Abstract
Geo-authentic herbs refer to medicinal materials produced in a specific region with superior quality. Stephania tetrandra S. Moore (S. tetrandra) is cultivated in many provinces of China, including Anhui, Zhejiang, Fujian, Jiangxi, Hunan, Guangxi, Guangdong, Hainan, and Taiwan, among which Jiangxi is the geo-authentic origin. To explore habitat-related chemical markers of herbal medicine, an integrated chromatographic technique including gas chromatography-mass spectrometry (GC-MS), ultra-high-performance liquid chromatography coupled with quadrupole time-of-flight mass spectrometry (UHPLC-Q-TOF-MS/MS) and ultra-high-performance liquid chromatography-mass spectrometry (UHPLC-MS/MS) combined with chemometric analysis was established. The established methods manifested that they were clearly divided into two groups according to non-authentic origins and geo-authentic origins, suggesting that the metabolites were closely related to their producing areas. A total of 70 volatile compounds and 50 non-volatile compounds were identified in S. tetrandra. Meanwhile, tetrandrine, fangchinoline, isocorydine, magnocurarine, magnoflorine, boldine, and higenamine as chemical markers were accurately quantified and suggested importance in grouping non-authentic origins and geo-authentic origins samples. The discriminatory analysis also indicated well prediction performance with an accuracy of 80%. The results showed that the multiple chromatographic and chemometric analysis technique could be used as an effective approach for discovering the chemical markers of herbal medicine to fulfill the evaluation of overall chemical consistency among samples from different producing areas.
1. Introduction
S. tetrandra is derived from the dried root of Stephania tetrandra S. Moore (S. tetrandra) [1], a perennial liana plant of the genus Stephania, belonging to the Menispermaceae family [2]. It was first recorded as medicine in Shen Nong’s Herbal Classic and suitable as a treatment for arthralgia associated with rheumatoid arthritis, wet beriberi, eczema, and inflamed sores, which acts as a diuretic, analgesic, and anti-inflammatory [2,3,4]. Currently, the compounds isolated and identified from S. tetrandra are mainly alkaloids which are critical for evaluating their therapeutic effects and quality [5]. However, only two characteristic components, tetrandrine and fangchinoline, were defined as the quantitative indexes recorded in the Chinese Pharmacopoeia 2020 edition [1]. Other major components with high content and extensive pharmacological properties, such as magnoflorine, magnocurarine, isocorydine, higenamine, and boldine, have been less studied so far [2]. Therefore, it was an urgent need to develop more potential chemical markers to better evaluate the holistic chemical features of S. tetrandra from different origins.
It is well accepted that medicinal herbs exert their efficacies through synergistic actions via “multi-components hitting multi-targets” of complex chemicals in the herbs [6,7], and integrating multiple methods simultaneously characterizing different kinds of components has been employed as a comprehensive strategy to evaluate the holistic quality, thus assure the efficacy of medicinal herbs [8]. Moreover, chemometrics provides a variety of good algorithms to explore and obtain more valuable chemical information [9,10,11,12,13,14,15]. Among these, discriminatory analysis is an effective tool for accurate prediction according to various characteristic values. The combination of multiple chromatographic techniques and chemometrics would provide a reliable method for the biomarker screening of herbal medicine.
In this study, an integrated chromatographic technique based on ultra-high-performance liquid chromatography coupled with quadrupole time-of-flight mass spectrometry (UHPLC-Q-TOF-MS/MS) and gas chromatography-mass spectrometry (GC-MS) was proposed to study the geo-herbalism of S. tetrandra. The habitat-related biochemical markers were further investigated using multiple pattern recognition models. Subsequently, seven main components were quantitatively compared using the validated ultra-high-performance liquid chromatography-mass spectrometry (UHPLC-MS/MS). A discriminant function equation was established to verify the accuracy of the chemical markers and achieve the prediction of the origin of S. tetrandra. The proposed strategy, which is comprehensive and effective, can be used to assist the application of S. tetrandra as well as related herbal medicine.
2. Results and Discussion
2.1. Method Validation
The relative standard deviations (RSDs) of retention time (Rt) and peak area for precision, repeatability, and stability were less than 1.0% and 7.7% using GC-MS and UHPLC-Q-TOF-MS/MS analysis (Supplementary Table S1), which validated that the established method was precise for differential component analysis of S. tetrandra from different origins.
The UHPLC-MS/MS method was verified in terms of linearity, lower limits of quantification (LLOQs), precision, repeatability, stability, and recovery. The results are shown in Supplementary Tables S2–S4, which revealed that the established method was precise enough for the simultaneous quantitative determination of seven compounds.
2.2. Identification of Volatile Components by GC-MS Analysis
GC-MS analysis was carried out to analyze the volatile components of S. tetrandra samples, and the relative contents of volatile compounds were calculated by peak area normalization. According to the database NIST08 and NIST08s, 70 volatile components, including alkanes, fatty acids, esters, carbonyls, alcohols, and phenols, were identified. Among them, the content of esters was the highest, accounting for 28.44%, of which methyl (9E)-9-octadecenoate, methyl linoleate, and methyl palmitate were 11.48%, 6.50%, and 4.50%, respectively. Alkanes have the most types, with a relative content of 14.36%, of which 2,4-dimethyl-1-heptene was the most abundant, reaching 1.80%. The relative content of fatty acids was 10.26%, among which oleic acid and palmitic acid were the highest, which were 4.54% and 2.94%, respectively. The identification results are shown in Table 1, and the corresponding peak of each compound was exhibited in the total ion chromatograms (TIC) diagrams (Supplementary Figure S1).

Table 1.
Volatile chemicals of S. tetrandra by GC-MS analysis.
2.3. Identification of Nonvolatile Components by UHPLC-Q-TOF-MS/MS Analysis
Chromatographic data collected from UHPLC-Q-TOF-MS/MS were imported into Agilent Masshunter Qualitative Workstation Analysis B.07.00 (Agilent Technologies Inc., Santa Clara, CA, USA) for the identification of nonvolatile components of S. tetrandra. The identification of compounds is based on accurate mass, Rt, ion pattern, and MS/MS information. The obtained mass spectrograms were verified by: (a) matching with the instrument-generated molecular formula; (b) analyzing the structural information of metabolites acquired from the Metlin database (http://metlin.scripps.edu, accessed on 29 June 2022); (c) comparing with the fragment information of the standard samples; (d) combining with the compound information of the previous reports. According to the above-mentioned data acquisition and mining strategies, a total of 50 compounds, including 25 alkaloids, six amino acids, four amides, four fatty acids, two phenols, two purine derivatives, one phospholipid, one nucleoside, and five other compounds, were identified in S. tetrandra. The detailed information on the compounds is shown in Table 2, and the TIC figures of S. tetrandra under positive and negative ion modes are illustrated in Supplementary Figure S2.

