Analysis of the Antimicrobial and Anti-Biofilm Activity of Natural Compounds and Their Analogues against Staphylococcus aureus Isolates
Abstract
1. Introduction
2. Results
2.1. Antibiotic Resistance/Susceptibility Profile of S. aureus Isolates
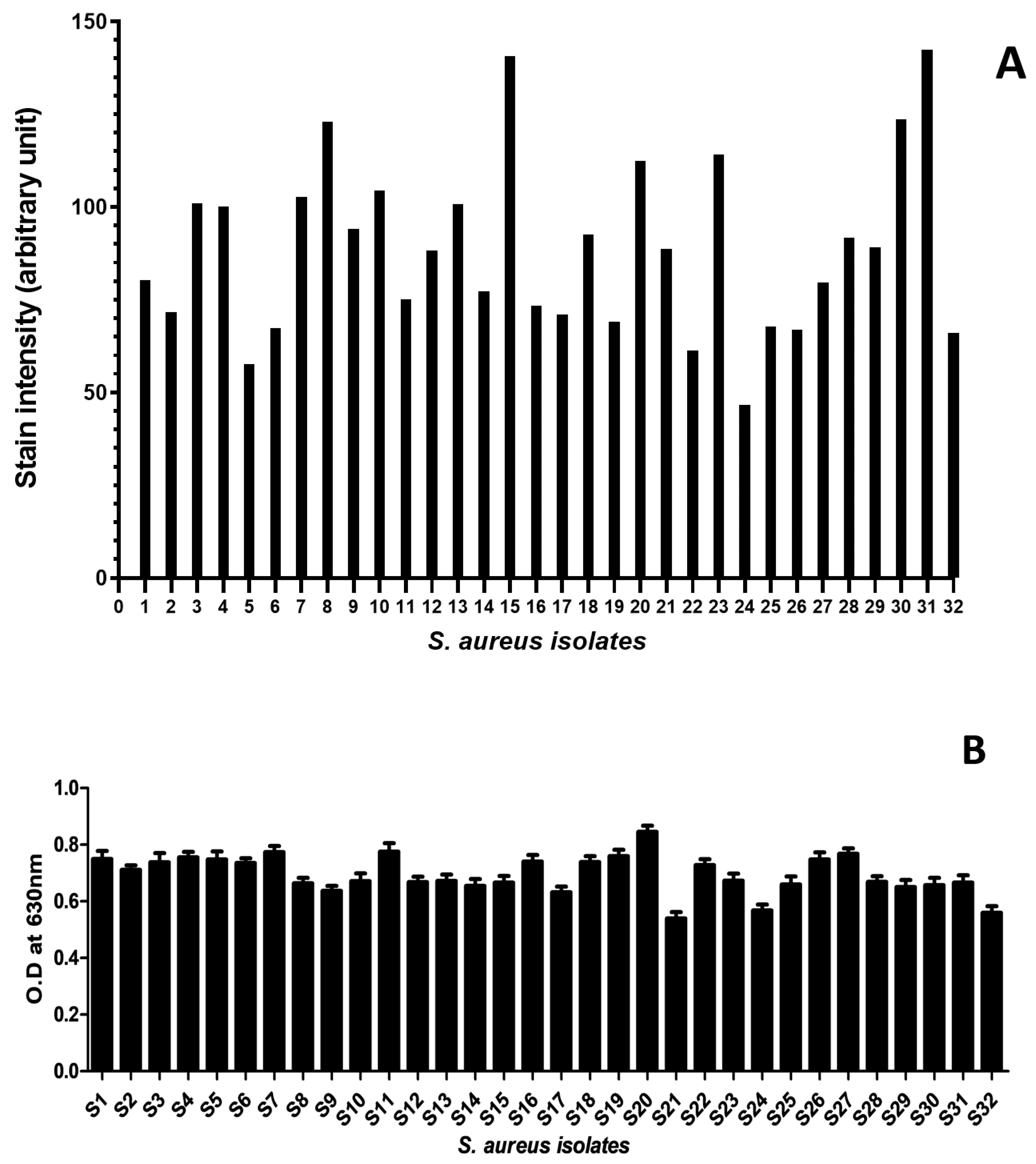
2.2. Analysis of Biofilm-Forming Capacity of S. aureus Isolates
2.3. Minimum Inhibitory Concentration (MIC) and Anti-Biofilm Activity of Test Compounds (I–XI) against S. aureus Isolates
2.4. Effect of Test Compounds on Expression of Biofilm-Associated Genes
3. Discussion
4. Materials and Methods
4.1. S. aureus Isolates
4.2. Kirby-Bauer Disc Diffusion Assay for Determination of Antibiotic Susceptibility/Resistance Pattern for S. aureus Isolates
4.3. Preparation of Stock Solutions of Natural Compounds
4.4. Determination of Biofilm Forming Potential of the S. aureus Isolates
4.4.1. Air–Liquid Interface Assay
4.4.2. Spectrophotometric Assay for Quantitative Analysis of Biofilm Formation
4.4.3. Analysis of Correlation
4.5. Determination of Minimum Inhibitory Concentration of Compounds (I–XI)
Determination of Bactericidal/Bacteriostatic Effect of Compounds (I–XI)
4.6. Determination of the Inhibitory Activity of the Test Compounds (I–XI) against S. aureus Biofilms
4.7. RNA Extraction, cDNA Synthesis, and qPCR Analysis of the Genes Involved in Biofilm Formation
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Skov, R.; Jensen, K. Community-associated meticillin-resistant Staphylococcus aureus as a cause of hospital-acquired infections. J. Hosp. Infect. 2009, 73, 364–370. [Google Scholar] [CrossRef] [PubMed]
- Hurley, M.N.; Smyth, A.R. Staphylococcus aureus in cystic fibrosis: Pivotal role or bit part actor? Curr. Opin. Pulm. Med. 2018, 24, 586–591. [Google Scholar] [CrossRef]
- Merghni, A.; Nejma, M.B.; Dallel, I.; Tobji, S.; Amor, A.B.; Janel, S.; Lafont, F.; Aouni, M.; Mastouri, M. High potential of adhesion to biotic and abiotic surfaces by opportunistic Staphylococcus aureus strains isolated from orthodontic appliances. Microb. Pathog. 2016, 91, 61–67. [Google Scholar] [CrossRef]
- Speziale, P.; Pietrocola, G.; Rindi, S.; Provenzano, M.; Provenza, G.; Di Poto, A.; Visai, L.; Arciola, C.R. Structural and functional role of Staphylococcus aureus surface components recognizing adhesive matrix molecules of the host. Future Microbiol. 2009, 4, 1337–1352. [Google Scholar] [CrossRef] [PubMed]
- Jefferson, K.K. What drives bacteria to produce a biofilm? FEMS Microbiol. Lett. 2004, 236, 163–173. [Google Scholar] [CrossRef] [PubMed]
