Abstract
Nardosinone, a sesquiterpene peroxide, is one of the main active constituents of the ethnomedicine Nardostachyos Radix et Rhizoma, and it has many bioactivities, such as antiarrhythmia and cardioprotection. To elucidate its in vivo existence forms, its metabolism is first studied using mice. All urine and feces are collected during the six days of oral dosing of nardosinone, and blood is collected at one hour after the last dose. Besides, to validate some metabolites, a fast experiment is performed, in which nardosinone was orally administered and the subsequent one-hour urine is collected and immediately analyzed by UHPLC-Q-TOF-MS. In total, 76 new metabolites are identified in this study, including 39, 51, and 12 metabolites in urine, plasma, and feces, respectively. Nardosinone can be converted into nardosinone acid or its isomers. The metabolic reactions of nardosinone included hydroxylation, hydrogenation, dehydration, glucuronidation, sulfation, demethylation, and carboxylation. There are 56 and 20 metabolites with the structural skeleton of nardosinone and nardosinone acid, respectively. In total, 77 in vivo existence forms of nardosinone are found in mice. Nardosinone is mainly excreted in urine and is not detected in the feces. These findings will lay the foundation for further research of the in vivo effective forms of nardosinone and Nardostachyos Radix et Rhizoma.
1. Introduction
Nardosinone is a sesquiterpene peroxide and exists in a variety of medicinal plants, such as Nardostachys jatamansi DC. [1], Millettia speciosa [2], Sargassum fusiforme [3], Taraxacum kok-saghyz Rodin [4], Chrysanthemum morifolium [5]. In addition, nardosinone was also detected in Mahuang Fuzi Xixin decoction [6]. Nardosinone is mainly derived from Nardostachys jatamansi DC. Nardostachyos Radix et Rhizoma (NRR), the root and rhizome of Nardostachys jatamansi DC., has the traditional pharmacological action of regulating qi, relieving pain, and relieving stagnation-syndrome, and is often used as the raw material for spices, food and cosmetics [7], and contains multiple chemical constituents, such as terpenes, flavonoids, coumarins, lignans, and volatile oils [8], of which sesquiterpenoids (e.g., nardosinone) are the main active components [9]. Pharmacological studies have shown that NRR has pharmacological effects of improving headaches, palpitations, and syncope [9], and it also has antioxidant, neuroprotective, antiepileptic, cardioprotective, anti-neuroinflammation, and other pharmacological activities [10,11,12]. Furthermore, NRR is mainly used in the form of compound prescription in clinical, e.g., Wenxin Granules can be used to treat premature ventricular contractions in patients with chronic heart failure [1].
Modern pharmacological studies have shown that nardosinone could exert anti-inflammatory, neuroprotective, cardioprotective, and more pharmacological actions [1]. For example, it could play a role in antiarrhythmia by hindering the calcium overload in the cAMP/PKA signaling pathway [13]. Nardosinone can regulate the proliferation and differentiation of neural stem cells, which implies that it might have therapeutic effects on brain injury and neurodegenerative diseases [14]. Nardosinone could also exert a cardioprotective effect, such as inhibiting the hypertrophy of H9c2 cells induced by Ang II [15]. Moreover, nardosinone, as the most effective anti-Parkinson’s disease compound in NRR, has been verified that it could alleviate Parkinson’s disease symptoms in mice [16]. It was manifested that nardosinone had good anti-trypanosomal activity in vitro [17]. However, nardosinone is easily degraded and generated other constitutes in the conditions of light and high temperature, while it was stable in alkaline conditions and low temperature. Therefore, it should be noted that the temperature should be controlled below 40 °C, the pH of the solution should be controlled between 7 and 12, and light should be avoided in the experimental process of extraction, separation, and administration of nardosinone [18].
Natural peroxides have multiple biological activities and are one of the important natural product types. For example, a large number of chemical components, such as polyketones with six-membered or five-membered peroxide rings, have been found in sponges [19,20,21], and the great majority of components have pharmacological effects such as anticancer, antibacterial, and antimalarial [21]; two diterpenoids with seven-membered peroxy rings were found in rhizomes of Hedychium coronarium [22]; a peroxydimer monoterpene was isolated from Amomum cardamomum, which has powerful antimalarial activity [23]. Furthermore, there were also a large number of studies on sesquiterpene peroxides, such as artemisinin compounds (artemisinin, dihydroartemisinin, and artemether). Studies have demonstrated that the peroxy-bond was the key pharmacophore of the antimalarial effect of artemisinin [24], and the peroxy-bond could be broken into monooxygen bridges based on the metabolism studies of artemisinin and dihydroartemisinin in vivo and in vitro [25]. Yingzhaosu A, as a sesquiterpene peroxide, also has significant antimalarial activity [26,27], but there are no reports on its metabolism research. The sesquiterpene peroxide (1R, 4R, 6S, 7S, 9S)-4α-hydroxy-1,9-peroxybisabola-2,10-diene was isolated from the rhizomes of Alpinia japonica [28]; the sesquiterpene peroxide 1α,8α-epidioxy-4α-hydroxy-5αH-guai-7(11),9-dien-12,8-olide, which has good anti-influenza virus activity was isolated from Curcuma wenyujin [29]. The activity of sesquiterpene peroxides has been studied in-depth, while the metabolism researches of them are less, hence their metabolic pathways and existence forms in vivo are still unclear.
Therefore, the metabolism of nardosinone in mice was carried out to (1) clarify the metabolites and metabolic pathways of nardosinone in mice; (2) elucidate whether the peroxy-bond of nardosinone is broken to form a mono-oxygen bridge in vivo and whether nardosinone can lose hydroxy-isopropyl in vivo to generate nardosinone acid or its isomers. This study will help with the following: (1) reveal the existing forms of nardosinone in vivo, and lay the foundation for further searching for the effective forms; (2) further illustrate the metabolic characteristics of sesquiterpene peroxide; (3) lay the foundation for exploring the effective forms of Nardostachyos Radix et Rhizoma.
2. Results
In this study, 39, 51, and 12 metabolites were respectively identified in urine, plasma, and feces in mice (Figures S1–S3), and a total of 76 new metabolites (Table 1) were detected and preliminarily identified and the metabolic pathway of nardosinone is shown in Figure 1. Nardosinone could lose hydroxy-isopropyl to generate nardosinone acid or its isomers in the metabolism in vivo, which is a rare metabolic reaction. There were 56 and 20 metabolites with the structural skeleton of nardosinone and nardosinone acid, respectively.

Table 1.
Identification of nardosinone and its 76 metabolites in mice urine, feces, and plasma samples by UHPLC-Q-TOF-MS.
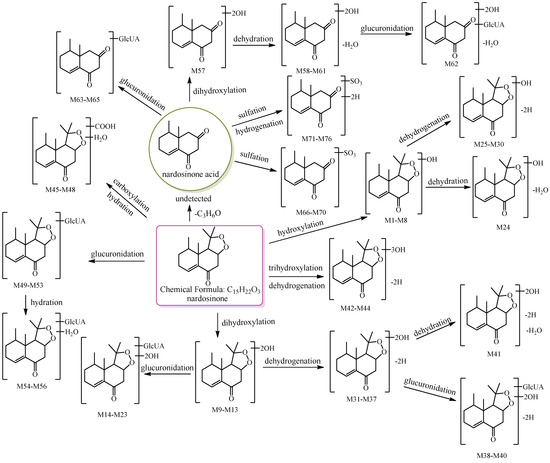
Figure 1.
The 76 new metabolites and proposed metabolic pathway of nardosinone in mice. The pink rim and olive circle represent nardosinone and nardosinone acid, respectively.
To conveniently describe the fragment characteristics of nardosinone and its metabolites, the three rings were named A, B, and C, respectively. The C-C bonds and carbon atoms in the skeleton were designated by the letters a-n and numbers 1–15 (Figure 2).
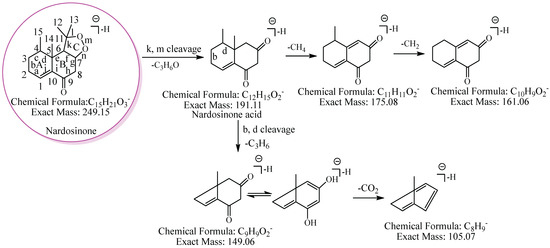
Figure 2.
The proposed fragmentation pathway of nardosinone. The pink circle represents nardosinone, and the C-C bonds and carbon atoms in the skeleton were designated by the letters a–n and numbers 1–15; A, B and C represent the three rings, respectively.