Table 2.
Possible non-volatile chemicals of S. tetrandra by UHPLC-Q-TOF-MS/MS analysis.
2.4. Exploration of Habitat-Related Chemical Markers Based on Global Components
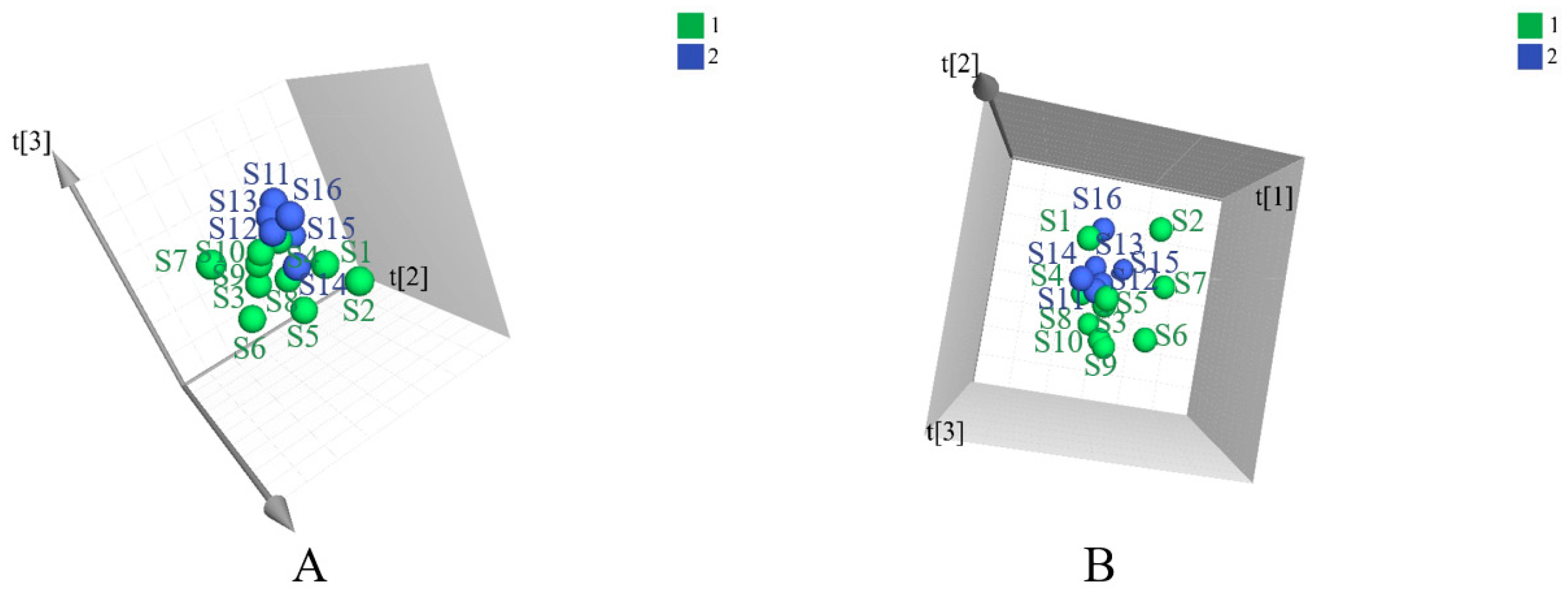
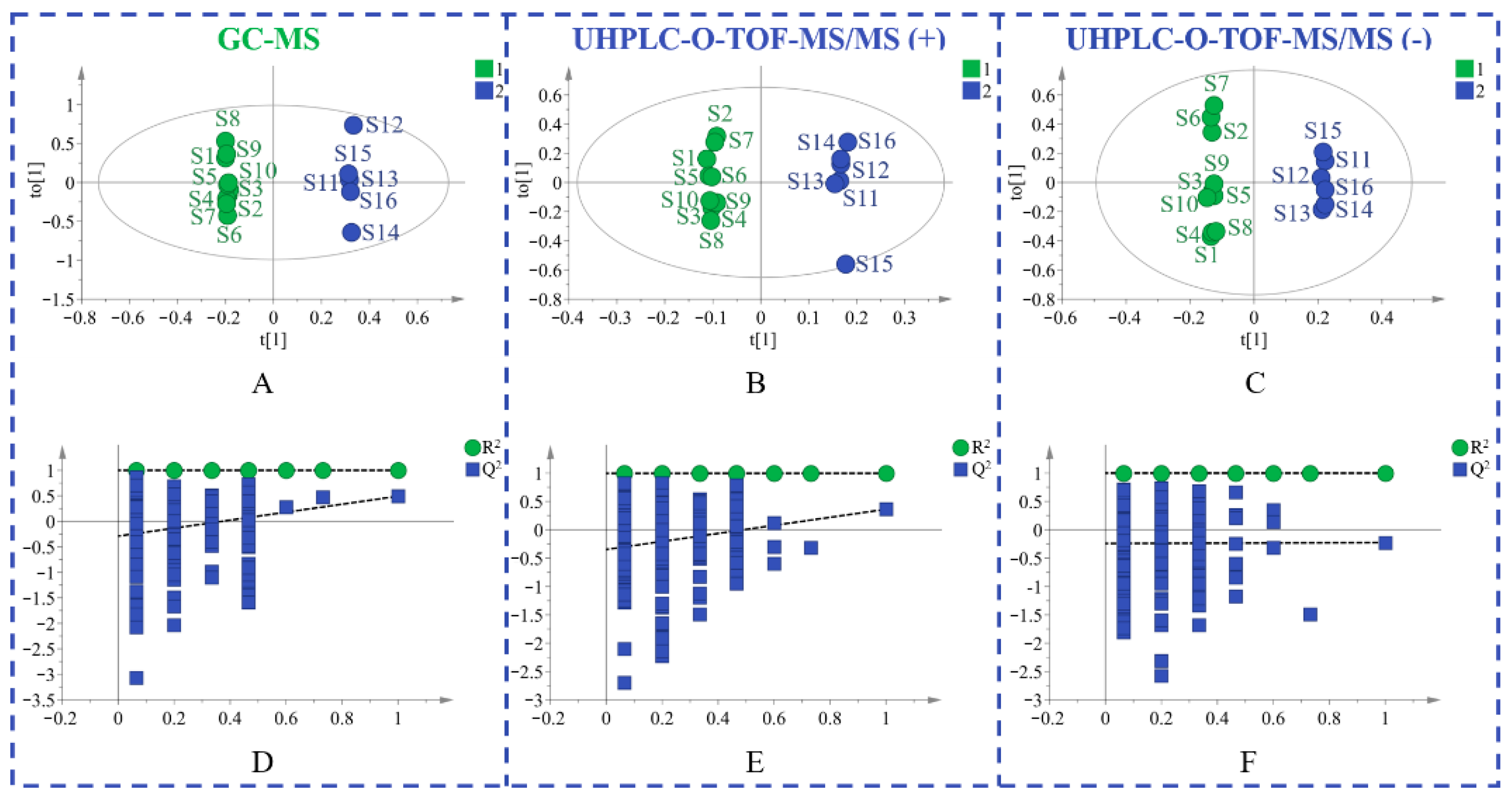
Principal component analysis (PCA) is an unsupervised pattern recognition method that is often used to sort unclassified samples into groups. As displayed in Figure 1, samples from different provinces were separated into two groups. The geo-authentic samples had a more clustered distribution and homogeneous quality, whereas the non-authentic samples had a wide range of interval distribution and large differences in quality. Supervised orthogonal partial least squares discriminant analysis (OPLS-DA) was subsequently used to filter out random noises, distinguish differences between groups, and improve the validity and analytical ability of the model. The results in Figure 2A–C showed that the medicinal materials were grouped into two groups according to the non-authentic and geo-authentic origins. The model was conducted using 7-fold cross-validation in this research. R2 describes the goodness of fit and the cross-validation parameter, Q2, represents the predictive ability of the model. The constructed model had good quality, with a cumulative R2Y of 0.997 and Q2Y of 0.491. Permutation tests were conducted 200 times to assess whether the model was overfitted. As shown in Figure 2D–F, the blue regression lines of the Q2 points intersected the vertical axis below zero, and the intercepts of all regression lines on the vertical axis were less than 0.5; therefore, the model was reliable and not overfitted.