- Boles, B.R.; Horswill, A.R. Staphylococcal biofilm disassembly. Trends Microbiol. 2011, 19, 449–455. [Google Scholar] [CrossRef] [PubMed]
- Uruén, C.; Chopo-Escuin, G.; Tommassen, J.; Mainar-Jaime, R.C.; Arenas, J. Biofilms as promoters of bacterial antibiotic resistance and tolerance. Antibiotics 2020, 10, 3. [Google Scholar] [CrossRef] [PubMed]
- Saravanan, M.; Nanda, A. Incidence of methicillin resistant Staphylococcus aureus (MRSA) from septicemia suspected children. Indian J. Sci. Technol. 2009, 2, 36–39. [Google Scholar] [CrossRef]
- Nikhat, S.; Fazil, M. History, phytochemistry, experimental pharmacology and clinical uses of honey: A comprehensive review with special reference to Unani medicine. J. Ethnopharmacol. 2022, 282, 114614. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Su, B.; Jiang, H.; Cui, N.; Yu, Z.; Yang, Y.; Sun, Y. Traditional uses, phytochemistry and pharmacological activities of the genus Cinnamomum (Lauraceae): A review. Fitoterapia 2020, 146, 104675. [Google Scholar] [CrossRef]
- Deryabin, D.; Galadzhieva, A.; Kosyan, D.; Duskaev, G. Plant-derived inhibitors of AHL-mediated quorum sensing in bacteria: Modes of action. Int. J. Mol. Sci. 2019, 20, 5588. [Google Scholar] [CrossRef] [PubMed]
- Narisawa, N.; Furukawa, S.; Ogihara, H.; Yamasaki, M. Estimation of the biofilm formation of Escherichia coli K-12 by the cell number. J. Biosci. Bioeng. 2005, 99, 78–80. [Google Scholar] [CrossRef]
- Ebert, C.; Tuchscherr, L.; Unger, N.; Pöllath, C.; Gladigau, F.; Popp, J.; Löffler, B.; Neugebauer, U. Correlation of crystal violet biofilm test results of Staphylococcus aureus clinical isolates with Raman spectroscopic read-out. J. Raman Spectrosc. 2021, 52, 2660–2670. [Google Scholar] [CrossRef]
- Nourbakhsh, F.; Namvar, A.E. Detection of genes involved in biofilm formation in Staphylococcus aureus isolates. GMS Hyg. Infect. Control 2016, 11, Doc07. [Google Scholar] [PubMed]
- Rajaduraipandi, K.; Mani, K.; Panneerselvam, K.; Mani, M.; Bhaskar, M.; Manikandan, P. Prevalence and antimicrobial susceptibility pattern of methicillin resistant Staphylococcus aureus: A multicentre study. Indian J. Med. Microbiol. 2006, 24, 34–38. [Google Scholar] [CrossRef]
- Gandara, A.; Mota, L.C.; Flores, C.; Perez, H.R.; Green, C.F.; Gibbs, S.G. Isolation of Staphylococcus aureus and antibiotic-resistant Staphylococcus aureus from residential indoor bioaerosols. Environ. Health Perspect. 2006, 114, 1859–1864. [Google Scholar] [CrossRef] [PubMed]
- Yılmaz, E.Ş.; Aslantaş, Ö. Antimicrobial resistance and underlying mechanisms in Staphylococcus aureus isolates. Asian Pac. J. Trop. Med. 2017, 10, 1059–1064. [Google Scholar] [CrossRef] [PubMed]