In this study, the usual neutral losses in mass spectrometry were 58.04 Da (C3H6O), 30.01 Da (CH2O), 14.02 Da (CH2), 16.03 Da (CH4), 42.05 Da (C3H6), 176.03 Da (C6H8O6), 79.95 Da (SO3), 43.99 Da (CO2), 27.99 Da (CO), 42.01 Da (C2H2O), 122.04 Da (C7H6O2), and 162.05 Da (C6H10O5), indicating that a molecule which shows these neutral losses may contain hydroxy-isopropyl, formaldehyde or methanol, methyl, methane, propyl group, glucuronyl, sulfonyl, carboxyl or lactone, carbonyl, acetyl, benzoyl, and hexosyl (more likely glucosyl) groups, respectively. In addition, the fragment ions at m/z 175.02 (C6H7O6), m/z 193.04 (C6H9O7), m/z 96.96 (SO4H), m/z 81.97 Da (H2SO3), and m/z 191.11 (C12H15O2) indicate that the molecule may contain glucuronyl, glucuronic acid, sulfate, sulfonyl, and nardosinone acid groups, respectively.
2.1. Mass Spectral Fragmentation Features of Nardosinone
The proposed fragmentation pathway of nardosinone is shown in Figure 2. Nardosinone (C15H22O3) showed [M−H]− at m/z 249.15. The fragment ion at m/z 191.11 (C12H15O2) was formed by a neutral loss of 58.04 Da (C3H6O) from the cleavage of the peroxy-bridge of the ion at m/z 249.15; the fragment ions at m/z 175.08 and m/z 161.06 were formed by the continuous neutral losses of 16.03 Da (CH4) and 14.02 Da (CH2) from the ion at m/z 191.11. And the ion at m/z 149.06, which could lose 43.99 Da (CO2) to form the fragment ion at m/z 105.07, and the fragmentation pathway of CO2 was reported [30], was formed by a neutral loss of 42.05 Da (C3H6) from the cleavage of b and d bonds of the A ring of the ion at m/z 191.11.
2.2. Identification of the Metabolites with the Skeleton of Nardosinone
2.2.1. Metabolites Formed by Monohydroxylation of Nardosinone
M1−M8 showed [M−H]− at m/z 265.14, and their molecular formulae were predicted as C15H22O4. There was an additional O atom in their molecular formulae in comparison with nardosinone. Therefore, they were identified as monohydroxylated nardosinones.
M1: The ions at m/z 207.10 (C12H15O3) and m/z 149.06 (C9H9O2) were formed by the consecutive losses of two 58.04 Da (C3H6O) from [M−H]− in the MS2 spectra. According to the fragmentation pathway of nardosinone, it was speculated that the cleavage of the peroxide bridge could lose C3H6O. In addition, C3H6O can be lost by the cleavage of the b and d bonds of the A ring when the hydroxylation occurs at C-3, C-4, or C-15. The ions at m/z 191.07 (C11H11O3) and m/z 177.06 (C10H9O3) were formed by the consecutive neutral losses of 16.03 Da (CH4) and 14.02 Da (CH2) from the ion at m/z 207.10, which could imply that hydroxylation did not occur at C-15. C-3 was a secondary carbon with parahydrogen, and C-4 was a tertiary carbon with tertiary hydrogen, which was more active than the parahydrogen of C-3, so it was inferred that the hydroxylation was more likely to occur at C-4. Thus, M1 was preliminarily identified as 4-hydroxyl nardosinone. The possible structure and fragmentation pathways of M1 are shown in Figure 3.
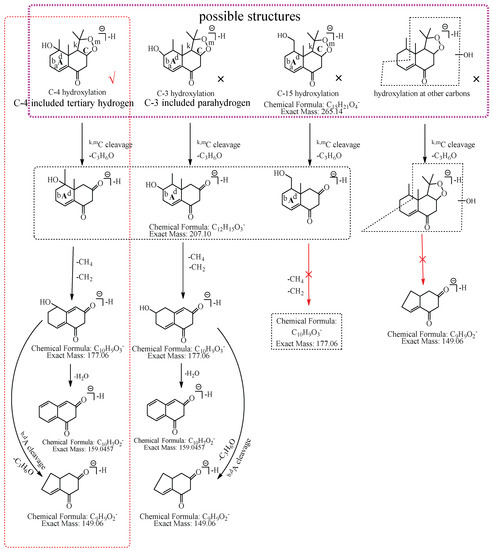
Figure 3.
The proposed fragmentation pathways of M1 (4-hydroxyl nardosinone). “√” represents the possible structure. “×” represents the impossible structure. The purple rim represents the possible structures; A and C represent the hexatomic rings, respectively. The red rim represents the most possible fragmentation pathway.
M2: The [M−H]− of M2 directly lost 72.06 Da (C4H8O) to produce m/z 193.09, while the ion at m/z 207.10 (C12H15O3), which was formed by the loss of 58.04 Da (C3H6O) from [M−H]− was not observed in MS2 spectra, demonstrating that the hydroxylation did not occur at C-3, C-4, or C-15 but it should occur at C-2. The ion at m/z 193.09 (C11H13O3) consecutively lost CO and 16.03 Da (CH4) to generate m/z 165.10 (C10H13O2) and m/z 149.06 (C9H9O2), and m/z 149.06 (C9H9O2) lost CO2 to generate m/z 105.07 (C8H10). Therefore, M2 was tentatively identified as 2-hydroxyl nardosinone. The possible structures and fragmentation pathways of M2 are shown in Figure 4.
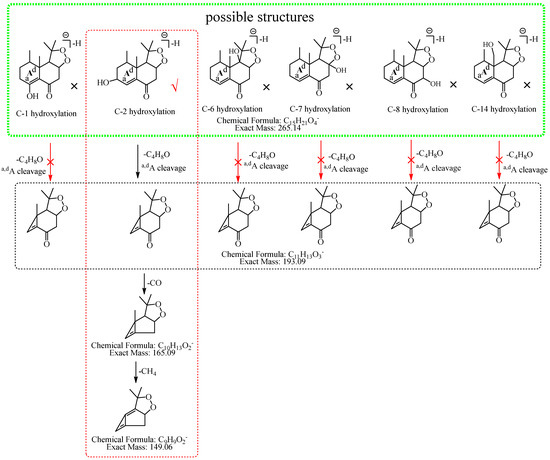
Figure 4.
The proposed fragmentation pathways of M2 (2-hydroxyl nardosinone). “√” represents the possible structure. “×” represents the impossible structure. The green rim represents the possible structures; A represents the hexatomic ring. The red rim represents the most possible fragmentation pathway.
M3: The ions at m/z 207.10 (C12H15O3) and m/z 149.06 (C9H9O2) were formed by the consecutive losses of two C3H6O from [M−H]− in the MS2 spectra, which inferred that the hydroxylation occurred at C-3, C-4, or C-15. The ion at m/z 177.06 (C10H9O3) was formed by the consecutive losses of two 14.02 Da (CH2) from the ion at m/z 207.10, which could imply that hydroxylation did not occur at C-15. As described in the identification of M1, C-3 was a secondary carbon with parahydrogen, and C-4 was a tertiary carbon with tertiary hydrogen, which was more active than the parahydrogen of C-3, so it was inferred that the hydroxylation was more likely to occur at C-4. Therefore, M3 was preliminarily identified as 4-hydroxyl nardosinone. The possible structure and fragmentation pathways of M3 are shown in Figure 5.
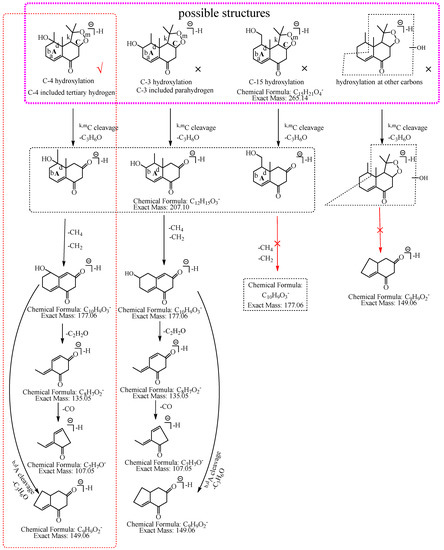
Figure 5.
The proposed fragmentation pathways of M3 (4-hydroxyl nardosinone). “√” represents the possible structure. “×” represents the impossible structure. The purple rim represents the possible structures; A and C represent the hexatomic rings, respectively. The red rim represents the most possible fragmentation pathway.
M4: The [M−H]− of M4 at m/z 265.14 directly lost C5H8O2 to produce m/z 165.09 (C10H13O2), while the ions at m/z 207.10 (C12H15O3) and m/z 193.09, which were respectively formed by the losses of 58.04 Da (C3H6O) and 72.06 Da (C4H8O) from [M−H]−, were not observed in MS2 spectra, implying that the hydroxylation did not occur at C-2, C-3, C-4, C-6, C-7, C-12, C-13, or C-15. The ion at m/z 165.09 (C10H13O2) consecutively lost 16.03 Da (CH4) and 43.99 Da (CO2) to generate m/z 149.06 (C9H9O2) and m/z 105.07 (C8H9), suggesting that the hydroxylation did not occur at C-1, C-2, and C-14. Thus, it was speculated that the hydroxylation occurred at C-8, and M4 was preliminarily identified as 8-hydroxyl nardosinone. The possible structure and fragmentation pathways of M4 are shown in Figure 6.