Figure 1.
The PCA figures of two groups of S. tetrandra samples using UHPLC-Q-TOF-MS/MS analysis in the positive (A) and negative (B) ion models. (1: non-authentic origins; 2: geo-authentic origins).
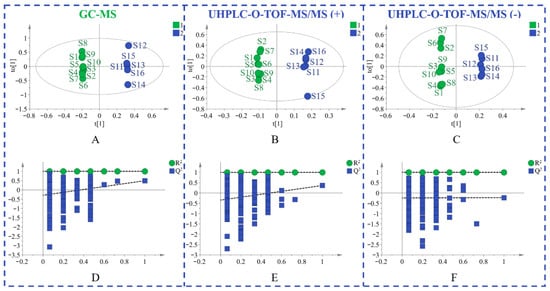
Figure 2.
The OPLS-DA figures of two groups of S. tetrandra samples using GC-MS analysis (A), UHPLC-Q-TOF-MS/MS analysis in positive (B) and negative (C) ion model, validation of the model by a permutation test of GC-MS data (D), UHPLC-Q-TOF-MS/MS data in positive (E) and negative (F) ion model. (1: non-authentic origins; 2: geo-authentic origins).
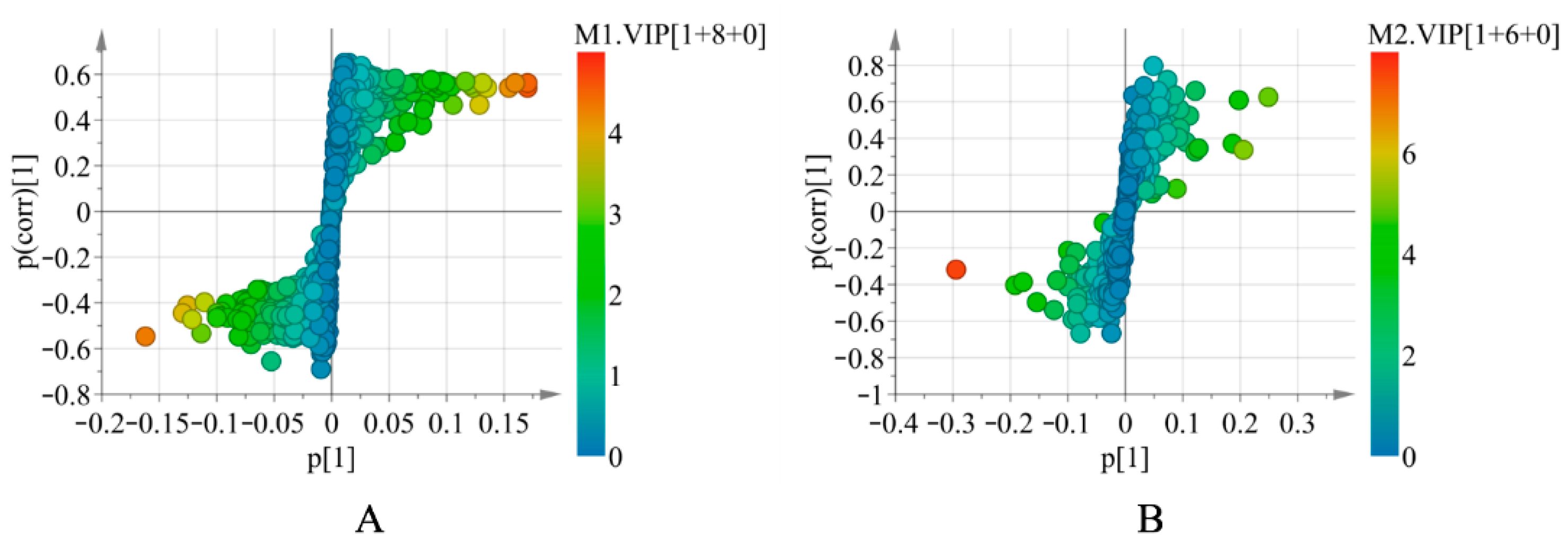
In addition, based on the above models, the variables with the variable importance in the projection (VIP) > 1 were explicitly detected. To further visualize the components with VIP > 1, OPLS-DA was performed to generate S-plots (Figure 3). Finally, 14 differential volatile components (Supplementary Table S5) and 14 nonvolatile markers (Supplementary Table S6) were characterized, all of which played a significant role in the differentiation of different origins of S. tetrandra.

Figure 3.
The OPLS-DA S-plots of S. tetrandra by GC-MS analysis (A) and UHPLC-Q-TOF-MS/MS analysis in positive ion mode (B).