- Bouchiat, C.; El-Zeenni, N.; Chakrakodi, B.; Nagaraj, S.; Arakere, G.; Etienne, J. Epidemiology of Staphylococcus aureus in Bangalore, India: Emergence of the ST217 clone and high rate of resistance to erythromycin and ciprofloxacin in the community. New Microbes New Infect. 2015, 7, 15–20. [Google Scholar] [CrossRef]
- Chakrakodi, B.; Prabhakara, S.; Nagaraj, S.; Etienne, J.; Arakere, G. High prevalence of ciprofloxacin resistance in community associated Staphylococcus aureus in a tertiary care Indian hospital. Adv. Microbiol. 2014, 2014, 42306. [Google Scholar]
- Merritt, J.H.; Kadouri, D.E.; O’Toole, G.A. Growing and analyzing static biofilms. Curr. Protoc. Microbiol. 2011, 22, 1B.1.1–1B.1.18. [Google Scholar] [CrossRef]
- Ahmed, K.; Ahmed, H.; Ahmed, F.A.; Ali, A.A.; Akbar, J.; Rana, J.; Tariq, U.; Abidi, S.H. Analysis of anti-microbial and anti-biofilm activity of hand washes and sanitizers against S. aureus and P. aeruginosa. J Pak Med Assoc 2020, 2019, 2776. [Google Scholar] [CrossRef] [PubMed]
- Abidi, S.H.; Sherwani, S.K.; Siddiqui, T.R.; Bashir, A.; Kazmi, S.U. Drug resistance profile and biofilm forming potential of Pseudomonas aeruginosa isolated from contact lenses in Karachi-Pakistan. BMC Ophthalmol. 2013, 13, 57. [Google Scholar] [CrossRef] [PubMed]
- Lima, J.L.d.C.; Alves, L.R.; Jacomé, P.R.L.d.A.; Bezerra Neto, J.P.; Maciel, M.A.V.; Morais, M.M.C.D. Biofilm production by clinical isolates of Pseudomonas aeruginosa and structural changes in LasR protein of isolates non biofilm-producing. Braz. J. Infect. Dis. 2018, 22, 129–136. [Google Scholar] [CrossRef] [PubMed]
- Kuehnast, T.; Cakar, F.; Weinhäupl, T.; Pilz, A.; Selak, S.; Schmidt, M.A.; Rüter, C.; Schild, S. Comparative analyses of biofilm formation among different Cutibacterium acnes isolates. Int. J. Med. Microbiol. 2018, 308, 1027–1035. [Google Scholar] [CrossRef] [PubMed]
- Magana, M.; Sereti, C.; Ioannidis, A.; Mitchell, C.A.; Ball, A.R.; Magiorkinis, E.; Chatzipanagiotou, S.; Hamblin, M.R.; Hadjifrangiskou, M.; Tegos, G.P. Options and limitations in clinical investigation of bacterial biofilms. Clin. Microbiol. Rev. 2018, 31, e00084-16. [Google Scholar] [CrossRef] [PubMed]
- Kord, M.; Ardebili, A.; Jamalan, M.; Jahanbakhsh, R.; Behnampour, N.; Ghaemi, E.A. Evaluation of biofilm formation and presence of ica genes in Staphylococcus epidermidis clinical isolates. Osong Public Health Res. Perspect. 2018, 9, 160. [Google Scholar] [CrossRef]
- Elkhashab, T.H.; Adel, L.A.; Nour, M.S.; Mahran, M.; Elkaffas, M. Association of intercellular adhesion gene A with biofilm formation in staphylococci isolates from patients with conjunctivitis. J. Lab. Physicians 2018, 10, 309–315. [Google Scholar] [CrossRef]