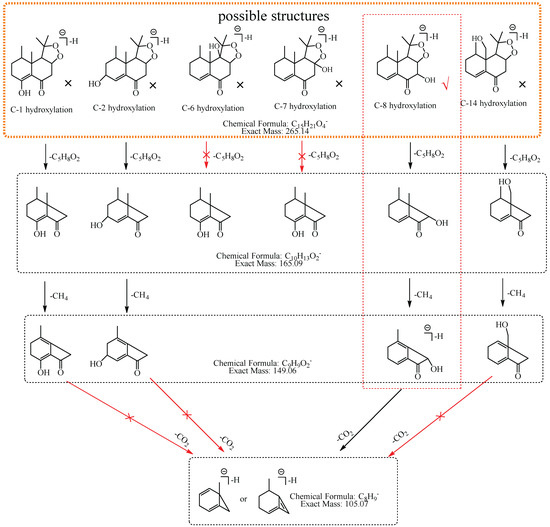
Figure 6.
The proposed fragmentation pathways of M4 (8-hydroxyl nardosinone). “√” represents the possible structure. “×” represents the impossible structure. The orange rim represents the possible structures. The red rim represents the most possible fragmentation pathway.
M5: The ions at m/z 207.10 (C12H15O3) and m/z 149.06 (C9H9O2) were formed by the consecutive losses of two C3H6O from [M−H]− in the MS2 spectra, which inferred that the hydroxylation occurred at C-3, C-4, or C-15. The ions at m/z 191.07 (C11H11O3) and m/z 177.06 (C10H9O3) formed by the consecutive losses of 16.03 Da (CH4) and 14.02 Da (CH2) from the ion at m/z 207.10 were observed, which could infer that hydroxylation did not occur at C-15. If hydroxylation occurred at C-3, C-4, or C-15, the ion at m/z 207.10 (C12H15O3) would directly lose 70.04 Da (C4H6O) to form the ion at m/z 137.06 (C8H9O2). In addition, the ions at m/z 165.09 (C10H13O2) and m/z 137.06 (C8H9O2) formed by the consecutive losses of 42.01 Da (C2H2O) and 28.03 Da (C2H4) from the ion at m/z 207.10 (C12H15O3) were observed in the MS2 spectra. Therefore, M5 was preliminarily identified as 3-hydroxyl nardosinone or 4-hydroxyl nardosinone. The possible structure and fragmentation pathways of M5 are shown in Figure 7.
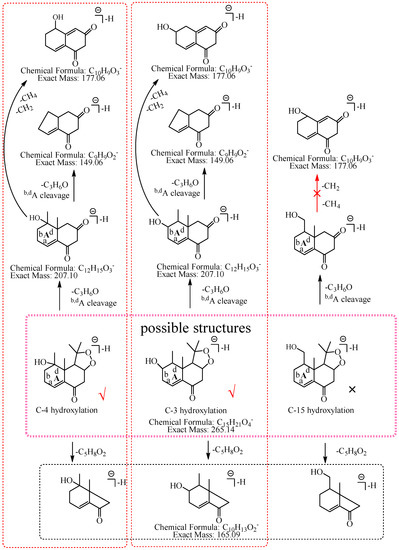
Figure 7.
The proposed fragmentation pathways of M5 (3-hydroxyl nardosinone or 4-hydroxyl nardosinone). “√” represents the possible structure. “×” represents the impossible structure. The pink rim represents the possible structures; A represents the hexatomic ring. The red rims represent the most possible fragmentation pathways.
M6: The [M−H]− at m/z 265.14 of M6 lost 58.04 Da (C3H6O) to generate m/z 207 in MS2 spectra, while the ion at m/z 149.06, which was formed by the losses of two C3H6O from [M−H]− was not observed, implying that the hydroxylation did not occur at C-3, C-4, C-12, C-13, or C-15. The ion m/z 265.14 ([M−H]−) lost 100.05 Da (C5H8O2) to generate m/z 165.09 (C10H13O2), indicating that the hydroxylation did not occur at C-6 and C-7. The characteristic fragment ion at m/z 177.06 (C10H9O3), which was formed by the losses of two 15.02 Da (CH3•) from m/z 207.10 (C12H15O3), was observed in MS2 spectra; indicating that the hydroxylation did not occur at C-14. The ion at m/z 177.06 (C10H9O3) lost 18.01 Da (H2O) to generate m/z 159.05 (C10H7O2), which implied that the hydroxylation did not occur at C-8. It also lost 40.03 Da (C3H4) to generate m/z 137.03 (C7H5O3), suggesting that the hydroxylation did not occur at C-2. Therefore, it was speculated that the hydroxylation should occur at C-1, and M6 was preliminarily identified as 1-hydroxyl nardosinone. The possible structure and fragmentation pathways of M6 are shown in Figure 8.
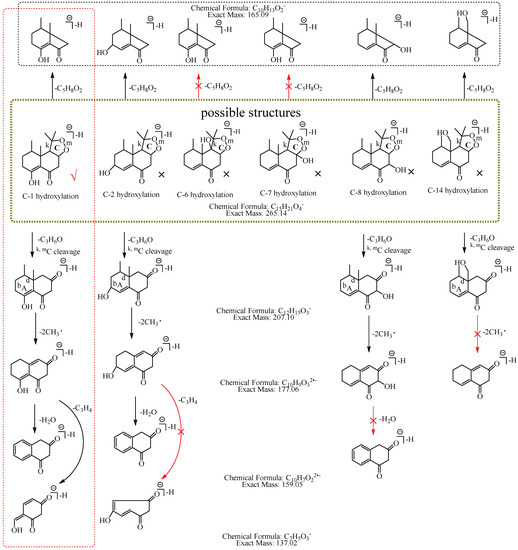
Figure 8.
The proposed fragmentation pathways of M6 (1-hydroxyl nardosinone). “√” represents the possible structure. “×” represents the impossible structure. The olive rim represents the possible structures; C represents the hexatomic ring. The red rim represents the most possible fragmentation pathway.
M7: In MS2 spectra, m/z 265.14 ([M−H]−) lost 58.04 Da (C3H6O) to generate m/z 207, while the ion at m/z 149.06, which was obtained by the losses of two C3H6O from [M−H]− was not observed, implying that the hydroxylation did not occur at C-3, C-4, C-12, C-13 or C-15. The [M−H]− could lose 100.05 Da (C5H8O2) to generate m/z 165.09 (C10H13O2), indicating that the hydroxylation did not occur at C-6 and C-7. m/z 191.07 (C11H11O3) lost 18.01 Da (H2O) to produce m/z 173.06 (C11H9O2), suggesting that the hydroxylation did not occur at C-8 and C-14. Therefore, the hydroxylation should occur at C-1 or C-2. C-2 has α-H of alkene, which is more active. Consequently, it was speculated that the hydroxylation more likely occurred at C-2, and M7 was identified as 2-hydroxyl nardosinone. The possible structure and fragmentation pathways of M7 are shown in Figure 9.
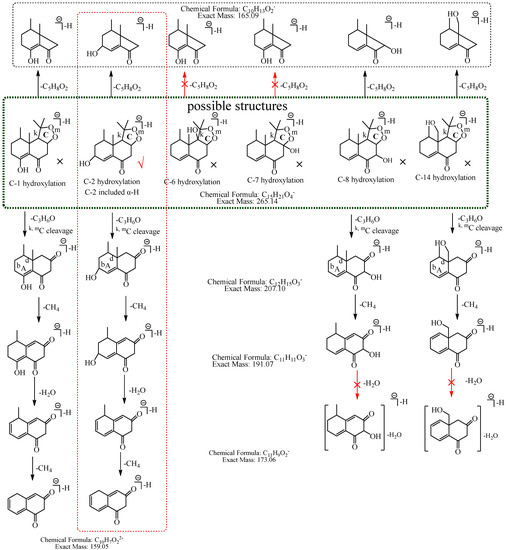
Figure 9.
The proposed fragmentation pathways of M7 (2-hydroxyl nardosinone). “√” represents the possible structure. “×” represents the impossible structure. The atrovirens rim represents the possible structures; A and C represent the hexatomic rings. The red rim represents the most possible fragmentation pathway.