2.5. Quantitative Analysis of Habitat-Related Chemical Markers Based on Nonvolatile Components
Alkaloids are the main active ingredients and pharmacodynamic substances in S. tetrandra. Among them, tetrandrine, fangchinoline, isocorydine, magnocurarine, magnoflorine, boldine, and higenamine have many pharmacological activities, such as antimicrobial effects, anti-inflammatory, anticancer, immunomodulatory effects, and antiplatelet effects [2,12,13,14,15]. Thus, they were picked as habitat-related chemical markers for further quantitative analysis.
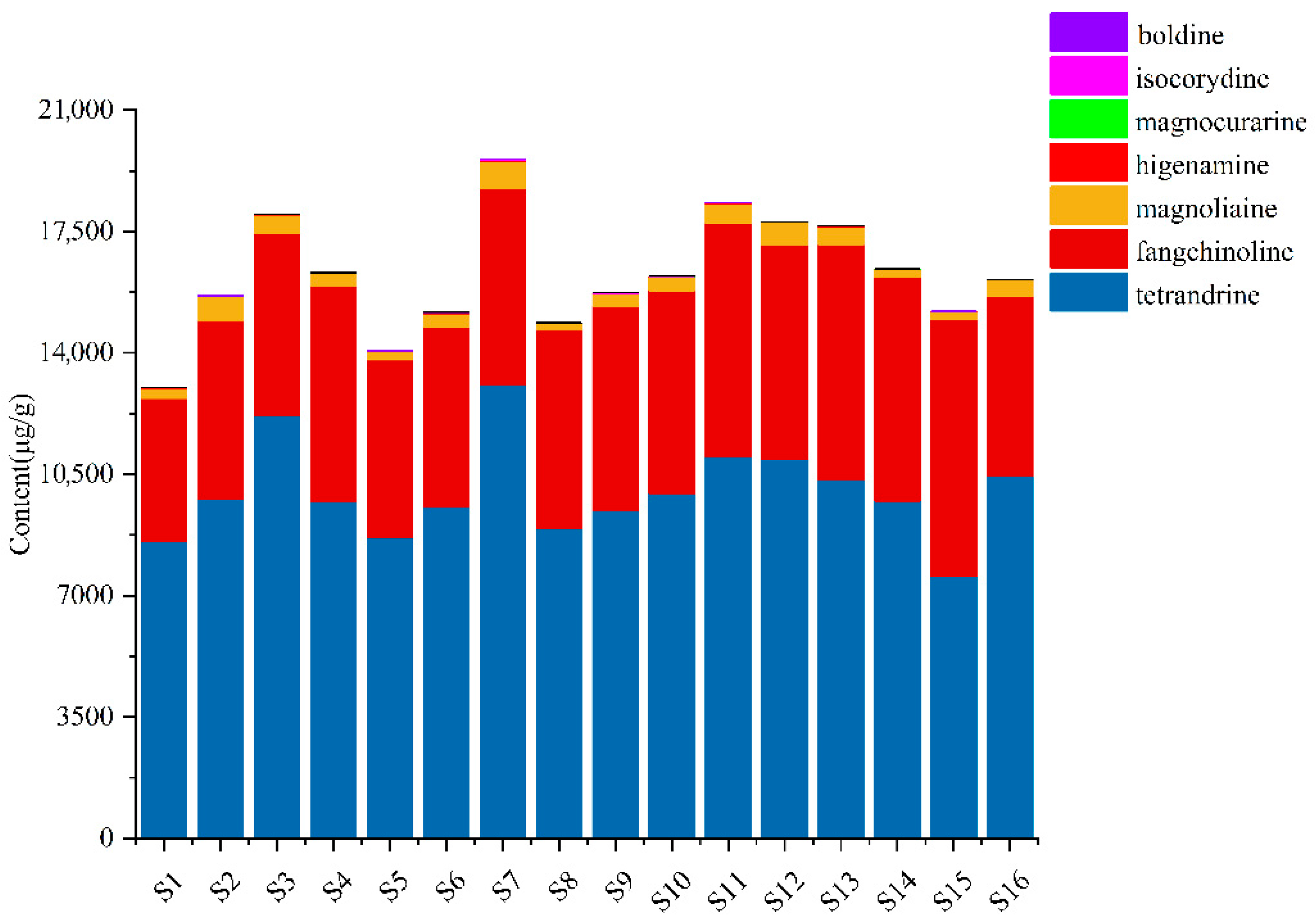
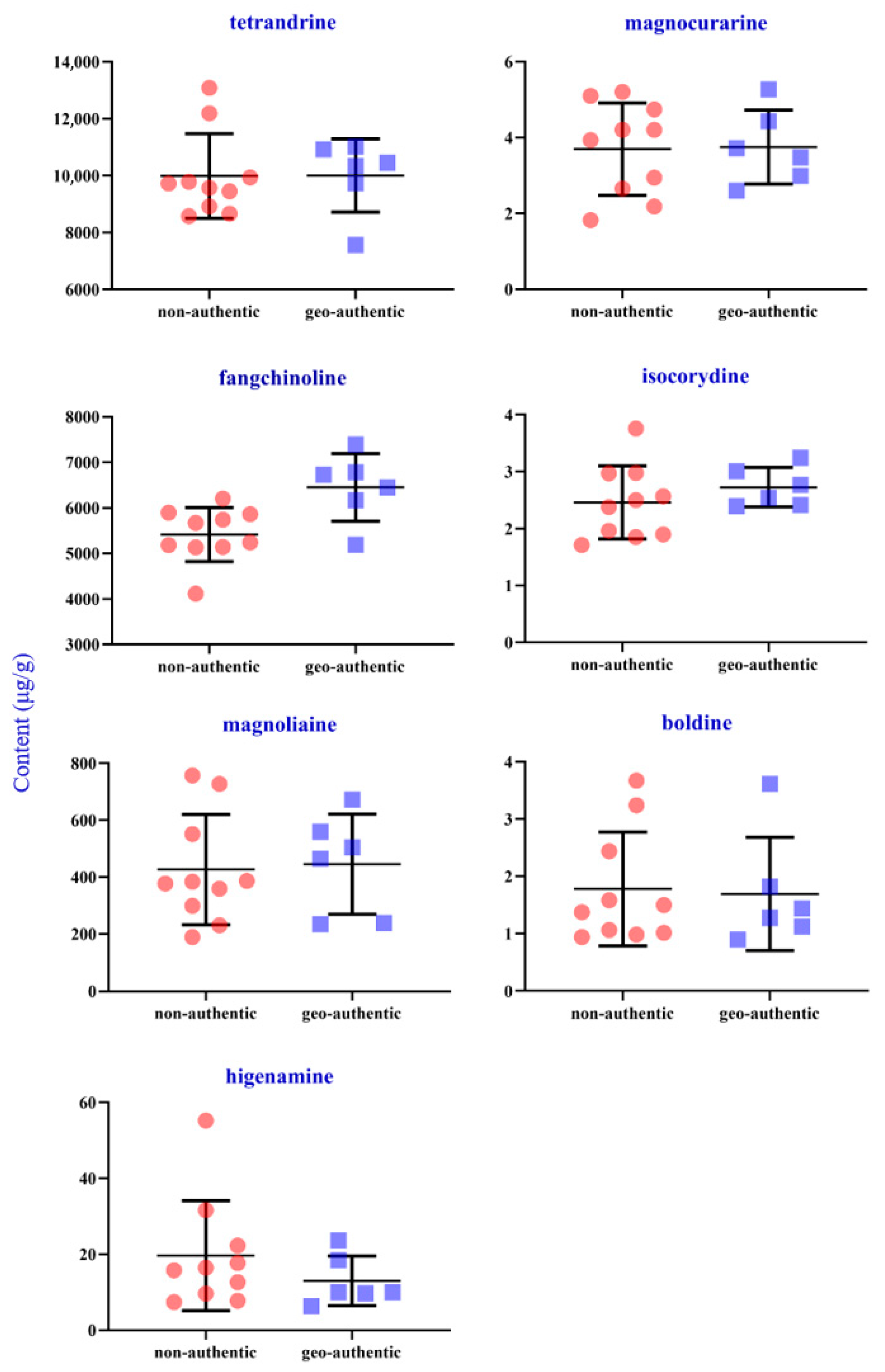
The structural formulas and detailed content of seven analytes in the 16 samples from different origins are exhibited in Figure S3 and Supplementary Table S7. The contents of seven compounds in sample extracts from different origins were inconsistent, which indicated that the different growth conditions, such as climate, and sunlight of different origins, may influence the quality of S. tetrandra. As shown in Figure 4, the total contents of analytes in samples from geo-authentic origins (S11–S16), especially the characteristic components, tetrandrine and fangchinoline, were higher. Besides, the contents of tetrandrine, fangchinoline, isocorydine, and higenamine in samples from geo-authentic origins were highly consistent (Figure 5). In contrast, the compound contents in non-authentic origin samples showed high fluctuation, and the contents of the S1 and S5 samples were low.
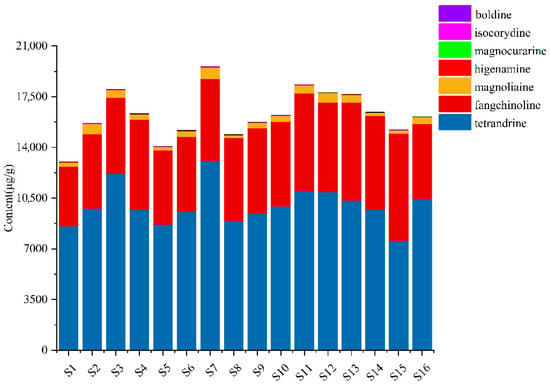
Figure 4.
The total contents of seven alkaloids of S. tetrandra from different batches.

Figure 5.
Contents of seven alkaloids of S. tetrandra from non-authentic origins and geo-authentic origins.
2.6. Discriminatory Analysis
Discriminant analysis is a multivariate statistical analysis method to determine the classification of research objects according to various characteristic values under the condition of a classification determination. In this study, establishing the domain U = {X1, X2, …, X11}, representing 11 randomly selected samples of S. tetrandra, and selecting the content of seven characteristic peaks such as tetrandrine, fangchinoline, magnoflorine, higenamine, magnocurarine, isocorydine, and boldine as the discriminant factors to form an 11 × 7 matrix. Then, by SPSS 21.0 (IBM, San Diego, CA, USA), Wilk’s lambda method was used for stepwise discriminant analysis, and a discriminant function for determining the origin of S. tetrandra was obtained. The regression estimation method was used to evaluate the superiority and inferiority of the discriminant function. Finally, the discriminant function equation of S. tetrandra was obtained as follows (S1: tetrandrine, S2: fangchinoline, S3: magnoflorine, S4: higenamine, S5: magnocurarine, S6: isocorydine, S7: boldine):