- He, S.; Zhan, Z.; Shi, C.; Wang, S.; Shi, X. Ethanol at subinhibitory concentrations enhances biofilm formation in Salmonella enteritidis. Foods 2022, 11, 2237. [Google Scholar] [CrossRef]
- De, P.; Baltas, M.; Bedos-Belval, F. Cinnamic acid derivatives as anticancer agents-a review. Curr. Med. Chem. 2011, 18, 1672–1703. [Google Scholar] [CrossRef]
- Chochkova, M.; Stoykova, B.; Petrova, P.; Gyoshkova, N.; Ivanova, G.; Štícha, M.; Milkova, T. Synthesis and radical scavenging activity of cinnamic acid esters. Bulg. Chem. Commun. 2017, 1, 68–73. [Google Scholar]
- Marchese, A.; Barbieri, R.; Coppo, E.; Orhan, I.E.; Daglia, M.; Nabavi, S.F.; Izadi, M.; Abdollahi, M.; Nabavi, S.M.; Ajami, M. Antimicrobial activity of eugenol and essential oils containing eugenol: A mechanistic viewpoint. Crit. Rev. Microbiol. 2017, 43, 668–689. [Google Scholar] [CrossRef] [PubMed]
- Chokpaiboon, S.; Unagul, P.; Nithithanasilp, S.; Komwijit, S.; Somyong, W.; Ratiarpakul, T.; Isaka, M.; Bunyapaiboonsri, T. Salicylaldehyde and dihydroisobenzofuran derivatives from the marine fungus Zopfiella marina. Nat. Prod. Res. 2018, 32, 149–153. [Google Scholar] [CrossRef]
- Fitzgerald, D.J.; Stratford, M.; Gasson, M.J.; Narbad, A. Structure−function analysis of the vanillin molecule and its antifungal properties. J. Agric. Food Chem. 2005, 53, 1769–1775. [Google Scholar] [CrossRef] [PubMed]
- El-Saber Batiha, G.; Alkazmi, L.M.; Wasef, L.G.; Beshbishy, A.M.; Nadwa, E.H.; Rashwan, E.K. Syzygium aromaticum L. (Myrtaceae): Traditional uses, bioactive chemical constituents, pharmacological and toxicological activities. Biomolecules 2020, 10, 202. [Google Scholar] [CrossRef] [PubMed]
- Sharma, A.; Shahzad, B.; Rehman, A.; Bhardwaj, R.; Landi, M.; Zheng, B. Response of phenylpropanoid pathway and the role of polyphenols in plants under abiotic stress. Molecules 2019, 24, 2452. [Google Scholar] [CrossRef]
- Ali, I.A.; Matinlinna, J.P.; Lévesque, C.M.; Neelakantan, P. Trans-cinnamaldehyde attenuates Enterococcus faecalis virulence and inhibits biofilm formation. Antibiotics 2021, 10, 702. [Google Scholar] [CrossRef] [PubMed]
- Sinha, A.K.; Sharma, U.K.; Sharma, N. A comprehensive review on vanilla flavor: Extraction, isolation and quantification of vanillin and others constituents. Int. J. Food Sci. Nutr. 2008, 59, 299–326. [Google Scholar] [CrossRef] [PubMed]
- Nićiforović, N.; Abramovič, H. Sinapic acid and its derivatives: Natural sources and bioactivity. Compr. Rev. Food Sci. Food Saf. 2014, 13, 34–51. [Google Scholar] [CrossRef]