M8: The ions at m/z 205.09 (C12H13O3) and m/z 149.06 (C9H9O2) were formed by the consecutive losses of 58.04 Da (C3H6O) and 56.03 Da (C3H4O) from [M−H]−. Compared with the fragmentation characteristics of M1 (consecutive neutral losses of two C3H6O were observed), M8 did not directly lose the second C3H6O but lost C3H4O. It was speculated that the hydroxylation more likely occurred at C-3, C-4, or C-15 and a carbon–carbon double bond existed among C-3, C-4, and C-15. Therefore, the neutral loss of 56.03 Da (C3H4O) was speculated to be generated by the hydroxylation that occurred at C-15 and the dehydrogenation that occurred at C-3 and C-4, or the hydroxylation that occurred at C-15 and the dehydrogenation that occurred at C-15 and C-4, or the hydroxylation and dehydrogenation that both occurred at C-15, or the hydroxylation that occurred at C-3 and the dehydrogenation that occurred at C-3 and C-4. Therefore, M8 was preliminarily identified as 3-hydroxyl nardosinone or 15-hydroxyl nardosinone. The possible structure and fragmentation pathways of M8 are shown in Figure 10.
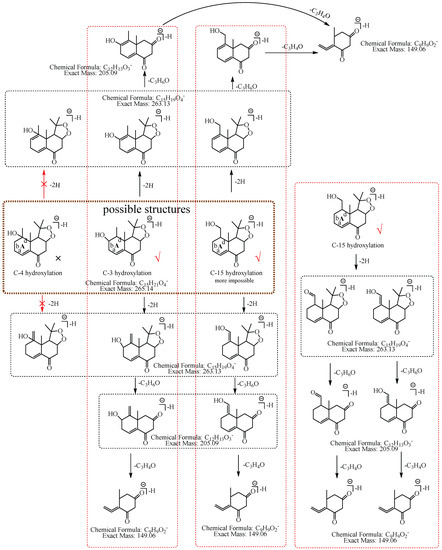
Figure 10.
The proposed fragmentation pathways of M8 (3-hydroxyl nardosinone or 15-hydroxyl nardosinone). “√” represents the possible structure. “×” represents the impossible structure. The brown rim represents the possible structures; A represents the hexatomic ring. The red rims represent the most possible fragmentation pathways.
2.2.2. Dihydroxylated Nardosinone and Their Glucuronides (M9−M13 and M14−M23)
M9−M13 showed [M−H]− at m/z 281.14, and their molecular formulae were predicted as C15H22O5. The characteristic fragment ion at m/z 223.10 (C12H15O4) was formed by a loss of 58.04 Da (C3H6O) from [M−H]− in the MS2 spectra. Compared to the nardosinone (C15H22O3), M9−M13 had two additional O atoms in their molecular formulae, thus, they were identified as dihydroxylated nardosinone isomers.
The molecular formulae of M14−M23 were verified to be C21H30O11 according to their [M−H]− at m/z 457.17. In the MS2 spectra of M14, M15, M16, M19, and M20, m/z 281.14 (C15H21O4) formed by the loss of 176.03 Da (C6H8O6) from [M−H]− was observed, and in the MS2 spectra of M17 and M23, the fragment ion at m/z 175.02 (C6H7O6) was detected. Therefore, M14, M15, M16, M17, M19, M20, and M23 were identified as dihydroxylated nardosinone glucuronides. The [M−H]− consecutively lost 18.01 Da (H2O), 58.04 Da (C3H6O), and 176.03Da (C6H8O6) to produce m/z 439.16 (C21H27O10), m/z 381.12 (C18H21O9), and m/z 205.09 (C12H13O3) in MS2 spectra of M22; the ion at m/z 205.09 (C12H13O3) formed by the loss of 176.03Da (C6H8O6) from [M−H]− was observed in MS2 spectra of M18 and M21. M18, M21, and M22, as the isomers of M14, M15, M16, M17, M19, M20, and M23, were identified as dihydroxylated nardosinone glucuronide isomers.
2.2.3. Metabolites Formed by Hydroxylation, and Dehydrogenataion or Dehydration of Nardosinone
M24 showed [M−H]− at m/z 247.13, and its molecular formula was predicted as C15H20O3. [M−H]− sequentially lost two C3H6O to generate m/z 189.09 (C12H13O3) and m/z 131.05 (C9H7O2), implying that the hydroxylation should occur at C-3, C-4 or C-15. Moreover, M24 had one fewer O atom and two fewer H atoms in its molecular formula in comparison with the M1−M8; thus, it was identified as a hydroxylated dehydrated nardosinone isomer.
M25−M30 had the molecular formulae of C15H20O4, which was predicted by their [M−H]− at m/z 263.13. The characteristic fragment ion at m/z 245.11 (C15H18O3) was formed by the loss of 18.02 Da (H2O) from [M−H]− in MS2 spectra of M25 and M26; the ion at m/z 205.09 (C12H13O3) was formed by the loss of 58.04 Da (C3H6O) from [M−H]− in MS2 spectra of M27−M30. Compared to the M1−M8, M25−M30 had two fewer H atoms in their molecular formulae, and the characteristic fragment ion at m/z 263.13 was observed in the MS2 spectra of M8. Thus, they were identified as hydroxylated dehydrogenated nardosinone isomers.
Dihydroxylated dehydrogenated nardosinone and their phase II metabolites (M31−M37 and M38−M40): M31−M37 showed [M−H]− at m/z 279.12, and their molecular formulae were predicted as C15H20O5. [M−H]− lost 58.04 Da (C3H6O) to produce m/z 221.08 (C12H13O4) in MS2 spectra of M31, M32, M35, and M37. M31−M37 had one more O atom than M25−M30; therefore, they were identified as dihydroxylated dehydrogenated nardosinone isomers. M38−M40 showed [M−H]− at m/z 455.16, and their molecular formulae were predicted as C21H28O11. The ion at m/z 279.13 (C15H19O5) was formed by the loss of 176.03 Da (C6H8O6) from [M−H]− in MS2 spectra of M38 and M39, and m/z 175.02 (C6H7O6) was observed in the MS2 spectra of M40. Thus, M38−M40 were speculated as dihydroxylated dehydrogenated nardosinone glucuronide isomers.
The molecular formula of M41 was predicted to be C15H18O4 based on its [M−H]− at m/z 261.11. [M−H]− lost 58.04 Da (C3H6O) to generate m/z 203.07 (C12H11O3). Compared to the M31−M37, M41 had one fewer O atom and two fewer H atoms in its molecular formula. Thus, M41 was speculated as dihydroxylated dehydrogenated dehydrated nardosinone.
M42−M44 showed [M−H]− at m/z 295.12, and their molecular formulae were predicted as C15H20O6. The characteristic fragment ions at m/z 277.11 (C15H19O6), m/z 233.12 (C14H19O4), and m/z 205.13 (C13H19O3) were formed by consecutive losses of 18.01 Da (H2O), 43.99 Da (CO2), and 27.99 Da (CO) from [M−H]− in MS2 spectra of M43. Compared to M31−M37, M42−M44 had one more O atom in their molecular formulae. Thus, M42−M44 was speculated as trihydroxylated dehydrogenated nardosinone isomers.
2.2.4. Metabolites Formed by Hydroxylation, Hydrogenation, and Carboxylation of Nardosinone
The molecular formulae of M45−M48 were predicted to be C16H24O6 based on their [M−H]− at m/z 311.15. [M−H]− lost 43.99 Da (CO2) to generate m/z 267.16 (C15H24O4) in the MS2 spectra of M47, and it had one more O atom and two more H atoms compared with nardosinone; thus, M47 was speculated as hydrogenated hydroxylated carboxylated nardosinone. M45, M46, and M48 were the isomers of M47 and were identified as hydrogenated hydroxylated carboxylated nardosinone isomers.
2.2.5. Nardosinone Glucuronides
M49−M53 showed [M−H]− at m/z 425.18, and their molecular formulae were predicted as C21H30O9. [M−H]− lost 250.16 Da (C15H22O3, nardosinone) to generate m/z 175.02 (C6H7O6) in MS2 spectra of M49−M52. Therefore, M49−M53 were identified as nardosinone glucuronides. The characteristic fragment ion at m/z 367.14 (C18H23O8) was formed by the loss of 58.04Da (C3H6O) from [M−H]− in the MS2 spectra of M50 and M51, suggesting that the glucuronidation did not occur at the hydroxyl, which was generated by the cleavage of the peroxide bridge, but probably occurred at the hydroxyl of C-7 or C-9. While the neutral loss of 58.04 Da (C3H6O) in MS2 spectra of M49, M52, and M53 was not observed, suggesting that the glucuronidation probably occurred at the hydroxyl, which was generated by the cleavage of the peroxide bridge.