Y1 = 0.069S1 + 0.073S2 − 0.157S3 − 0.957S4 − 4.415S5 − 79.374S6 − 30.520S7 − 376.515 (non-authentic origins)
Y2 = 0.069S1 + 0.075S2 − 0.155S3 − 1.056S4 − 7.327S5 − 70.871S6 − 30.181S7 − 399.405 (geo-authentic origins)
By taking the content of each characteristic peak after screening into the function equation and comparing the Y values of the function equations from different origins, the value of which is the largest belonging to the origin represented by this equation. Through the test of the regression estimation method, we tested another five batches of S. tetrandra of known origin, the discriminant analysis of the source origin was compared with the actual results, and the correct rate was 80% (Table 3). This showed that the discriminant function equation established was relatively stable, can achieve the prediction and identification of the origin of S. tetrandra, and is valuable in promotion and application.

Table 3.
Effect evaluation of discriminant function equations of two groups of S. tetrandra samples.
3. Materials and Methods
3.1. Chemical Reagents and Materials
Tetrandrine, fangchinoline, magnoflorine, magnocurarine, isocorydine, higenamine, and boldine were collected from Chengdu DeSiTe Biological Technology Co., Ltd. (Chengdu, China). Chromatographic grade methanol and acetonitrile were provided by Thermo Fisher Scientific Co., Ltd. (Shanghai, China). Formic acid was purchased from ROE Co., Ltd. (St. Louis, MO, USA). The Milli-Q system (Millipore, Bedford, MA, USA) was used to obtain purified water.
Sixteen batches of S. tetrandra were collected from seven different provinces (Guangxi, Guangdong, Sichuan, Neimeng, Anhui, Zhejiang, and Jiangxi) in China. The sample information is shown in Supplementary Table S8.
3.2. Preparation of Standard and Sample Solutions
Stock solutions of tetrandrine, fangchinoline, magnoflorine, magnocurarine, isocorydine, higenamine, and boldine were dissolved with methanol at a concentration of 10 mg/mL and serially diluted to plot the standard curves.
All dried samples were grounded, and the powder was passed through a 50-mesh sieve. Powdered samples (1 g) were ultrasonically extracted in 10 mL of n-hexane for 30 min. After cooling, the resulting mixture was centrifuged, and the supernatant filtered through 0.22 µm nylon membranes was collected for GC-MS analysis.
Pulverized samples (0.5 g) were sonicated in 20 mL of 70% methanol for 30 min. After centrifugation, the supernatants were filtered through a 0.22 µm nylon membrane to obtain the sample solutions for LC-MS/MS analysis.