- Osman, M.F.; Mohd Hassan, N.; Khatib, A.; Tolos, S.M. Antioxidant activities of Dialium indum L. Fruit and gas chromatography-mass spectrometry (GC-MS) of the active fractions. Antioxidants 2018, 7, 154. [Google Scholar] [CrossRef]
- Kot, B.; Wicha, J.; Piechota, M.; Wolska, K.; Gruzewska, A. Antibiofilm activity of trans-cinnamaldehyde, p-coumaric, and ferulic acids on uropathogenic Escherichia coli. Turk. J. Med. Sci. 2015, 45, 919–924. [Google Scholar] [CrossRef]
- Kikuzaki, H.; Hisamoto, M.; Hirose, K.; Akiyama, K.; Taniguchi, H. Antioxidant properties of ferulic acid and its related compounds. J. Agric. Food Chem. 2002, 50, 2161–2168. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.; Kim, S.; Cho, K.-H.; Lee, J.-H.; Lee, J. Antibiofilm activities of cinnamaldehyde analogs against Uropathogenic Escherichia coli and Staphylococcus aureus. Int. J. Mol. Sci. 2022, 23, 7225. [Google Scholar] [CrossRef] [PubMed]
- Shi, L.; Ge, H.-M.; Tan, S.-H.; Li, H.-Q.; Song, Y.-C.; Zhu, H.-L.; Tan, R.-X. Synthesis and antimicrobial activities of Schiff bases derived from 5-chloro-salicylaldehyde. Eur. J. Med. Chem. 2007, 42, 558–564. [Google Scholar] [CrossRef] [PubMed]
- More, P.G.; Karale, N.N.; Lawand, A.S.; Narang, N.; Patil, R.H. Synthesis and anti-biofilm activity of thiazole Schiff bases. Med. Chem. Res. 2014, 23, 790–799. [Google Scholar] [CrossRef]
- Bilcu, M.; Grumezescu, A.M.; Oprea, A.E.; Popescu, R.C.; Mogoșanu, G.D.; Hristu, R.; Stanciu, G.A.; Mihailescu, D.F.; Lazar, V.; Bezirtzoglou, E. Efficiency of vanilla, patchouli and ylang ylang essential oils stabilized by iron oxide@ C14 nanostructures against bacterial adherence and biofilms formed by Staphylococcus aureus and Klebsiella pneumoniae clinical strains. Molecules 2014, 19, 17943–17956. [Google Scholar] [CrossRef]
- Nostro, A.; Scaffaro, R.; D’arrigo, M.; Botta, L.; Filocamo, A.; Marino, A.; Bisignano, G. Study on carvacrol and cinnamaldehyde polymeric films: Mechanical properties, release kinetics and antibacterial and antibiofilm activities. Appl. Microbiol. Biotechnol. 2012, 96, 1029–1038. [Google Scholar] [CrossRef]
- Malheiro, J.F.; Maillard, J.-Y.; Borges, F.; Simões, M. Evaluation of cinnamaldehyde and cinnamic acid derivatives in microbial growth control. Int. Biodeterior. Biodegrad. 2019, 141, 71–78. [Google Scholar] [CrossRef]
- Knobloch, J.K.-M.; Horstkotte, M.A.; Rohde, H.; Mack, D. Evaluation of different detection methods of biofilm formation in Staphylococcus aureus. Med. Microbiol. Immunol. 2002, 191, 101–106. [Google Scholar] [CrossRef]
