sci. 2002, 59, 1872–1891. [Google Scholar] [CrossRef]
- Lu, J.Y.; Verkruyse, L.A.; Hofmann, S.L. Lipid thioesters derived from acylated proteins accumulate in infantile neuronal ceroid lipofuscinosis: Correction of the defect in lymphoblasts by recombinant palmitoyl-protein thioesterase. Proc. Natl. Acad. Sci. USA 1996, 93, 10046–10050. [Google Scholar] [CrossRef]
- Lawrence, R.E.; Zoncu, R. The lysosome as a cellular centre for signalling, metabolism and quality control. Nat. Cell Biol. 2019, 21, 133–142. [Google Scholar] [CrossRef]
- Régnier, C.H.; Song, H.Y.; Gao, X.; Goeddel, D.V.; Cao, Z.; Rothe, M. Identification and Characterization of an IκB Kinase. Cell 1997, 90, 373–383. [Google Scholar] [CrossRef]
- Buttgereit, F.; Burmester, G.-R.; Brand, M.D. Bioenergetics of immune functions: Fundamental and therapeutic aspects. Immunol. Today 2000, 21, 194–199. [Google Scholar] [CrossRef]
- Kawai, T.; Akira, S. TLR signaling. Cell Death Differ. 2006, 13, 816–825. [Google Scholar] [CrossRef]
- Toney, M.D. Aspartate aminotransferase: An old dog teaches new tricks. Arch. Biochem. Biophys. 2014, 544, 119–127. [Google Scholar] [CrossRef]
- Chang, Y.-C.; Yang, Y.-C.; Tien, C.-P.; Yang, C.-J.; Hsiao, M. Roles of Aldolase Family Genes in Human Cancers and Diseases. Trends Endocrinol. Metab. 2018, 29, 549–559. [Google Scholar] [CrossRef] [PubMed]
- Broberg, S.; Sahlin, K. Adenine nucleotide degradation in human skeletal muscle during prolonged exercise. J. Appl. Physiol. 1989, 67, 116–122. [Google Scholar] [CrossRef] [PubMed]
- Garay-Arroyo, A.; Covarrubias, A.A. Three genes whose expression is induced by stress inSaccharomyces cerevisiae. Yeast 1999, 15, 879–892. [Google Scholar] [CrossRef]
- Hille, B. Ionic channels in excitable membranes. Current problems and biophysical approaches. Biophys. J. 1978, 22, 283–294. [Google Scholar] [CrossRef]
- Eisenreich, W.; Heesemann, J.; Rudel, T.; Goebel, W. Metabolic Adaptations of Intracellullar Bacterial Pathogens and their Mammalian Host Cells during Infection (“Pathometabolism”). In Metabolism and Bacterial Pathogenesis; John Wiley & Sons: Hoboken, NJ, USA, 2015; pp. 27–58. [Google Scholar]
- Waltman, W.D.; Mallinson, E.T. Isolation of Salmonella from Poultry Tissue and Environmental Samples: A Nationwide Survey. Avian Dis. 1995, 39, 45–54. [Google Scholar] [CrossRef] [PubMed]
- Fasina, Y.O.; Bowers, J.B.; Hess, J.B.; McKee, S.R. Effect of dietary glutamine supplementation on Salmonella colonization in the ceca of young broiler chicks. Poult. Sci. 2010, 89, 1042–1048. [Google Scholar] [CrossRef]
- Scheltema, R.A.; Hauschild, J.-P.; Lange, O.; Hornburg, D.; Denisov, E.; Damoc, E.; Kuehn, A.; Makarov, A.; Mann, M. The Q Exactive HF, a Benchtop Mass Spectrometer with a Pre-filter, High-performance Quadrupole and an Ultra-high-field Orbitrap Analyzer. Mol. Cell. Proteom. 2014, 13, 3698–3708. [Google Scholar] [CrossRef]