The molecular formulae of M54−M56 were verified to be C21H32O10 based on their [M−H]− at m/z 443.19. M54−M56 had one more O atom and two more H atoms than M49−M53; therefore, they were identified as hydrogenated hydroxylated nardosinone glucuronides. In MS2 spectra of M55, [M−H]− consecutively lost 58.04 Da (C3H6O) and 18.01 Da (H2O) to generate m/z 385.15 (C18H25O9) and m/z 367.14 (C18H23O8), and m/z 385.15 (C18H25O9) further lost 44.03 Da (C2H4O) to generate m/z 341.12 (C16H21O8). These implied that it was impossible that hydroxylation occurred at C-1, C-2, C-6, C-8, and C-14 and glucuronidation occurred at the hydroxyl of C-7, and hydroxylation, hydrogenation, and glucuronidation simultaneously occurred at C-7 of M55. In addition, m/z 193.03 (C6H9O7, glucuronic acid ion) was also observed in MS2 of M55, and it was speculated that the compound easily lost glucuronic acid and formed conjugated systems in the aglycone, and the possibility that hydroxylation and glucuronidation simultaneously occurred at C-4, C-14, and C-15 was basically excluded because it could not form conjugated systems after losing glucuronic acid of M55. Moreover, the possibility that hydroxylation and glucuronidation simultaneously occurred at C-1 was also excluded, owing to the glycosidic bond of enol form hydroxyl at C-1 being difficult to break. In conclusion, it was speculated that M55 was generated by the hydrogenation of the peroxide bridge and simultaneous hydroxylation and glucuronidation at C-2, C-3, C-6, or C-8.
2.3. Identification of Metabolites with the Skeleton of Nardosinone Acid
2.3.1. Metabolites Formed by Dihydroxylation, Dehydration, and Glucuronidation of Nardosinone Acid
M57 had the molecular formula of C12H16O4 predicted by its [M−H]− at m/z 223.10. The characteristic fragment ions at m/z 191.11 (C12H15O2) and m/z 161.10 (C11H13O) were formed by the consecutive losses of 31.99 Da (2O, didehydroxylation) and 30.01 Da (CH2O) from [M−H]−. M57 had two additional O atoms in its molecular formula by comparison with nardosinone. Therefore, it was speculated as dihydroxylated nardosinone acid.
C12H14O3 and its phase II metabolites (M58−M61 and M62). M58−M61 showed [M−H]− at m/z 205.09, and their molecular formulae were predicted as C12H14O3. [M−H]− continuously lost two CH3• to generate m/z 190.07 (C11H10O3•) and m/z 175.05 (C10H7O3) in MS2 spectra of M58−M61. M58−M61 had one fewer O atom and two fewer H atoms than M57. Therefore, they were speculated as dehydrated dihydroxylated nardosinone acid isomers. M62 had the molecular formula of C18H22O9 predicted by its [M−H]− at m/z 381.12. The [M−H]− of M62 lost 176.03 Da (C6H8O6) to produce m/z 205.09 (C12H13O3), and the ions at m/z 190.06 and m/z 175.04 were identical with those of M58−M61. Therefore, M62 was identified as a dehydrated double hydroxylated nardosinone acid glucuronide.
2.3.2. Glucuronides and Sulfates of Nardosinone Acid
M63−M65 had the molecular formulae of C18H24O8 predicted by their [M−H]− at m/z 367.14. In their MS2 spectra, [M−H]− lost 176.03 Da (C6H8O6) to produce the m/z 191.11 (C12H15O2). Therefore, they were identified as nardosinone acid glucuronide isomers. The ion at m/z 193.04 (C6H9O7) was observed in the MS2 spectra of M63−M64, implying that the glycosidic bond broke easily and there were no conjugated systems near the glycosidic bond. Because enol form hydroxyl of C-7 and carbonyl of C-9 both had a stable conjugated system, glucuronidation should occur at hydroxyl of C-6 (the rearrangement of C-7 hydroxyl to C-6) for M63−M64. The ion at m/z 193.04 was not observed in the MS2 spectra of M65. Considering the energy difference required for bond cleavage, it was speculated that the glucuronidation occurred at the enol form hydroxyl of C-7 or the carbonyl of C-9, but the carbonyl of C-9 has already formed a stable conjugated system with the carbon–carbon double bond between C-10 and C-1, so the possibility that glucuronidation occurred at the carbonyl of C-9 was less. Therefore, it was speculated that the glucuronidation occurred at the enol form hydroxyl of C-7 in M65.
M66−M70 showed [M−H]− at m/z 271.06, and their molecular formulae were predicted as C12H16O5S. In the MS2 spectra of M66, its [M−H]− lost 79.96 Da (SO3) to produce m/z 191.11 (C12H15O2), while the ions at m/z 96.96 (HSO4) and m/z 80.96(HSO3) were not observed. Considering the energy difference required for bond cleavage, it was speculated that the sulfation occurred at enol form hydroxyl. However, the carbonyl of C-9 already formed a stable conjugated system with the carbon–carbon double bond between C-10 and C-1, so the possibility that sulfation occurred at the carbonyl of C-9 was less. Therefore, it was speculated that the sulfation occurred at the enol hydroxyl of C-7 in M66. In the MS2 spectra of M67, its [M−H]− lost 81.97 Da (H2SO3) to produce m/z 189.10 (C12H13O2); the ion at m/z 80.96 (HSO3) was observed in the MS2 spectra of M69; the ions at m/z 96.96 (HSO4) and m/z 80.96 (HSO3) were both observed in the MS2 spectra of M68 and M70. The sulfate ion at m/z 96.96 (HSO4), the sulfonyl ion at m/z 80.96 (HSO3), and the neutral loss of 81.97 Da (H2SO3) all indicated that the sulfate ester and the vicinal H were simultaneously lost, suggesting that the hydroxyl, which was sulfated was not enol form hydroxyl and did not in the conjugated system. Thus, it was speculated that the sulfation should occur at the hydroxyl of C-6. Therefore, M66−M70 were identified as nardosinone acid sulfates and the possible structures and the fragmentation pathways are shown in Figure S4.
The molecular formulae of M71−M76 were verified to be C12H18O5S based on their [M−H]− at m/z 273.08. The characteristic fragment ions at m/z 191.11 (C12H15O2) and m/z 80.96 (HSO3) were observed in the MS2 spectra of M73, M74, and M76, and the characteristic fragment ion at m/z 80.96 (HSO3) was observed in the MS2 spectra of M71, M72, and M75. As mentioned above, it was speculated that the sulfation did not occur at the enol form hydroxyl of M71−M76, and the possible sulfation sites were deduced as follows: (1) if sulfation occurred at hydroxyl of C-6 and the hydrogenation occurred at carbon–carbon double bond between C-7 and C-8, the conjugated system could not be formed after losing H2SO3, thus, it was less likely to occur; (2) if sulfation occurred at hydroxyl of C-6 and the hydrogenation occurred at carbon–carbon double bond between C-10 and C-1, the conjugated system could be formed after losing H2SO3, thus, it was more likely to occur; (3) if sulfation occurred at hydroxyl of C-6 and the hydrogenation occurred at carbonyl of C-9, the conjugated system could not be formed after losing H2SO3, thus, it was less likely to occur; (4) if sulfation occurred at hydroxyl of C-7 (reduction of carbonyl), the conjugated system could be formed after losing H2SO3, thus, it was more likely to occur; (5) if the sulfation occurred at hydroxyl of C-9 (reduction of carbonyl), the conjugated system could be formed with the enol hydroxyl of C-7 after losing H2SO3, but the carbonyl of C-9 was less likely to be hydrogenated because it already formed a conjugated system, therefore, the possibility that sulfation and hydrogenation simultaneously occurred at carbonyl of C-9 was less. Since the sulfation could not occur at the enol form hydroxyl (because no conjugated system could be formed after losing H2SO3) for M71−M76, when the hydrogenation occurred at the carbon–carbon double bond between C-10 and C-1, the sulfation should not occur at the enol form hydroxyl of C-7 and C-9. In conclusion, it was speculated that the sulfation should occur at the hydroxyl of C-6 and the hydrogenation occur at the carbon–carbon double bond between C-10 and C-1, or that the sulfation and hydrogenation simultaneously occur at the enol form hydroxyl of C-7. Compared to M66−M70, M71−M76 had two additional H atoms in their molecular formulae, therefore, M71−M76 were identified as hydrogenated nardosinone acid sulfates, and their possible structures and fragmentation pathways are shown in Figure S5.
2.4. Identification of Several Metabolites of Nardosinone with the Skeleton of Nardosinone Acid in the Fast Validation Experiment
We detected and identified three metabolites (F1-F3, the extracted ion chromatogram of them is shown in Figure S6). The molecular formulae of F1-F3 were predicted to be C12H18O5S according to their [M−H]− at m/z 273.08. The ion at m/z 79.96 (SO3) was observed in the MS2 spectra of F1, implying that the sulfation occurred at the enol form hydroxyl (C-7 or C-9); the ion at m/z 80.96 Da (HSO3) was observed in the MS2 spectra of F2 and F3, indicating that the sulfation occurred at the hydroxyl of C-6 or C-7 (reduction of carbonyl). F1-F3 showed [Aglycon−H]− (C12H17O2) at m/z 193.12, which had two more H atoms than the molecular formula of nardosinone acid, thus F1-F3 were speculated as hydrogenated nardosinone acid sulfates. The larger CLogP value means a longer retention time in reversed-phase UHPLC, thus, the possible structures of F2 (ClogP = 1.52 and tR = 12.89 min) and F3 (ClogP = 1.78 and tR = 15.82 min) are shown in Figure 11.
