Recent Advances in Detection for Breast-Cancer-Derived Exosomes
Abstract
:1. Introduction
1.1. Breast Cancer
1.2. Exosomes
1.3. Exosomes and Breast Cancer
2. Common Methods of Exosomes Separation
3. Detection of Breast-Cancer-Derived Exosomes
3.1. Optical Method
3.1.1. Colorimetric Method
General Material
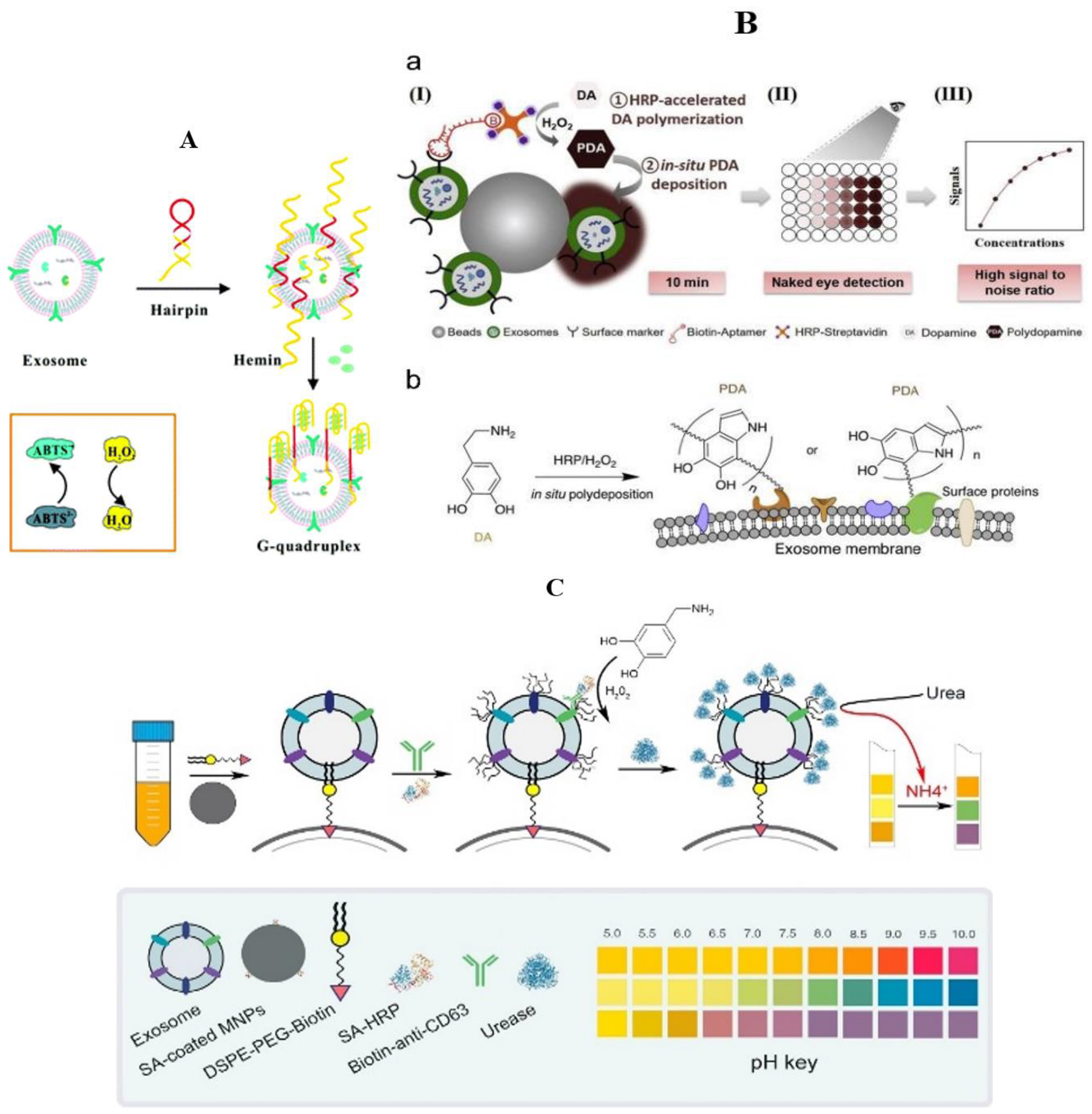
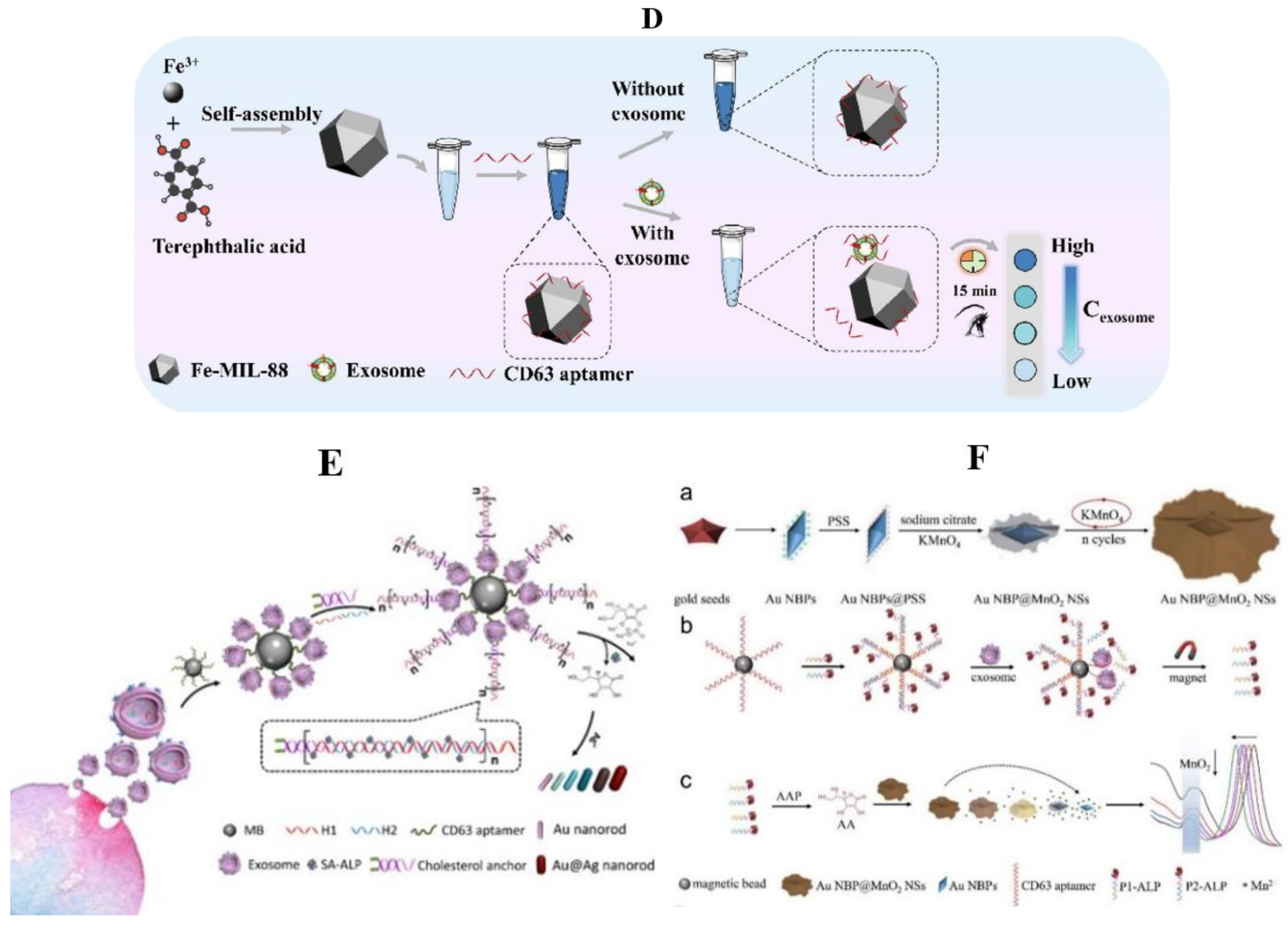
Nanometer Material
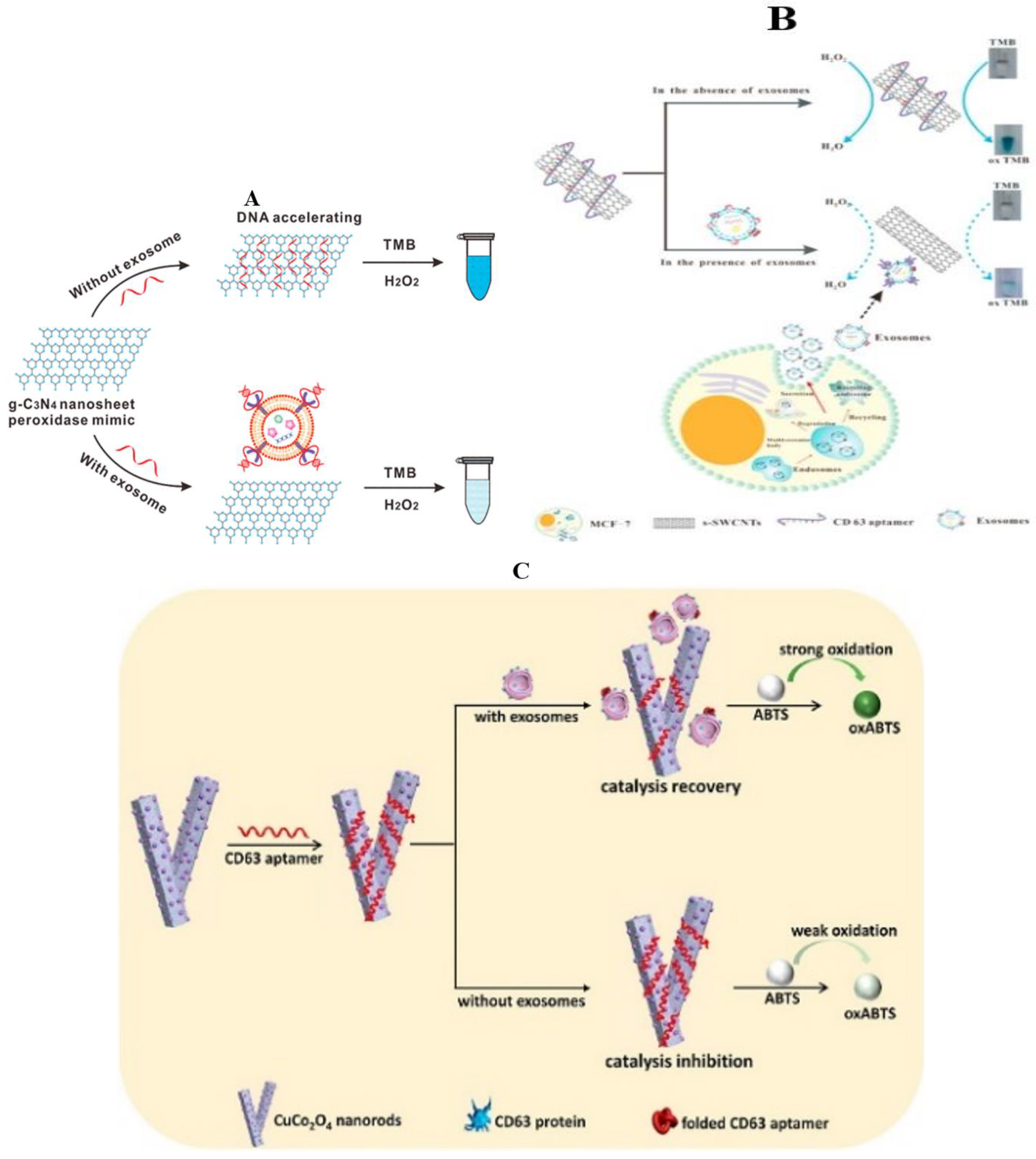
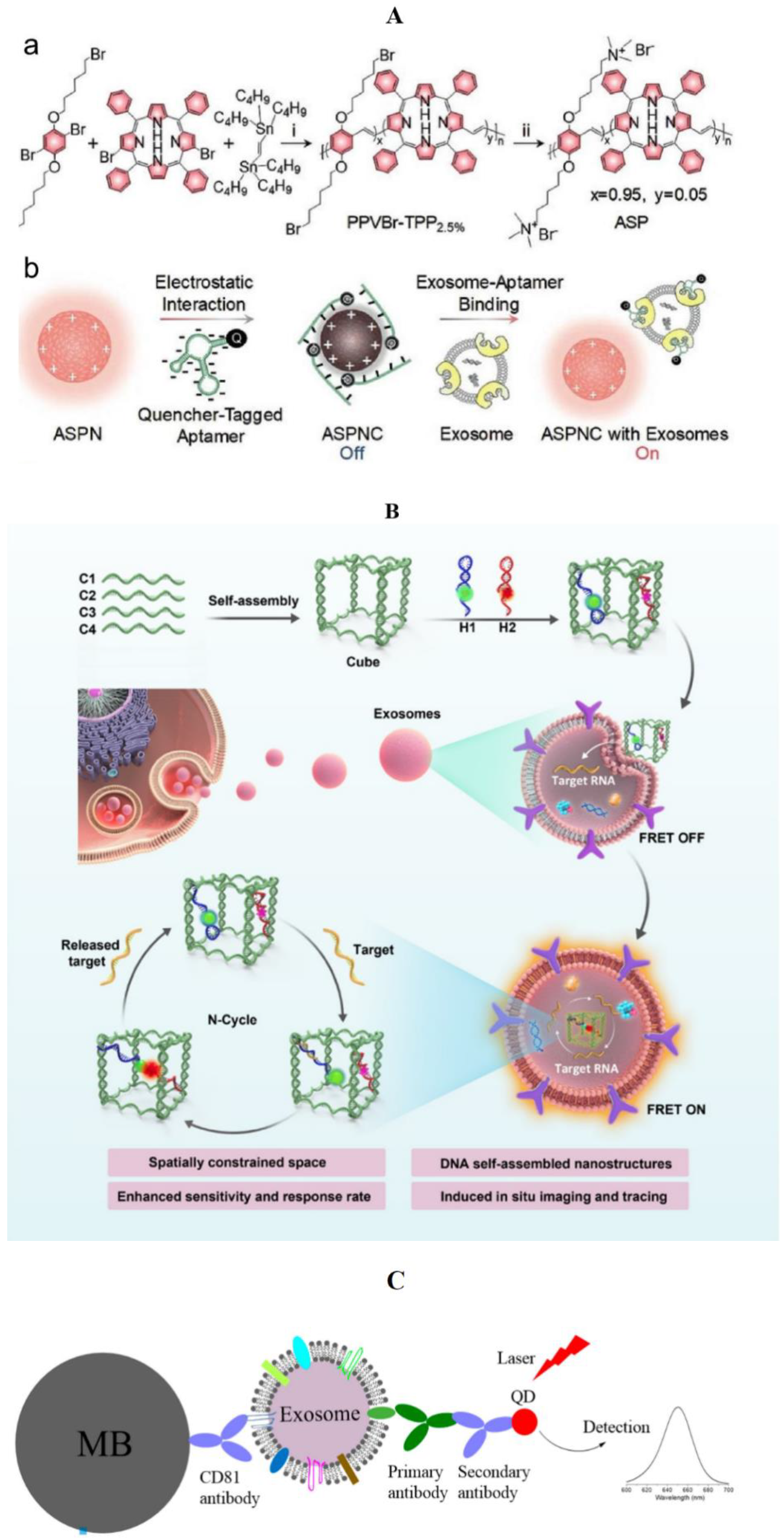