- Mi, H.; Ebert, D.; Muruganujan, A.; Mills, C.; Albou, L.-P.; Mushayamaha, T.; Thomas, P.D. PANTHER version 16: A revised family classification, tree-based classification tool, enhancer regions and extensive API. Nucleic Acids Res. 2021, 49, D394–D403. [Google Scholar] [CrossRef]
- Safran, M.; Rosen, N.; Twik, M.; BarShir, R.; Stein, T.I.; Dahary, D.; Fishilevich, S.; Lancet, D. The GeneCards Suite. In Practical Guide to Life Science Databases; Springer: Singapore, 2021; pp. 27–56. [Google Scholar]

| Log10 CFU/g Cecal Contents | ||
|---|---|---|
| Treatment | Day 3 PC | Day 7 PC |
| CON | ND | ND |
| CON−SE | 5.68 ± 0.36 a | 4.17 ± 0.18 b |
| p-value | <0.0001 | <0.0001 |
| Comparison | Total | Increased | Decreased |
|---|---|---|---|
| Timepoints (D3-D7) | 360 | 235 | 125 |
| CON-CON−SE | 216 | 105 | 111 |
| CONSE@D3 | 106 | 110 | |
| CONSE@D7 | 104 | 112 |
| Term | % | p-Value | Genes |
|---|---|---|---|
| TIME | |||
| Proteins that increased between time points (D3 and D7) | |||
| Spliceosome | 5.5 | 2.2 × 10−5 | SRSF6, EFTUD2, SF3B1, RBM25, FUS, ALYREF, SRSF7, HNRNPK, SF3A3, DHX15, SF3B3, FUSSNW1, LSM5 |
| Actin filament binding | 5.5 | 7.5 × 10−5 | CORO1C, BIN1, ACTR2, FMNL1, TLN1, CAPB, TPM1, SCIN, TPM3, MYH9, ACTR2, MYO1F, HCLS1 |
| ATP binding | 13.1 | 1.7 × 10−2 | DDX17, TOP1, DDX1, CAMK2D, TOP2B, DDX18, ACTR2, ATP2A3, SMC3, EHD4, UBE2N, SYK, PRKCB, SMARCA5, SWAP70, DARS, PAK2, PKN2, RPS6KA3, NSF, LONP1, VPS4B, MYH9, DHX15, CAMK2D, ATP2A2, PIP4K2A, DNAJA4, MYO1F, LOC107051177 |
| Proteins that decreased between time points (D3 and D7) | |||
| Cell cycle | 3.2 | 1.9 × 10−1 | MCM6, MCM4, CDK1, MCM5 |
| Heme binding | 4.0 | 2.4 × 10−2 | HBE1, HBE, CAT, CYGB, HMOX1 |
| Salmonella Challenge | |||
| Proteins that increased due to Salmonella Challenge | |||
| Biosynthesis of amino acids | 5.7 | 5.9 × 10−4 | GOT1, ALDOC, ASS1, ASL, TKTL1, GOT2 |
| Glutathione metabolism | 5.7 | 2.5 × 10−4 | RRM1, GSTO2, GCLC, PRDX6, MGST1, GPX1 |
| Response to oxidative stress | 3.8 | 6.6 × 10−3 | GCLC, PRDX6, SLC25A4, GPX1 |
| D-threo-aldose 1-dehydrogenase activity | 2.9 | 6.4 × 10−3 | KCNAB2, LOC418170, AKR1E2 |
| Lysosome | 7.6 | 8.0 × 10−5 | PPT1, CTSS, PRDX6, CTSH, IFI30, HSPA8, CTSB, NPC2 |
| Unfolded protein binding | 5.7 | 2.4 × 10−4 | HSPA8, CALR, CALR3, CALR, PTGES3, HSP90AA1 |
| Proteins that decreased due to Salmonella Challenge | |||
| Actin filament binding | 5.4 | 1.3 × 10−2 | NUR2L, TPM1, MYH9, LASP1, TPM4, TPM3 |
| RNA binding | 18.0 | 2.7 × 10−9 | HNRNPM, POLDIP3, RBM23, RPL19, PCBP2, HNRNPAB, HTATSF1, SRSF6, SFPQ, HNRNPK, HNRNPA3, RBMXL1, NONO, RPL11, HNRNPR, HNRNPH3, HNRNPU, MBNL1, ATXN2, EWSR1, FUBP1 |
| Proteins increased in the CON−SE group at D3 | |||
| Biosynthesis of amino acids | 5.7 | 5.9 × 10−4 | GOT1, ALDOC, ASS1, ASL, TKTL1, GOT2 |
| Response to oxidative stress | 3.8 | 6.6 × 10−3 | BCLC, PRDX6, SLC25A4, GPX1 |
| Glutathione metabolism | 5.7 | 2.5 × 10−4 | RRM1, GSTO2, GCLC, PRDX6, MGST1, GPX1 |
| D-threo-aldose 1-dehydrogenase activity | 2.9 | 6.4 × 10−3 | KCNAB2, LOC418170, AKR1E2 |