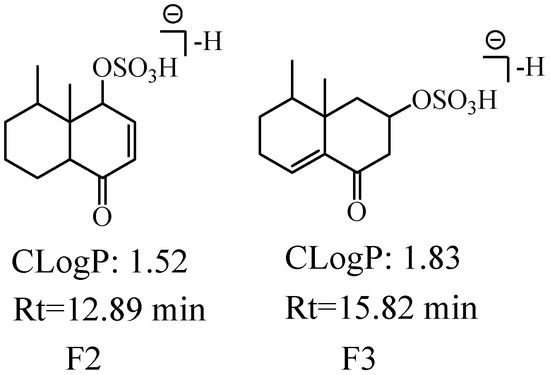
Figure 11.
The possible structures of F2 and F3. The CLogP represents CLogP values, Rt represents retention time.
3. Discussion
3.1. Comparative Analysis of the Metabolic Characteristics of Nardosinone and Other Sesquiterpene Peroxides
Artemisinin compounds are typical representatives of sesquiterpene peroxides, and many researchers have carried out the metabolism studies of them owing to their unique structures and antimalarial activity. It was inferred that the peroxide bridge was broken into a mono-oxygen bridge based on the metabolic researches of dihydroartemisinininin derived-dimer and dihydroartemisinin. Moreover, the main metabolic reactions included hydroxylation, dehydration, glucuronidation, carbonylation, dehydrogenation, etc. It was worth noting that the metabolic reactions of mono-hydroxylation, dihydroxylation, trihydroxylation, and quahydroxylation were not found in the metabolism of dihydroartemisinin but dihydroartemisinininin derived-dimer. Furthermore, carboxylated metabolites were found in the in vitro metabolism of artemether [31,32]. The in vivo and in vitro metabolism of artemisinin and dihydroartemisinin manifested that the peroxide bridge could be broken into a mono-oxygen bridge and the metabolic reactions mainly included dihydroxylation, deoxidation, hydroxylation, and glucuronidation, and 25 metabolites of artemisinin and 16 metabolites of dihydroartemisinin were identified, respectively [25]. The metabolites of hydroxylated dihydroartemisinin (DHA+O), dehydro-hydroxylated dihydroartemisinin (DHA−H2+O), and dehydration-hydroxylated dihydroartemisinin (DHA−H2O+O) were identified in the in vivo metabolism of dihydroartemisinin [33], which resembled the metabolites of hydroxylated nardosinone, dehydro-hydroxylated nardosinone, and dehydra-hydroxylated nardosinone in this study. Compared with the metabolism of other sesquiterpene peroxides, combined with the metabolic pathway and metabolites of nardosinone, we speculated that the peroxide bridge cleavage (loss of C3H6O) of nardosinone was more likely to generate enol hydroxyl or carbonyl other than the mono-oxygen bridge, which was inconsistent with the cleavage of the peroxide bridge and the formation of a mono-oxygen bridge in the metabolism of artemisinin. Furthermore, the common metabolic reactions reported in other sesquiterpene peroxides were also discovered in the metabolism of nardosinone, e.g., hydroxylation, carboxylation, sulfation, glucuronidation, etc.
3.2. Discussion on the Origin of Metabolites with the Nardosinone Acid Skeleton
It was reasonable that the hydroxy-isopropyl (C3H6O) could be lost in the fragmentation pathway of nardosinone, while we found many metabolites with the nardosinone acid skeleton (e.g., M57−M76) in biological samples. The loss of hydroxy-isopropyl involved the cleavage of the C-C bond, which is a rare metabolic reaction. Therefore, we designed a fast validation experiment to confirm it. If metabolites with a nardosinone acid skeleton are detected in this experiment, it means that they are indeed generated by in vivo metabolism.
According to the literature, nardosinone is stable under the conditions of an alkaline, low temperature, and away from light [18]. Therefore, the environmental factors should be strictly controlled to inhibit the degradation of nardosinone during the experimental processes.
In our fast validation experiment, nardosinone was suspended in a 0.5% carboxymethyl cellulose sodium (CMC-Na) solution (alkaline) and stored at −20 °C in a dark place. Then the mice were orally administered with nardosinone once, and in the first-hour urine samples of mice were collected and immediately filtered through a 0.22-μm membrane and analyzed by using UHPLC-Q-TOF-MS as quickly as possible. However, the metabolites with a nardosinone acid skeleton were still detected, which indicated that nardosinone acid and its metabolites were not generated by experimental operations such as ultrasonic extraction but generated by the in vivo metabolism of nardosinone. Furthermore, the results of the liver metabolism of the Cistanche deserticola total glycosides showed that the Cistanche deserticola total glycosides could lose the C3H5O group (m/z 57.06) to generate the metabolite methylated laricin [34]. Therefore, we consider that it is reasonable that nardosinone can lose hydroxy-isopropyl (C3H6O) to generate nardosinone acid in the in vivo metabolism.
But we still do not know where nardosinone loses the hydroxy-isopropyl (C3H6O) to generate nardosinone acid in vivo, and it could be the stomach, the liver, the intestinal tract, or somewhere else. We speculate that nardosinone is transformed into nardosinone acid by the intestinal microflora. Therefore, it deserves further research to explore the in vivo formation mechanism of nardosinone acid.
4. Materials and Methods
4.1. Reagent
The purity of nardosinone was greater than 98% (UHPLC, 254 nm) and it was purchased from Chengdu Push Bio-technology (Chengdu, China, Lot No. PS010660). HPLC-grade formic acid (Lot No. 212271), UHPLC-grade acetonitrile (Lot No. 207296), and HPLC-grade methanol (Lot No. 203511) were purchased from Thermo Fisher Scientific (Waltham, MA, USA); HPLC-grade ethanol (Lot No. 32061) was purchased from Beijing Tong Guang Fine Chemicals Company (Beijing, China). Sodium carboxymethyl cellulose (Lot No. A18105) was purchased from Sinopharm Chemical Reagents Co., Ltd. (Shanghai, China). Ultrapure water was prepared using a Milli-Q Integral 3 ultrapure water machine (Millipore, Billerica, MA, USA).
4.2. Animal Experiments
Nine ICR mice (male, 30 ± 2 g) were purchased from the Department of Laboratory Animal Sciences at the Peking University Health Science Center. They were randomized into three groups (test group I, test group II, and blank group) with three mice in each group. The experiment lasted for 10 d for the test group I and blank group, and 4 d for group II. All mice were housed in mouse metabolism cages with water and food ad libitum for the first three days, then the mice were dosed by gavage once per day for the following 7 d for the test group I and blank group, and 1 d for the test group II. The mice of test groups were orally administered with a dose of 80 mg/kg mouse-weight nardosinone suspended in 2.0 mL 0.5% carboxymethyl cellulose sodium (CMC-Na) solution, and the mice of blank group were orally administered with the same volume of 0.5% carboxymethyl cellulose sodium (CMC-Na) solution. The mice were allowed to eat and drink ad libitum. All animal experiments were approved by the Animal Ethics Committee of Peking University Health Science Center (approval number: LA2019117).
4.3. Samples Collection and Preparation
4.3.1. Collection of Samples
In total, 8 mL ethanol was added into each urine collection tube for bacteriostasis before each urine collection. All the urine and feces were collected during the six days of dosing for the test group I and blank group, and one hour after the last oral administration, the blood was collected into 1.5 mL heparin sodium-containing tubes by excising the eyeballs. All samples were kept at −80 °C.
The first-hour urine of mice after orally administered with nardosinone was collected for the test group II, and was immediately filtered through 0.22-μm membrane, and analyzed by UHPLC-Q-TOF-MS directly. It took 2.5 h from drug administration to obtain mass spectra data.
4.3.2. Preparation of Samples
All urine samples from the test group I and blank group were merged into two samples (test group I and blank group), respectively. Each sample was centrifuged at 8000 rpm at 4 °C for 15 min, and the supernatant was harvested, concentrated, and dried at 55 °C. Then, the urine residue was ultrasonically extracted with 10 times volume of methanol for 30 min and filtered to get the supernatant. The supernatant was evaporated to dryness at 55 °C. Finally, 0.5 g residue was dissolved in 1.0 mL methanol, and was filtered through 0.22-μm membrane, and stored at −80 °C before further analysis.