3.1.2. Fluorescence Method
General Material
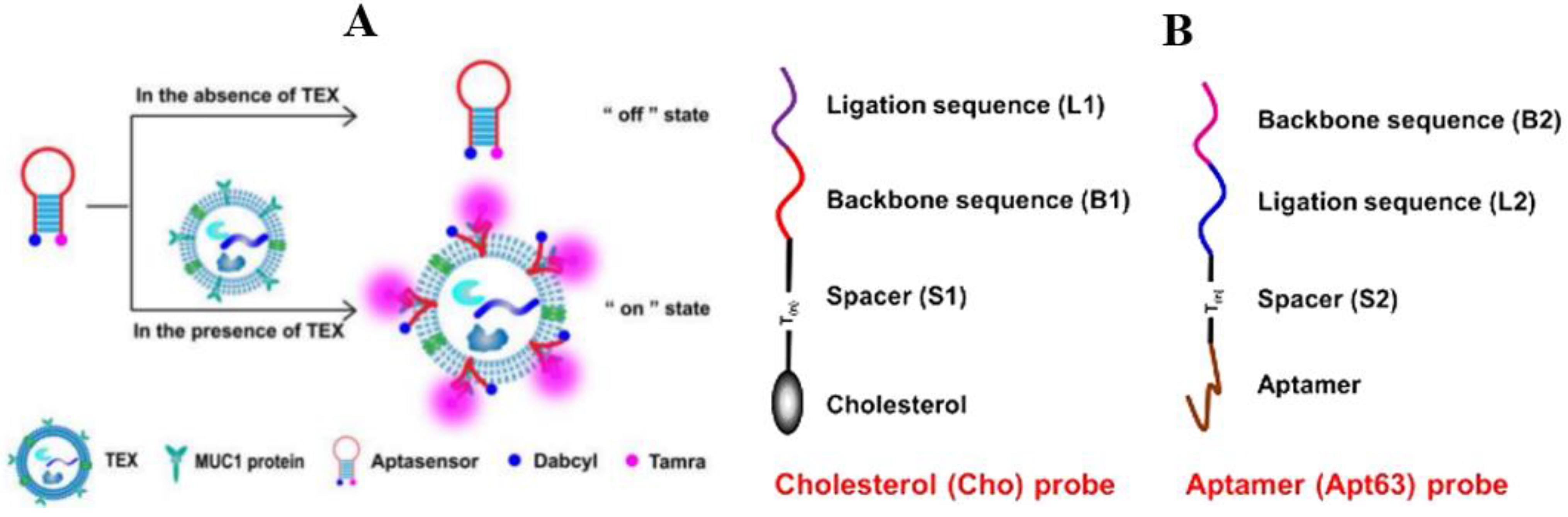


Nanometer Material
3.1.3. Surface Plasmon Resonance
3.1.4. Surface-Enhanced Raman Spectroscopy
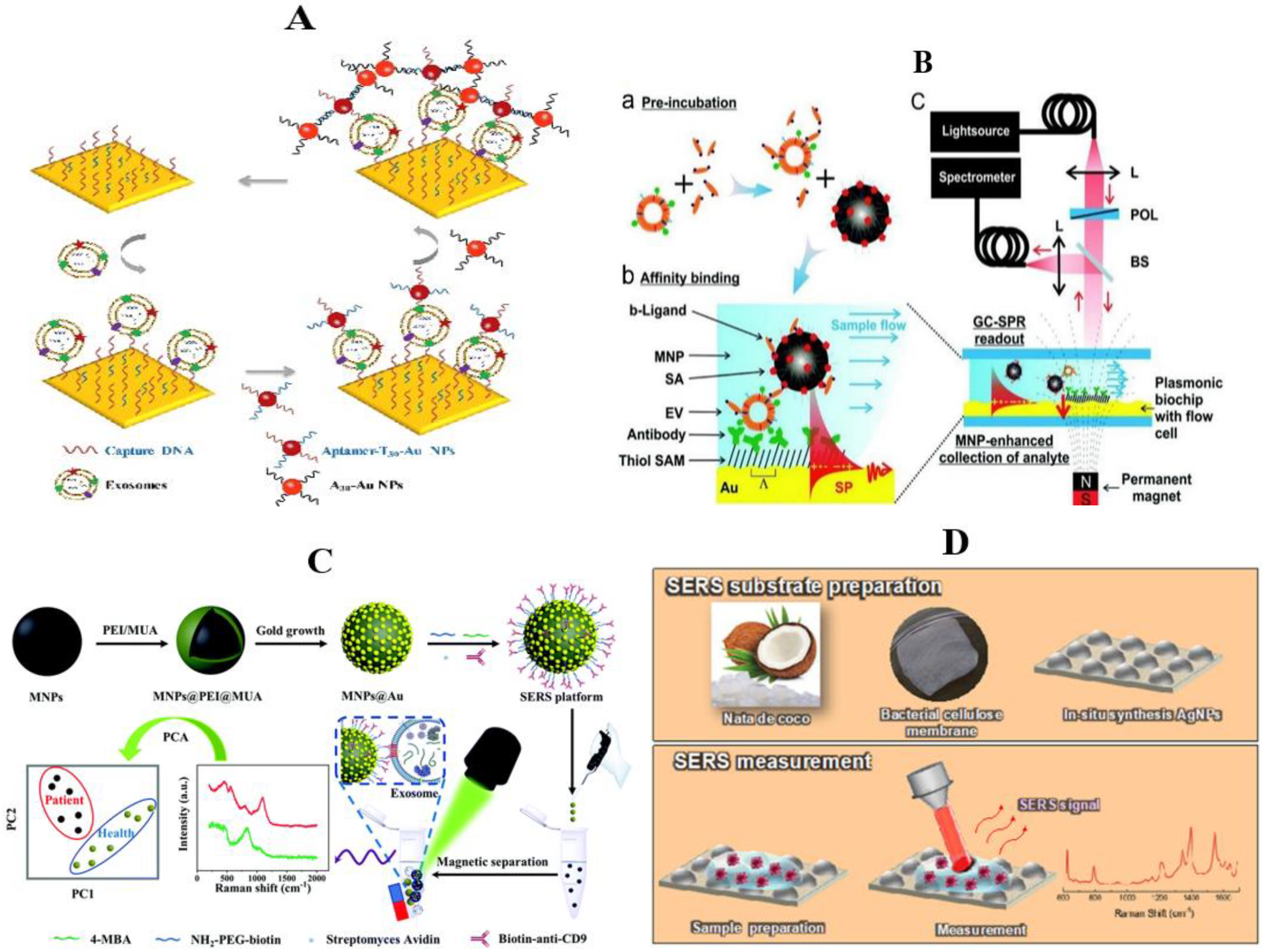
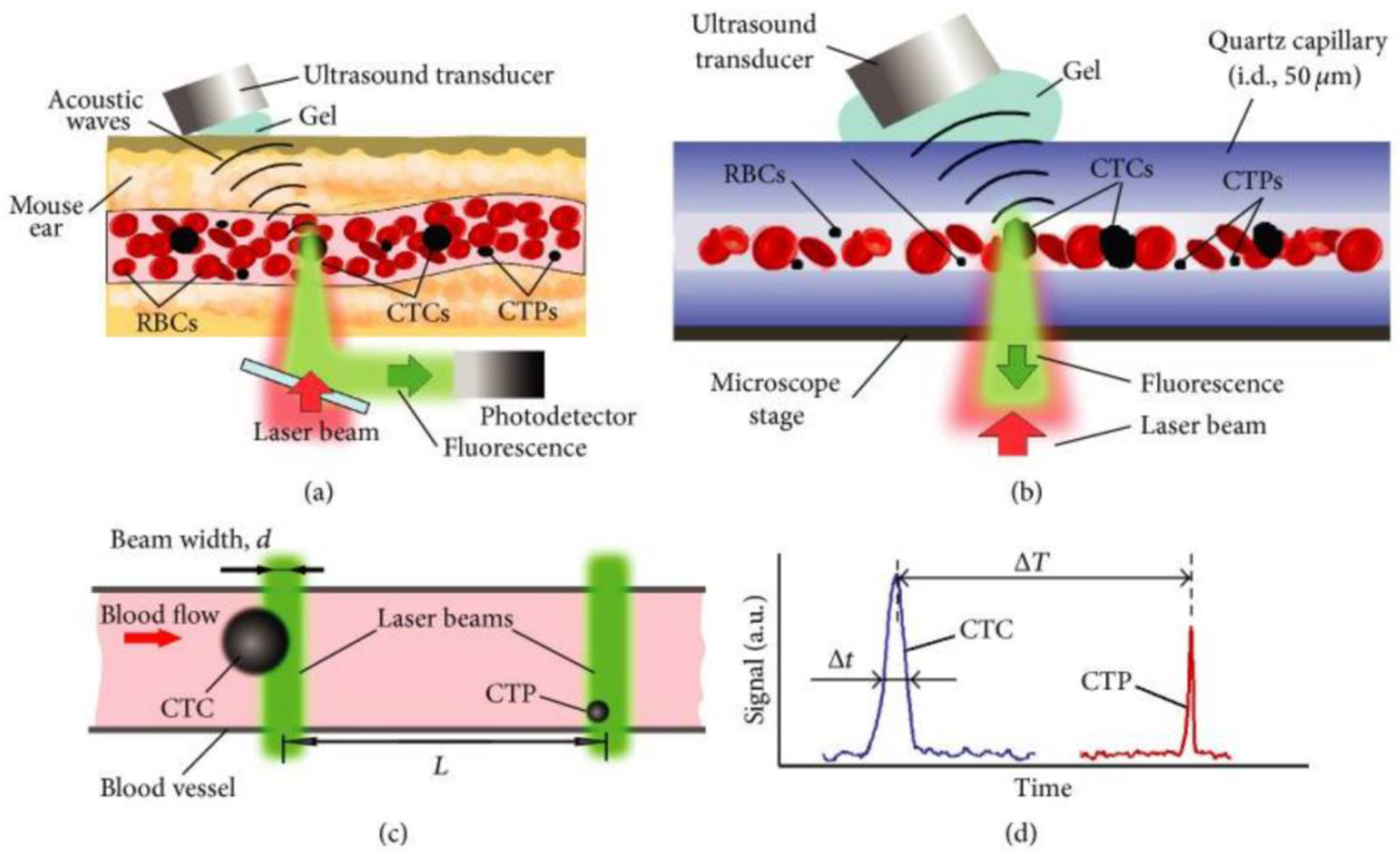
3.1.5. Photoacoustic Imaging
3.2. Electrochemical Method
3.2.1. General Material

3.2.2. Nanometer Material
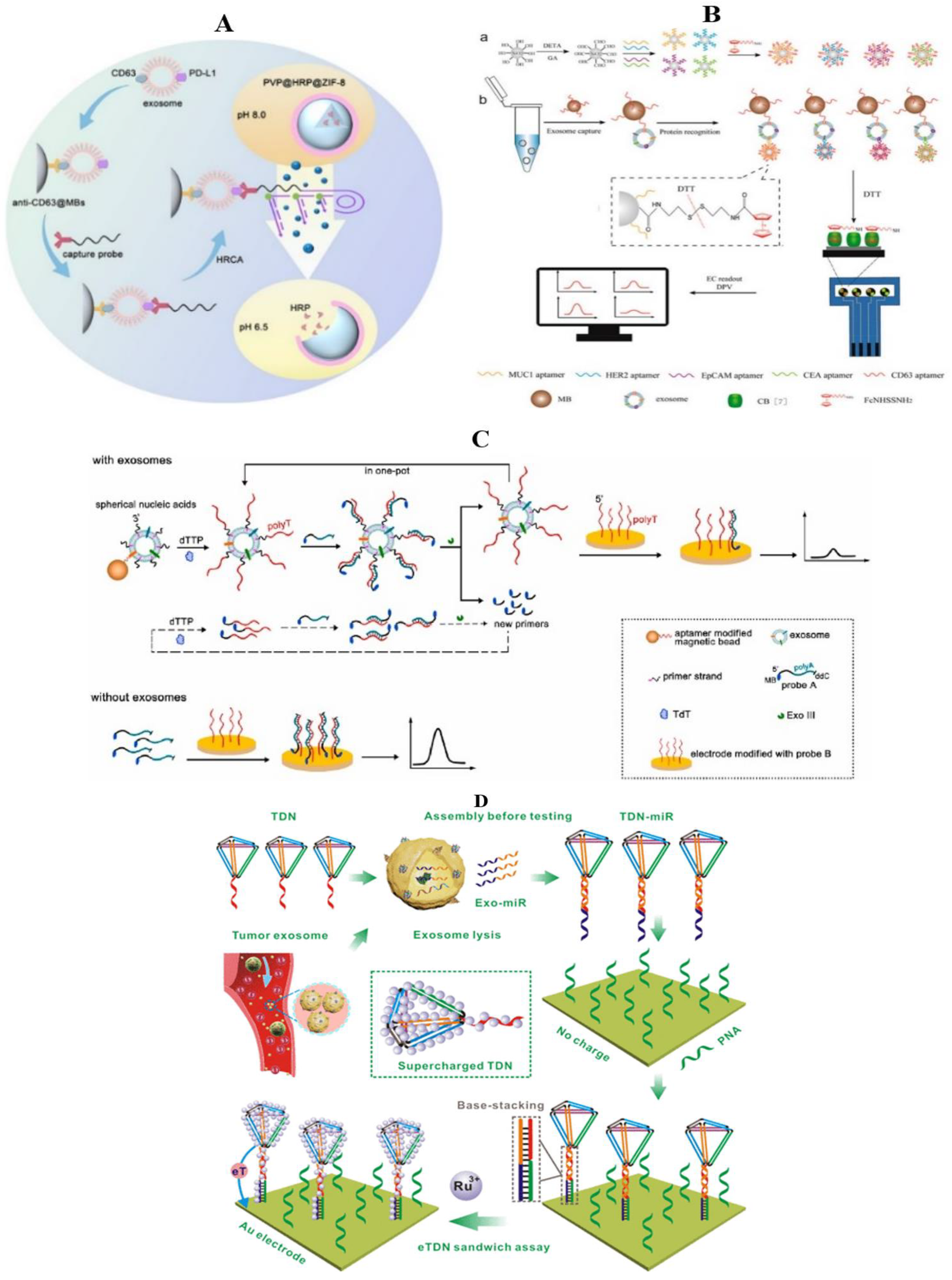
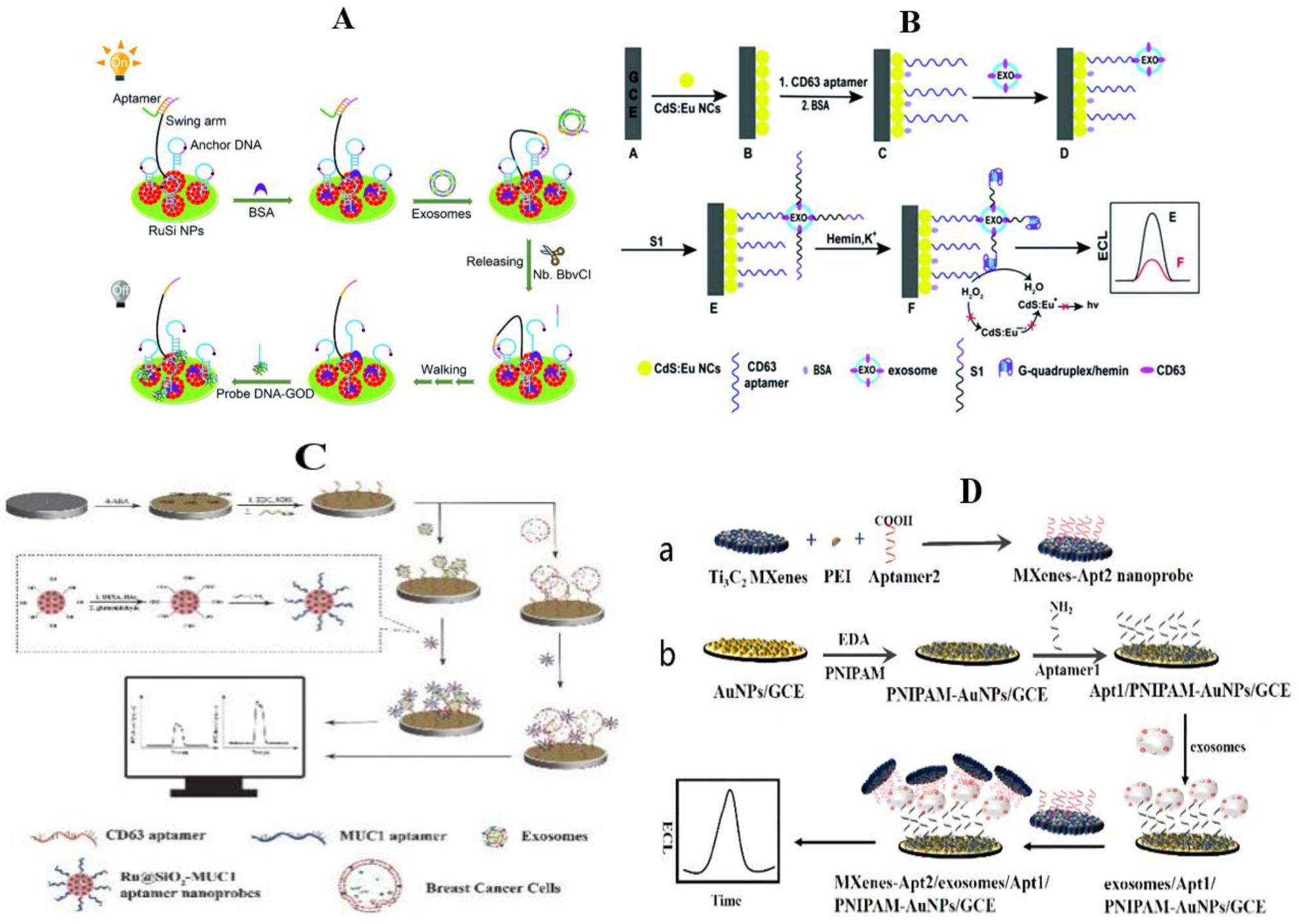
3.3. Electrochemiluminescence Method
3.4. Other Methods