- Achek, R.; Hotzel, H.; Nabi, I.; Kechida, S.; Mami, D.; Didouh, N.; Tomaso, H.; Neubauer, H.; Ehricht, R.; Monecke, S. Phenotypic and molecular detection of biofilm formation in Staphylococcus aureus isolated from different sources in Algeria. Pathogens 2020, 9, 153. [Google Scholar] [CrossRef]
- Drożdż, K.; Ochońska, D.; Ścibik, Ł.; Gołda-Cępa, M.; Biegun, K.; Brzychczy-Włoch, M. The Frequency of Occurrence of Resistance and Genes Involved in the Process of Adhesion and Accumulation of Biofilm in Staphylococcus aureus Strains Isolated from Tracheostomy Tubes. Microorganisms 2022, 10, 1210. [Google Scholar] [CrossRef] [PubMed]
- Haddad, O.; Merghni, A.; Elargoubi, A.; Rhim, H.; Kadri, Y.; Mastouri, M. Comparative study of virulence factors among methicillin resistant Staphylococcus aureus clinical isolates. BMC Infect. Dis. 2018, 18, 560. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Rivas, J.M.; Brown, E.L.; Liang, X.; Höök, M. Virulence potential of the staphylococcal adhesin CNA in experimental arthritis is determined by its affinity for collagen. J. Infect. Dis. 2004, 189, 2323–2333. [Google Scholar] [CrossRef] [PubMed]
- Kırmusaoğlu, S. The methods for detection of biofilm and screening antibiofilm activity of agents. In Antimicrobials, Antibiotic Resistance, Antibiofilm Strategies and Activity Methods; IntechOpen: London, UK, 2019; pp. 1–17. [Google Scholar]
- Mulat, M.; Khan, F.; Pandita, A. Chemical composition and antibacterial, anti-biofilm and anti-virulence activities of plant extracts against human pathogenic bacteria. Nat. Prod. J. 2022, 12, 54–68. [Google Scholar] [CrossRef]

| Antibiotics | Sensitive % (N) | Intermediate % (N) | Resistant % (N) |
|---|---|---|---|
| Ampicillin 10 µg (AMP) | 34.3 (11) | 31.2 (10) | 34.3 (11) |
| Gentamicin 120 µg (CN) | 93.7 (30) | 0 | 6.2 (2) |
| Chloramphenicol 30 µg (C) | 90.6 (29) | 3.1 (1) | 6.2 (2) |
| Ciprofloxacin 5 µg (CIP) | 34.3 (11) | 12.5 (4) | 53.1 (17) |
| Amikacin 30 µg (AK) | 81.2 (26) | 12.5 (4) | 6.2 (2) |
| Clindamycin 2 µg (DA) | 65.6 (21) | 21.8 (7) | 12.5 (4) |
| Streptomycin 10 µg (S) | 84.3 (27) | 0 | 15.6 (5) |
| Cephalothin 30 µg (KF) | 81.2 (26) | 0 | 18.5 (6) |
| Isolates | S11 | S12 | S31 | S24 | S25 | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Natural Compounds | Biofilm Inhibition (%) | MIC (mg/mL) | Biofilm Inhibition (%) | MIC (mg/mL) | Biofilm Inhibition (%) | MIC (mg/mL) | Biofilm Inhibition (%) | MIC (mg/mL) | Biofilm Inhibition (%) | MIC (mg/mL) |
| SALI | 70.66 | 1 | 75.07 | 1 | 92.52 | 1 | 72.67 | 30 | 88.59 | 12.5 |
| VAN | 72.99 | 1 | 73.37 | 1 | 87.38 | 55 | 70.15 | 40 | 83.77 | 55 |