All feces samples from the test group I and blank group were combined into two samples (test group I and blank group) and were dried at 50 °C for 48 h and mashed. Each feces sample was ultrasonically extracted 30 min with 10 times volume of methanol for three times. The three extracts were filtered and merged, and dried at 55 °C. Next, the residue was ultrasonically extracted by 10 times volume of methanol for 30 min and centrifuged at 8000 rpm at 4 °C for 15 min to get the supernatant, which was further evaporated to dryness at 55 °C to obtain the residue. Finally, 0.5 g residue was dissolved in 1.5 mL methanol, and was filtered through 0.22-μm membrane, and stored at −80 °C before further analysis.
All plasma samples from the test group I and blank group were mixed into two samples (test group I and blank group) and centrifuged at 5000 rpm at 4 °C for 15 min to collect 1.0 mL supernatant, and it was mixed with 5 mL methanol and centrifuged at 5000 rpm at 4 °C for 15 min. The supernatant was separated and dried by nitrogen blow at 40 °C. Finally, 0.2 mg residue was dissolved in 0.3 mL methanol, and filtered through 0.22-μm membrane, and stored at −80 °C before further analysis.
4.4. Instrumentation and Analytical Conditions
Ultra-performance liquid chromatography–quadrupole time-of-flight mass spectrometry (UHPLC-Q-TOF-MS) analysis was performed on the SCIEX Triple TOF 6600 and SCIEX Exion LC AD System (UHPLC). All data were analyzed by PeakView v.1.2 software. Chromatographic separations were performed on an ACQUITY UPLC BEH C18 column (2.1 × 150 mm, 1.7 μm, Waters, USA) with a VanGuard Pre-Column (2.1 × 5 mm, 1.7 μm, Waters, USA). The column temperature was 35 °C; the injection volume was 2 μL, and the flow rate was 0.3 mL/min. The mobile phase was 0.1% aqueous solution of formic acid (A) and acetonitrile (B). The gradient elution program was set as follows: 0.01−3 min, 0.5% B; 3−6 min, 0.5%−8% B; 6−20 min, 8%−15% B; 20−33 min, 15%−60% B; 33−35 min, 60%−100% B; 35−38 min, 100% B; 38−39 min, 100%−0.5% B; 39−43 min, 0.5% B. The MS parameters were the following: electrospray ionization, negative ion (ESI) mode; full-scan mass spectra, m/z 100−1000 (MS) and m/z 50−1000 (MS2); Gas1, 60 psi; Gas2, 60 psi; Curtain gas, 35 psi; source temperature, 600 °C; Ion spray voltage, −4500 V (negative ion); Declustering potential, 60/−60 V; Collision energy, 35 ± 15 V.
4.5. Identification of the Existence Forms of Nardosinone In Vivo (Original Constituents and Metabolites)
The base peak chromatograms (BPCs) of the drug-containing group and blank group samples were compared to find the distinguishing peaks and tentatively determine the in vivo existence forms of nardosinone. Afterward, the distinguishing peaks were confirmed by comparing the corresponding extracted ion chromatograms (EICs) of the drug-containing group and blank group. If a compound could be detected by the extracted ion chromatogram (EIC) of the drug-containing group but did not exist in blank group, it was preliminarily verified as a metabolite. Furthermore, the MS data of reference substances, the MS fragmentation information reported in the literature, and the information obtained by searching the SciFinder database could all be used to identify the in vivo forms [35,36].
5. Conclusions
The in vivo metabolism of nardosinone was studied for the first time in mice. A total of 76 new metabolites were identified by UHPLC-Q-TOF-MS technology, and the metabolic reactions mainly included hydroxylation, dehydration, hydrogenation, sulfation, glucuronidation, demethylation, carboxylation, etc. There were 56 and 20 metabolites with the skeletons of nardosinone and nardosinone acid, respectively. It was confirmed that nardosinone could be biotransformed into nardosinone acid or isomer in vivo based on the results of the fast validation experiment. These results will be conducive to an in-depth study of the in vivo effective forms of nardosinone and NRR and will have certain reference values for the in vivo metabolism studies of other sesquiterpene peroxides.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/molecules27217267/s1, Figure S1: Extracted ion chromatograms (EICs) of 39 metabolites of nardosinone in mice urine. A, B, C, and D represent the EICs of 39 metabolites of nardosinone group, the magnified EICs of 39 metabolites of nardosinone group, the EICs of 39 metabolites of blank group, the magnified EICs of 39 metabolites of blank group, respectively. Figure S2: Extracted ion chromatograms (EICs) of 51 metabolites of nardosinone in mice plasma. A, B, C, and D represent the EICs of 51 metabolites of nardosinone group, the magnified EICs of 51 metabolites of nardosinone group, the EICs of 51 metabolites of blank group, the magnified EICs of 51 metabolites of blank group, respectively. Figure S3: Extracted ion chromatograms (EICs) of 12 metabolites of nardosinone in mice feces. A and B represent the EICs of 12 metabolites of nardosinone group and blank group, respectively. Figure S4: The possible structures and proposed fragmentation pathways of M66-M70. The red rim represents the proposed fragmentation pathways of M67-M70 and the blue rim represents the proposed fragmentation pathway of M66. Figure S5: The possible structures and proposed fragmentation pathways of M71-M76. The olive rim represents the possible structures. The dull purple rim represents the proposed fragmentation pathways. Figure S6: Extracted ion chromatograms (EICs) of m/z 273.08.
Author Contributions
F.X. designed the study. J.Z. (Jing Zhang 1), Y.L. and J.Z. (Jing Zhang 2) performed the experiment. Y.-S.B., M.-Y.L. and S.-Q.W. participated in animal experiments. J.Z. (Jing Zhang 1) analyzed the data. J.Z. (Jing Zhang 1), F.X., L.-L.W., G.-X.L., M.-Y.S. and S.-Q.C. revised the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This study was financially supported by the National Science and Technology Major Project for Significant New Drugs Development (2019ZX09201004), the National Natural Science Foundation of China (No. 81973472, 81573593).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Acknowledgments
We are grateful to Wen Ma for her routine management and careful maintenance of the UHPLC−Q−TOF−MS instrument.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Wen, J.W.; Liu, L.Q.; Li, J.J.; He, Y. A review of nardosinone for pharmacological activities. Eur. J. Pharmacol. 2021, 908, 174343. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.W.; Chen, G.Y.; Song, X.P.; Chen, W.H.; Shu, H.M.; Han, C.R.; Ji, M.H. Chemical constituents from roots of Millettia speciosa. Chin. Tradit. Herb. Drugs 2014, 45, 1515–1520. [Google Scholar]
- Peng, H.; Huang, P.Z.; Song, Y.G.; Xu, H.H.; Zhou, M.Y.; Zhu, G.H.; Yang, M.; Ai, Z.F.; Su, D. Full spectrum analysis of chemical constituents of Sargassum fusiforme and its in vitro anti-neuroinflammatory activity. China Pharm. 2022, 33, 800–807+812. [Google Scholar]
- Kong, J.Q.; Meng, X.S.; Shang, Y.H.; Dong, Y.Y.; Ma, Q. Identification of Chemical Components in Taraxacum kok-saghyz Rodin and Investigation of Mass Spectrometric Fragmentation Pathways. J. Chin. Mass Spectrom. Soc. 2022, 43, 278–286. [Google Scholar]
- Xu, X.Y.; Li, C.X.; Zheng, K.B.; Li, A.P. Chemical Constituents of Water Extract from Chrysanthemum morifolium Flowers by UHPLC-ESI-Orbitrap MS. J. Trop. Subtrop. Bot. 2020, 29, 96–104. [Google Scholar]