Meanwhile, all equivalent volumes (100 μL) of sample solutions for GC-MS and LC-MS/MS analysis were respectively mixed as quality control (QC) samples. These sample solutions were all stored at 4 °C until analysis.
3.3. GC-MS Analysis
The volatile components were analyzed by a QP 2010 GC-MS (Shimadzu, Kyoto, Japan). Chromatographic separation was performed on a DB-5MS column (0.25 µm × 0.25 mm × 30 m). The temperature program was set as follows: 40 °C for 0 min, 40–190 °C at 10 °C/min, 190–210 °C at 3 °C/min, 210–215 °C at 1 °C/min, 215–240 °C at 6 °C/min.
Mass spectrometry was performed in electron impact (EI) mode and full scan mode at a mass-to-charge ratio (m/z) of 50–1000. The temperatures of the ion source and interface were 230 °C and 250 °C, respectively.
3.4. UHPLC-Q-TOF-MS/MS Analysis
The UHPLC-Q-TOF-MS/MS system is comprised of an Agilent 1290 UHPLC instrument (Agilent Technologies Inc., Palo Alto, CA, USA) and Agilent 6520 Q-TOF mass spectrometer (Agilent Corporation, Santa Clara, CA, USA). Chromatographic separation was performed on a Waters ACQUITY UPLC®®CSHTM C18 column (2.1 × 100 mm, 1.7 µm) at a temperature of 30 °C. The mobile phase consisted of 0.1% formic acid in water (solvent A) and acetonitrile (solvent B) at a flow rate of 0.2 mL/min, with a gradient elution program of 5–14% B at 0–5 min, 14–18% B at 5–12 min, 18–34% B at 12–14 min, 34–72% B at 14–22 min, 72–86% B at 22–23 min, 86–95% B at 23–31 min, and 95–95% B at 31–35 min. The injection volume for each sample was 5 μL.
The mass spectrometer was operated in both positive and negative modes with a scanning range of m/z 50–1500 and a scanning rate of 1 spectra/s. High resolution (4 GHz, High Res Mode) was used. The optimized instrumental parameters were as follows: capillary temperature, 350 °C; drying gas (N2) flow rate, 8 L/min; nebulizer pressure, 25 psi; collision energy, 30 V; fragment voltage, 135 V.
3.5. UHPLC-MS/MS Analysis
Quantitative analysis was performed on an Agilent 1290 UHPLC system (Agilent Technologies Inc., Palo Alto, CA, USA) equipped with an Agilent 6470 Triple quadrupole tandem mass spectrometer (Agilent Technologies, Singapore) with electrospray ionization (ESI) source. A Waters ACQUITY UPLC®® BEH C18 column (2.1 × 100 mm, 1.7 µm) was used for chromatographic separation, and the column temperature was maintained at 20 °C. The binary gradient elution system consisted of 0.05% formic acid in water (A) and acetonitrile (B). The gradient profile started from 10% B, increased linearly to 23% B within 2 min, and then increased to 50% B within 3 min.
The mass spectrometer was operated in positive mode. The optimized mass conditions were as follows: gas temperature, 320 °C; gas flow rate, 8 L/min; nebulizer, 35 psi; sheath gas temperature, 250 °C; sheath gas flow, 12 L/min; capillary voltage, 4000 V. Multiple reaction monitoring (MRM) mode was applied for the quantitative analysis of different compounds. An MRM diagram is shown in Supplementary Figure S4. The optimal mass spectral parameters and ion patterns are presented in Table 4.

Table 4.
Mass spectrometry parameters of seven target alkaloids.
3.6. Method Validation
The GC-MS and UHPLC-Q-TOF-MS/MS methods were validated in terms of precision, repeatability, and stability. The precision was evaluated by observing the intraday variations of the QC sample six consecutive times. The repeatability was accessed by preparing six replicate QC samples. The stability was obtained by detecting one QC sample at 0, 6, 12, 18, and 24 h. Fifteen chromatographic peaks were randomly selected to calculate the RSDs of peak area and RT to investigate precision, repeatability, and stability.
The methodology of UHPLC-MS/MS analysis was verified by determining linearity, LLOQs, precision, repeatability, stability, and recovery. A calibration curve for each alkaloid standard was constructed using a linear regression model, and linearity was verified using correlation coefficients (r). The LLOQs were estimated as the minimum concentration giving signal-to-noise ratios (S/N) of 10. Instrument precision was determined by analyzing six replicates. Repeatability was evaluated by performing six replicate analyses on the same QC sample. In the stability test, the QC sample solutions were stored at room temperature and then analyzed by replicate injections at 0, 2, 4, 8, 12, and 24 h. The recoveries for spiked samples were applied to examine the effect of the extraction method and matrix effect. Blank S. tetrandra samples (0.25 g) and certain amounts of mixed standard solution were dissolved in 20 mL of 70% methanol and then processed by the optimized method, which were regarded as the spiked samples. The recovery rate was calculated using the following formula: recovery rate (%) = (observed amount − original amount)/spiked amount × 100%. Pulverized samples (0.5 g) were sonicated in 20 mL of 70% methanol for 30 min
3.7. Data Preprocessing
The GC-MS and UHPLC-Q-TOF-MS/MS data of S. tetrandra from different origins were exported in MZ format using the GC-MS Postrun (Shimadzu, Kyoto, Japan) and Agilent Masshunter Qualitative Analysis software packages, respectively. The peak finding, alignment, and filtering of the raw data were preprocessed using R 2.7.2 software (R Foundation for Statistical Computing, Vienna, Austria) to obtain the Rt, m/z, and peak strength of each compound. Finally, the obtained data were imported into Simca-P 14.1 (Umetrics, Umea, Sweden) for OPLS-DA. Potential chemical markers to differentiate the S. tetrandra from different origins were screened according to the VIP value. R2 and Q2 values were used to validate the model. R2 implied the explanation capability towards original data, and Q2 indicated the prediction ability of the model. The discriminant analysis function equation was established by SPSS 21.0 software.