3.5. Comparison and Summary of Various Methods
4. Summary and Prospects
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Sample Availability
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef]
- Chen, W.; Zheng, R.; Baade, P.D.; Zhang, S.; Zeng, H.; Bray, F.; Jemal, A.; Yu, X.Q.; He, J. Cancer Statistics in China, 2015. CA Cancer J. Clin. 2016, 66, 115–132. [Google Scholar] [CrossRef] [Green Version]
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424, Erratum in CA Cancer J. Clin. 2020, 70, 313. [Google Scholar] [CrossRef] [Green Version]
- Sant, M.; Allemani, C.; Capocaccia, R.; Hakulinen, T.; Aareleid, T.; Coebergh, J.W.; Coleman, M.P.; Grosclaude, P.; Martinez, C.; Bell, J.; et al. Stage at diagnosis is a key explanation of differences in breast cancer survival across Europe. Int. J. Cancer 2003, 106, 416–442, Erratum in Int. J. Cancer 2003, 107, 1058. [Google Scholar] [CrossRef] [Green Version]
- Allemani, C.; Sant, M.; Weir, H.K.; Richardson, L.C.; Baili, P.; Storm, H.; Siesling, S.; Torrella-Ramos, A.; Voogd, A.C.; Aareleid, T.; et al. Breast cancer survival in the US and Europe: A CONCORD high-resolution study. Int. J. Cancer 2013, 132, 1170–1181. [Google Scholar] [CrossRef]
- Nelson, R.A.; Guye, M.L.; Luu, T.; Lai, L.L. Survival Outcomes of Metaplastic Breast Cancer Patients: Results from a US Population-based Analysis. Ann. Surg. Oncol. 2015, 22, 24–31. [Google Scholar] [CrossRef] [PubMed]