- Liang, X.; Liu, C.S.; Xia, T.; Tang, Q.F.; Tan, X.M. Identification of Active Compounds of Mahuang Fuzi Xixin Decoction and Their Mechanisms of Action by LC-MS/MS and Network Pharmacology. Evid. Based Compl Alt. 2020, 2020, 3812180. [Google Scholar] [CrossRef]
- Yu, S.L.; Ye, X.; Jia, G.F.; He, Z.J.; Sun, P.; Zhang, C.B.; Zhao, W.J. Nardostachys jatamansi, A Medicinal Plant from Qinghai-Tibet Plateau: A Review. Chin. J. Exp. Tradit. Med. Formulae 2021, 27, 243–250. [Google Scholar]
- Rao, Y.; Li, R.; Wang, X.W.; Xue, B.X.; Li, S.W.; Zhao, Y.; Zhang, L.H.; Xu, Y.T.; Wu, H.H. Data mining of compatibility characteristics for Chinese medicinal prescriptions containing Nardostachyos Radix et Rhizoma. Chin. Tradit. Herb. Drugs 2021, 52, 3331–3343. [Google Scholar]
- Rehman, T.; Ahmad, S. Nardostachys chinensis Batalin: A review of traditional uses, phytochemistry, and pharmacology. Phytother. Res. 2019, 33, 2622–2648. [Google Scholar] [CrossRef]
- Luo, J.F.; Shen, X.Y.; Lio, C.K.; Dai, Y.; Cheng, C.S.; Liu, J.X.; Yao, Y.D.; Yu, Y.; Xie, Y.; Luo, P.; et al. Activation of Nrf2/HO-1 Pathway by Nardochinoid C Inhibits Inflammation and Oxidative Stress in Lipopolysaccharide-Stimulated Macrophages. Front. Pharm. 2018, 9, 911. [Google Scholar] [CrossRef]
- Li, M.; Xu, X.; Yang, X.Y.; Kwong, J.S.W.; Shang, H.C. The cardioprotective and antiarrhythmic effects of Nardostachys chinensis in animal and cell experiments. BMC Complem. Altern. M. 2017, 17, 1–10. [Google Scholar] [CrossRef]
- Park, S.Y.; Kim, Y.H.; Park, G. Anti-neuro-inflammatory effects of Nardostachys chinensis in lipopolysaccharide-and lipoteichoic acid-stimulated microglial cells. Chin. J. Nat. Med. 2016, 14, 343–353. [Google Scholar]
- Jian, P.; Qing-Hai, L.I.; Fan, L.H. Experimental study on the inhibitory effect of nardosinone on myocardial cell in rats with tachyarrhythmia. Chin. J. Clin. Pharmacol. 2015, 31, 2240. [Google Scholar]
- Li, Z.H.; Li, W.; Shi, J.L.; Tang, M.K. Nardosinone Improves the Proliferation, Migration and Selective Differentiation of Mouse Embryonic Neural Stem Cells. PLoS ONE 2014, 9, e91260. [Google Scholar] [CrossRef]
- Du, M.; Huang, K.; Gao, L.; Yang, L.; Wang, W.S.; Wang, B.; Huang, K.; Huang, D. Nardosinone Protects H9c2 Cardiac Cells from Angiotensin II-induced Hypertrophy. J. Huazhong Univ. Sci. Med. 2013, 33, 822–826. [Google Scholar] [CrossRef]
- Bian, L.H.; Yao, Z.W.; Zhao, C.B.W.; Li, Q.Y.; Shi, J.L.; Guo, J.Y. Nardosinone Alleviates Parkinson’s Disease Symptoms in Mice by Regulating Dopamine D2 Receptor. Evid. Based Compl. Alt. 2021, 2021, 6686965. [Google Scholar] [CrossRef]
- Otoguro, K.; Iwatsuki, M.; Ishiyama, A.; Namatame, M.; Nishihara-Tukashima, A.; Kiyohara, H.; Hashimoto, T.; Asakawa, Y.; Omura, S.; Yamada, H. In vitro antitrypanosomal activity of plant terpenes against Trypanosoma brucei. Phytochemistry 2011, 72, 2024–2030. [Google Scholar] [CrossRef]
- Liu, G.L.; Liu, Y.; Shi, J.L.; Fang, N.; Li, J.; Lu, Y.W.; Wu, M.X. Stability of nardosinone. Chin. J. Pharm. Anal. 2015, 35, 360–363. [Google Scholar]
- Fattorusso, E.; Taglialatela-Scafati, O. Marine endoperoxides as antimalarial lead compounds. Phytochem. Rev. 2010, 9, 515–524. [Google Scholar] [CrossRef]
- Norris, M.D.; Perkins, M.V. Structural diversity and chemical synthesis of peroxide and peroxide-derived polyketide metabolites from marine sponges. Nat. Prod. Rep. 2016, 33, 861–880. [Google Scholar] [CrossRef]
- Chen, J.H.; Zhang, X.M.; Huang, Z.P.; Wang, W.H.; Zuo, X.F.; Zhao, Q. Advances in Resources of Natural Peroxides and Their Pharmacological Activities. J. Yunnan Univ. Tradit. Chin. Med. 2018, 41, 96–102. [Google Scholar]
- Nakatani, N.; Kikuzaki, H.; Yamaji, H.; Yoshio, K.; Kitora, C.; Okada, K.; Padolina, W.G. Labdane Diterpenes from Rhizomes of Hedychium-Coronarium. Phytochemistry 1994, 37, 1383–1388. [Google Scholar] [CrossRef]
- Kamchonwongpaisan, S.; Nilanonta, C.; Tarnchompoo, B.; Thebtaranonth, C.; Thebtaranonth, Y.; Yuthavong, Y.; Kongsaeree, P.; Clardy, J. An Antimalarial Peroxide from Amomum Krervanh Pierre. Tetrahedron Lett. 1995, 36, 1821–1824. [Google Scholar] [CrossRef]
- Cheng, S.J.; Fu, L.C. Research progress of artemisinins in the treatment of malaria. China Trop. Med. 2018, 18, 670–674+681. [Google Scholar]
- Liu, T.; Du, F.Y.; Wan, Y.K.; Zhu, F.P.; Xing, J. Rapid identification of phase I and II metabolites of artemisinin antimalarials using LTQ-Orbitrap hybrid mass spectrometer in combination with online hydrogen/deuterium exchange technique. J. Mass. Spectrom. 2011, 46, 725–733. [Google Scholar] [CrossRef]
- Szpilman, A.M.; Korshin, E.E.; Rozenberg, H.; Bachi, M.D. Total syntheses of yingzhaosu A and of its C(14)-epimer including the first evaluation of their antimalarial and cytotoxic activities. J. Org. Chem. 2005, 70, 3618–3632. [Google Scholar] [CrossRef]
- Tokuyasu, T.; Kunikawa, S.; Abe, M.; Masuyama, A.; Nojima, M.; Kim, H.S.; Begum, K.; Wataya, Y. Synthesis of antimalarial yingzhaosu a analogues by the peroxidation of dienes with Co(II)/O-2/Et3SiH. J. Org. Chem. 2003, 68, 7361–7367. [Google Scholar] [CrossRef]
- Li, Q.M.; Luo, J.G.; Yang, M.H.; Kong, L.Y. Terpenoids from Rhizomes of Alpinia japonica Inhibiting Nitric Oxide Production. Chem. Biodivers. 2015, 12, 388–396. [Google Scholar] [CrossRef]
- Dong, J.Y.; Ma, X.Y.; Cai, X.Q.; Yan, P.C.; Yue, L.; Lin, C.; Shao, W.W. Sesquiterpenoids from Curcuma wenyujin with anti-influenza viral activities. Phytochemistry 2013, 85, 122–128. [Google Scholar] [CrossRef]
- Liu, G.Q.; Dong, J.; Wang, H.; Wang, L.R.; Duan, Y.S.; Chen, S.Z. ESl Fragmentation Studies of Four Tea Catechins. Chem. J. Chin. Univ. 2009, 30, 1566–1570. [Google Scholar]
- Dai, H.L. Synthesis, Antimalarial Activity and Primary Study on Metabolism In Vivo of Dihydroartemisinin Derived-Dimer. Master’s Dissertation, Shanxi Medical University, Taiyuan, China, 2017. [Google Scholar]
- Liu, T. Biotransformation and Time-Dependent Pharmacokinetic Studies of Artemisinin Antimalarials. Master’s Dissertation, Shandong University, Jinan, China, 2012. [Google Scholar]
- Zhao, Y.F. Study on the Metabolites of Dihydroartemisinin and Their Distribution in Mice. Ph.D. Dissertation, China Academy of Chinese Medical Sciences, Beijing, China, 2021. [Google Scholar]
- Liu, Y. Study on the Action Process of Cistanche Deserticola Total Glycosides Based on Liver Metabolism. Master’s Dissertation, Harbin University of Commerce, Harbin, China, 2018. [Google Scholar]
- Liang, J.; Xu, F.; Zhang, Y.Z.; Huang, S.; Zang, X.Y.; Zhao, X.; Zhang, L.; Shang, M.Y.; Yang, D.H.; Wang, X.; et al. The profiling and identification of the absorbed constituents and metabolites of Paeoniae Radix Rubra decoction in rat plasma and urine by the HPLC-DAD-ESI-IT-TOF-MSn technique: A novel strategy for the systematic screening and identification of absorbed constituents and metabolites from traditional Chinese medicines. J. Pharm. Biomed. 2013, 83, 108–121. [Google Scholar]
- Xu, F.; Li, D.P.; Huang, Z.C.; Lu, F.L.; Wang, L.; Huang, Y.L.; Wang, R.F.; Liu, G.X.; Shang, M.Y.; Cai, S.Q. Exploring in vitro, in vivo metabolism of mogroside V and distribution of its metabolites in rats by HPLC-ESI-IT-TOF-MSn. J. Pharm. Biomed. 2015, 115, 418–430. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).