4. Conclusions
In conclusion, by combing the volatile and nonvolatile components based on multiple chromatographic analyses, the habitat-related chemical markers of S. tetrandra were discovered. Through integrated chemometrics analysis, 14 volatile components and 14 non-volatile oils were screened out as the important contributors to the chemical difference between geo-authentic and non-authentic origins samples. Among these, tetrandrine, fangchinoline, isocorydine, magnocurarine, magnoflorine, boldine, and higenamine as chemical markers with abundant pharmacological activities were quantitatively analyzed by UHPLC-MS/MS. The results showed that the total content of analytes in samples from geo-authentic origins was higher and more consistent. Finally, discriminant analysis was used to simulate the function equation representing the origin of S. tetrandra to trace the origin of medicinal materials, and it also verified the accuracy of the differential components obtained by OPLS-DA. The proposed method could aid the exploration of habitat-related chemical markers for other herbal medicines.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/molecules27217224/s1, Figure S1: TIC diagram of S. tetrandra based on GC-MS analysis; Figure S2: TICs of S. tetrandra in positive (A) and negative (B) ions model using UHPLC-Q-TOF-MS/MS analysis; Figure S3: The structures of seven investigated alkaloids; Figure S4: MRM chromatograms of higenamine (1), magnocurarine (2), boldine (3), magnoflorine (4), isocorydine (5), fangchinoline (6), tetrandrine (7). (A) standard solution; (B) S. tetrandra sample; Table S1: The RSDs of precision, repeatability, and stability in GC-MS and UHPLC-Q-TOF-MS/MS analysis; Table S2: Linear equation, linear range, correlation coefficients (r) and lower LOQ of seven alkaloids in UHPLC-MS/MS analysis (n = 6); Table S3: RSDs of precision, repeatability, and stability of seven alkaloids in UHPLC-MS/MS analysis (n = 6); Table S4: The results of recovery test of seven alkaloids in UHPLC-MS/MS analysis (n = 6); Table S5: Potential volatile markers responsible for differentiation of S. tetrandra from non-authentic and geo-authentic origins; Table S6: Differential nonvolatile compounds responsible for differentiation of S. tetrandra from non-authentic and geo-authentic origins; Table S7: The contents of seven alkaloids in S. tetrandra from different origins (µg/g); Table S8: Source information of 16 batches S. tetrandra samples.
Author Contributions
Conceptualization, J.H. and H.O.; formal analysis, X.C.; data curation, Z.L. and W.W.; writing—original draft preparation, X.C. and X.M.; writing—review and editing, M.G., M.Z. and Y.C.; supervision, J.H. and H.O. All authors have read and agreed to the published version of the manuscript.
Funding
This study was supported by the Science and Technology Project of Haihe Laboratory of Modern Chinese Medicine (22HHZYJC00005).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
All datasets presented in this study are included in the article/Supplementary Material.
Conflicts of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Sample Availability
The Stephania tetrandra samples are available from the authors.
References
- State Pharmacopoeia Commission. Pharmacopoeia of People’s Republic of China; China Medical Science and Technology Press: Beijing, China, 2020; p. 155.
- Jiang, Y.P.; Liu, M.; Liu, H.T.; Liu, S. A critical review: Traditional uses, phytochemistry, pharmacology and toxicology of Stephania tetrandra S. Moore (Fen Fang Ji). Phytochem Rev. 2020, 19, 449–489. [Google Scholar] [CrossRef] [PubMed]
- Jiangsu New Medical College. Dictionary of Traditional Chinese Medicine; Shanghai Science and Technology Publishing House: Shanghai, China, 2006; pp. 1182–1185. [Google Scholar]
- Cao, J.; Yang, Z.W.; Li, F.; Zhao, H.Y.; Zhang, Y.C. Advances on the clinical application of tetrandrine. World Clinical Drugs 2013, 34, 75–79. [Google Scholar]
- Zhang, Y.L.; Qi, D.L.; Gao, Y.Q.; Liang, C.X.; Zhang, Y.K.; Ma, Z.; Liu, Y.T.; Peng, H.; Zhang, Y.; Qin, H.; et al. History of uses, phytochemistry, pharmacological activities, quality control and toxicity of the root of Stephania tetrandra S. Moore: A review. J. Ethnopharmacol. 2020, 260, 112995. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Zhou, G.B.; Liu, P.; Song, J.H.; Liang, Y.; Yan, X.J.; Xu, F.; Wang, B.S.; Mao, J.H.; Shen, Z.X.; et al. Dissection of mechanisms of Chinese medicinal formula Realgar-Indigo naturalis as an effective treatment for promyelocytic leukemia. Proc. Natl. Acad. Sci. USA 2008, 105, 4826–4831. [Google Scholar] [CrossRef]
- Liu, X.J.; Wei, F.X.; Liu, H.L.; Zhao, S.J.; Du, G.H.; Qin, X.M. Integrating hippocampal metabolomics and network pharmacology deciphers the antidepressant mechanisms of Xiaoyaosan. J. Ethnopharmacol. 2021, 268, 113549. [Google Scholar] [CrossRef]
- Xu, F.; Kong, M.; Xu, J.D.; Xu, J.; Jiang, Y.; Li, S.L. Effects of sulfur fumigation and heating desulfurization on quality of medicinal herbs evaluated by metabolomics and glycomics: Codonopsis Radix, a pilot study. J. Pharm. Biomed. Anal. 2020, 191, 113581. [Google Scholar] [CrossRef] [PubMed]
- Ziegel, E.R. Statistics and chemometrics for analytical chemistry. Technometrics 2004, 46, 498–499. [Google Scholar] [CrossRef]
- Kumar, N.; Bansal, A.; Sarma, G.S.; Rawal, R.K. Chemometrics tools used in analytical chemistry: An overview. Talanta 2014, 123, 186–199. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.D.; Li, J.; Yang, X.J.; Wang, H.; He, J.; Liu, E.W.; Gao, X.M.; Chang, Y.X. A Metabolomics Coupled With Chemometrics Strategy to Filter Combinatorial Discriminatory Quality Markers of Crude and Salt-Fired Eucommiae Cortex. Front. Pharmacol. 2020, 11, 838. [Google Scholar] [CrossRef]