| A-MT | 70.15 | 27.5 | 73.44 | 30 | 84.58 | 25 | 70.3 | 100 | 85.53 | 30 |
| A-BT | 72.34 | 1 | 58.31 | 1 | 85.98 | 0.75 | 72.03 | 1 | 89.91 | 5 |
| NNDC | 68.91 | 2.5 | 66.25 | 5 | 69.16 | 21 | 67.84 | 150 | 58.33 | 100 |
| 4A3M | 72.48 | 1 | 68.92 | 1 | 85.05 | 12.5 | 55.8 | 1 | 89.47 | 20 |
| A-MCA | 66.57 | 1 | 67.43 | 1 | 15.88 | 5 | 53.17 | 2.5 | 87.38 | 5 |
| 3H4M | 68.91 | 2.5 | 72.55 | 2.5 | 91.59 | 17.5 | 63.09 | 7.5 | 81.58 | 160 |
| 4H35 | 67.52 | 5 | 72.77 | 2.5 | 92.99 | 15 | 69.07 | 25 | 90.35 | 25 |
| T4N | 72.99 | 1 | 72.03 | 1 | 88.31 | 1 | 71.16 | 250 | 87.28 | 250 |
| EUG | 70.51 | 7.5 | 72.63 | 7.5 | 38.32 | 100 | 58.26 | 100 | 16.22 | 100 |
| Ethanol (+ve control) | 100 | - | 100 | - | 100 | - | 100 | - | 100 | - |
| Blank media (−ve control) | 0 | - | 0 | - | 0 | - | 0 | - | 0 | - |
| Gene | Forward Primer (5′ to 3′) | Reverse Primer (5′ to 3′) |
|---|---|---|
| 16S rRNA | GGGACCCGCACAAGCGGTGG | GGGTTGCGCTCGTTGCGGGA |
| icaA (intercellular adhesion gene) | GAGGTAAAGCCAACGCACTC | CCTGTAACCGCACCAAGTTT |
| fnbA (fibronectin-binding protein A) | AAATTGGGAGCAGCATCAGT | GCAGCTGAATTCCCATTTTC |
| clfA (clumping factor A) | ACCCAGGTTCAGATTCTGGCAGCG | TCGCTGAGTCGGAATCGCTTGCT |
| cna (collagen binding protein) | AATAGAGGCGCCACGACCGT | GTGCCTTCCCAAACCTTTTGAGC |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mastoor, S.; Nazim, F.; Rizwan-ul-Hasan, S.; Ahmed, K.; Khan, S.; Ali, S.N.; Abidi, S.H. Analysis of the Antimicrobial and Anti-Biofilm Activity of Natural Compounds and Their Analogues against Staphylococcus aureus Isolates. Molecules 2022, 27, 6874. https://doi.org/10.3390/molecules27206874
Mastoor S, Nazim F, Rizwan-ul-Hasan S, Ahmed K, Khan S, Ali SN, Abidi SH. Analysis of the Antimicrobial and Anti-Biofilm Activity of Natural Compounds and Their Analogues against Staphylococcus aureus Isolates. Molecules. 2022; 27(20):6874. https://doi.org/10.3390/molecules27206874
Chicago/Turabian StyleMastoor, Sobia, Fizza Nazim, Syed Rizwan-ul-Hasan, Khalid Ahmed, Shabnam Khan, Syed Nawazish Ali, and Syed Hani Abidi. 2022. "Analysis of the Antimicrobial and Anti-Biofilm Activity of Natural Compounds and Their Analogues against Staphylococcus aureus Isolates" Molecules 27, no. 20: 6874. https://doi.org/10.3390/molecules27206874
APA StyleMastoor, S., Nazim, F., Rizwan-ul-Hasan, S., Ahmed, K., Khan, S., Ali, S. N., & Abidi, S. H. (2022). Analysis of the Antimicrobial and Anti-Biofilm Activity of Natural Compounds and Their Analogues against Staphylococcus aureus Isolates. Molecules, 27(20), 6874. https://doi.org/10.3390/molecules27206874