| Lysosome | 7.6 | 8.0 × 10−5 | PPT1, CTSS, PRDX6, CTSH, IFI30, HSPA8, CTSB, NPC2 |
| Apoptosis | 5.7 | 1.5 × 10−2 | TUBA4AL, CTSS, CTSH, CTSB, FADD, CHUK |
| Proteins decreased in the CON−SE group at D3 | |||
| RNA binding | 17.7 | 3.8 × 10−9 | HNRNPM, POLDIP3, RBM23, RPL19, PCBP2, HNRNPAB, HTATSF1, SRSF6, SFPQ, HNRNPK, HNRNPA3, RBMXL1, NONO, RPL11, HNRNPR, HNRNPH3, HNRNPU, MBNL1, ATXN2, EWSR1, FUBP1 |
| Stress fiber | 4.4 | 5.9 × 10−4 | TPM1, MYH9, PDLIM7, TPM4, TPM3 |
| Proteins increased in the CON−SE group at D7 | |||
| Biosynthesis of amino acids | 5.9 | 5.0 × 10−4 | GOT1, ALDOC, ASS1, ASL, TKTL1, GOT2 |
| Glutathione metabolism | 5.9 | 2.1 × 10−4 | RRM1, GSTO2, GCLC, PRDX6, MGST1, GPX1 |
| Lysosome | 7.8 | 6.5 × 10−5 | PPT1, CTSS, PRDX6, CTSH, IFI30, HSPA8, CTSB, NPC2 |
| Apoptosis | 5.9 | 1.3 × 10−2 | TUBA4AL, CTSS, CTSH, CTSB, FADD, CHUK |
| Proteins decreased in the CON−SE group at D7 | |||
| RNA binding | 17.7 | 3.8 × 10−9 | HNRNPM, POLDIP3, RBM23, RPL19, PCBP2, HNRNPAB, HTATSF1, SRSF6, SFPQ, HNRNPK, HNRNPA3, RBMXL1, NONO, RPL11, HNRNPR, HNRNPH3, HNRNPU, MBNL1, ATXN2, EWSR1, FUBP1 |
| Stress fiber | 4.4 | 5.9 × 10−4 | TPM1, MYH9, PDLIM7, TPM4, TPM3 |
| Composition of Starter Diets (D1 to 14) | |
|---|---|
| Ingredients | Quantity |
| Corn (7.5% Crude protein) | 51.46 |
| Soybean meal (47.5% Crude Protein) | 40.39 |
| Poultry fat | 3.64 |
| Limestone | 1.07 |
| Mono-Dicalcium phosphate | 2.03 |
| Salt NaCl | 0.40 |
| Sodium bicarbonate | 0.02 |
| L-Lysine HCl 98% | 0.13 |
| DL-Methionine 99.0% | 0.34 |
| L-Threonine 98.5% | 0.11 |
| NCSU Poultry Vitamin Premix2 | 0.05 |
| NCSU Poultry Mineral Premix3 | 0.20 |
| Choline chloride 60% | 0.10 |
| Selenium Premix | 0.05 |
| Analyzed nutrient composition | |
| Metabolizable energy (Kcal/kg) | 3,117 |
| Crude Protein, % | 24.63 |
| Crude Fat, % | 4.74 |
| Crude Fiber, % | 2.3 |
| Ash, % | 6.32 |
| Calculated nutrient composition | |
| Total Sulfur Amino Acids, % | 1.03 |
| Lysine, % | 1.42 |
| Calcium, % | 0.96 |
| Available phosphorus, % | 0.48 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Adetunji, A.; Casey, T.; Franco, J.; Shah, D.; Fasina, Y. Proteomic Analysis of the Effect of Salmonella Challenge on Broiler Chicken. Molecules 2022, 27, 7277. https://doi.org/10.3390/molecules27217277
Adetunji A, Casey T, Franco J, Shah D, Fasina Y. Proteomic Analysis of the Effect of Salmonella Challenge on Broiler Chicken. Molecules. 2022; 27(21):7277. https://doi.org/10.3390/molecules27217277
Chicago/Turabian StyleAdetunji, Adedeji, Theresa Casey, Jackeline Franco, Devendra Shah, and Yewande Fasina. 2022. "Proteomic Analysis of the Effect of Salmonella Challenge on Broiler Chicken" Molecules 27, no. 21: 7277. https://doi.org/10.3390/molecules27217277
APA StyleAdetunji, A., Casey, T., Franco, J., Shah, D., & Fasina, Y. (2022). Proteomic Analysis of the Effect of Salmonella Challenge on Broiler Chicken. Molecules, 27(21), 7277. https://doi.org/10.3390/molecules27217277