- Pan, J.X.; Chen, G.; Li, J.J.; Zhu, Q.D.; Li, J.J.; Chen, Z.J.; Yu, Z.P.; Ye, L.Y. Isocorydine suppresses doxorubicin-induced epithelial-mesenchymal transition via inhibition of ERK signaling pathways in hepatocellular carcinoma. Am. J. Cancer. Res. 2018, 8, 154–164. [Google Scholar]
- Arbain, D.; Ahmad, A.; Nelli, Z.; Sargent, M.V. (R)-Magnocurarine from Evodia cf. trichotoma. Planta. Med. 1993, 59, 290. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.Y.; Xu, H.H.; Qiao, S.Y.; Lu, C.; Wang, G.; Liu, M.J.; Guo, B.S.; Tan, Y.; Ju, D.H.; Xiao, C. Boldine isolated from Litsea cubeba inhibits bone resorption by suppressing the osteoclast differentiation in collagen-induced arthritis. Int. Immunopharmacol. 2017, 51, 114–123. [Google Scholar] [CrossRef] [PubMed]
- Wu, M.P.; Zhang, Y.S.; Zhou, Q.M.; Xiong, J.; Dong, Y.R.; Yan, C. Higenamine protects ischemia/reperfusion induced cardiac injury and myocyte apoptosis through activation of β2-AR/PI3K/AKT signaling pathway. Pharmacol. Res. 2016, 104, 115–123. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.X.; Luo, Y.Y.; Liu, X.H.; Song, J.P.; Hua, Y.J.; Wang, S.N.; Zhao, H.; Yan, Y. Difference of chemical compositions in Xanthii Herba and Xanthii Fructus by UPLC-Triple TOF-MS/MS. Zhong Cao Yao 2016, 47, 3951–3958. [Google Scholar] [CrossRef]
- Kong, J.; Liu, C.X.; Zhang, N.; Li, W.; Zhang, Y.C.; Jiang, W.Q.; Qu, Y.X.; Li, Y.Z.; Huang, J.M. Qualitative analysis of chemical composition of Xiaoxianxiong Decoction based on UPLC-Q-TOF/HRMSE. Drug Eval. Res. 2020, 43, 1273–1282. [Google Scholar] [CrossRef]
- Zhai, X.Y.; Zhang, L.; Li, B.T.; Feng, Y.L.; Xu, G.L.; Ouyang, H.; Yang, S.L.; Chen, J. Discrimination of toxic ingredient between raw and processed Pinellia ternata by UPLC/Q-TOF-MS/MS with principal component analysis and T-test. Chin. Herb. Med. 2019, 11, 200–208. [Google Scholar] [CrossRef]
- Dong, F.; Li, Z.X.; Jia, C.M.; Sun, Y.Z.; Sun, Z.H.; Song, X.Y.; Wang, M.Q. Component analysis of sesame oil based on UPLC/Q-TOF MS/MS. China Oils Fats 2014, 29, 120–124. Available online: https://kns.cnki.net/kcms/detail/61.1099.TS.20211129.1053.008.html (accessed on 3 October 2022).
- Zhao, X.M.; Cheng, Y.X.; Liang, C.X.; Guo, J.; Liu, X.Q.; Feng, W.H.; Zhao, Z.B.; Yan, L.H.; Wang, Z.M. Analysis of Chemical Constituents in Euodiae Fructus by UPLC-Q-TOF-MS/MS. Zhongguo Shi Yan Fang Ji Xue Za Zhi 2021, 27, 113–126. [Google Scholar] [CrossRef]
- Qin, W.H.; Hua, L.; Guo, Y.L.; Wang, Y.H.; Ran, J.C.; Yang, Y. Differentiation of Different Parts of Cordyceps sinensis Based on UPLC-Q-TOF-MS Combined with Metabolomics Methods. Zhongguo Shi Yan Fang Ji Xue Za Zhi 2018, 24, 69–76. [Google Scholar] [CrossRef]
- Deng, G.M.; Xiang, B.; Xiao, X.Q.; Ge, J.W.; Chen, Z.; Yang, L.P.; Wei, F. Study on Chemical Constituents of Lindera aggregate by GC-MS and UPLC-ESI-MS/MS. Zhong Yao Cai 2016, 39, 2229–2236. [Google Scholar] [CrossRef]
- Wei, C.H.; Zeng, J.X.; Shi, Y.F.; Luo, G.M.; Gao, Y.P.; Zhong, G.Y.; Zhu, J.X. Analytical Studies on Corydalis hendersonii Hemsl by UHPLC-Q-TOF-MS. J. JiangxiI Univ. TCM 2020, 26, 131–139. [Google Scholar] [CrossRef]
- Tian, X.M.; Yan, L.H.; Jiang, L.Y.; Xiang, G.F.; He, Y.J. Chemical composition analysis of Schima superba root based on UPLC/Q-TOF-MS metabonomics. Zhongguo Nong Xue Tong Bao 2018, 34, 76–83. [Google Scholar]
- Liu, C.M.; Hua, Z.D.; Bai, Y.P. Determination of Opium Alkaloids and Discrimination of Sample Origins by UPLC/Q-TOF and PLS-DA. Res. Pap. 2014, 30, 65–68. [Google Scholar] [CrossRef]
- Chen, M. Study on the chemical components and metabolism of Fangji Huangqi Tang. Shanxi Univ. 2018. [Google Scholar]
- Ye, J.; Yang, M.J.; Yang, X.Y.; Zhang, H.; Zan, L.F. Analysis of chemical constituents in Ziziphus jujuba var. spinosa folium by UPLC-QTOF-MS. Nat. Prod. Res. Dev. 2019, 31, 1183–1191. [Google Scholar] [CrossRef]
- Xiao, J.; Song, N.N.; Lu, T.; Pan, Y.N.; Song, J.Y.; Chen, G.; Sun, L.; Li, N. Rapid characterization of TCM Qianjinteng by UPLC-QTOF-MS and its application in the evaluation of three species of Stephania. J. Pharm. Biomed. Anal. 2018, 156, 284–296. [Google Scholar] [CrossRef] [PubMed]
- Sim, H.J.; Yoon, S.H.; Kim, M.S.; Kim, B.; Park, H.M.; Hong, J. Identification of alkaloid constituents from Fangchi species using pH control liquid-liquid extraction and liquid chromatography coupled to quadrupole time-of-flight mass spectrometry. Rapid. Commun. Mass. Spectrom. 2015, 29, 837–854. [Google Scholar] [CrossRef]
- Hu, H.L.; Wang, Z.G.; Hua, W.; You, Y.; Zou, L. Effect of Chemical Profiling Change of Processed Magnolia officinalis on the Pharmacokinetic Profiling of Honokiol and Magnolol in Rats. J. Chromatogr. Sci. 2016, 54, 1201–1212. [Google Scholar] [CrossRef]
- Zheng, X.J.; Zheng, W.L.; Zhou, J.J.; Gao, X.; Liu, Z.H.; Han, N.; Yin, J. Study on the discrimination between Corydalis Rhizoma and its adulterants based on HPLC-DAD-Q-TOF-MS associated with chemometric analysis. J. Chromatogr. B. Analyt. Technol. Biomed. Life Sci. 2018, 1090, 110–121. [Google Scholar] [CrossRef]
- Qin, W.H.; Ran, J.C.; Ye, L.H.; Hua, L.; Wang, Y.H.; Guo, Y.L.; Yang, Y. UPLC-Q/TOF method for simultaneous qualitative and quantitative analysis of main chemical constituents in medicinal materials of Sinodielsia yunnanensis. Zhong Cao Yao 2018, 49, 3576–3582. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).