- Ali, A.M.G.; Greenberg, D.; Wishart, G.C.; Pharoah, P. Patient and tumour characteristics, management, and age-specific survival in women with breast cancer in the East of England. Br. J. Cancer 2011, 104, 564–570. [Google Scholar] [CrossRef] [PubMed]
- Ugnat, A.M.; Xie, L.; Morriss, J.; Semenciw, R.; Mao, Y. Survival of women with breast cancer in Ottawa, Canada: Variation with age, stage, histology, grade and treatment. Br. J. Cancer 2004, 90, 1138–1143. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nechuta, S.; Lu, W.; Zheng, Y.; Cai, H.; Bao, P.-P.; Gu, K.; Zheng, W.; Shu, X.O. Comorbidities and breast cancer survival: A report from the Shanghai Breast Cancer Survival Study. Breast Cancer Res. Treat. 2013, 139, 227–235. [Google Scholar] [CrossRef] [Green Version]
- Kalluri, R.; LeBleu, V.S. The biology, function, and biomedical applications of exosomes. Science 2020, 367, eaau6977. [Google Scholar] [CrossRef]
- Zhang, J.; Li, S.; Li, L.; Li, M.; Guo, C.Y.; Yao, J.; Mi, S.L. Exosome and Exosomal MicroRNA: Trafficking, Sorting, and Function. Genom. Proteom. Bioinform. 2015, 13, 17–24. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- He, C.J.; Zheng, S.; Luo, Y.; Wang, B. Exosome Theranostics: Biology and Translational Medicine. Theranostics 2018, 8, 237–255. [Google Scholar] [CrossRef] [PubMed]
- Wen, S.W.; Lima, L.G.; Lobb, R.J.; Norris, E.L.; Hastie, M.L.; Krumeich, S.; Moller, A. Breast Cancer-Derived Exosomes Reflect the Cell-of-Origin Phenotype. Proteomics 2019, 19, 1800180. [Google Scholar] [CrossRef]
- Nolte-‘t Hoen, E.N.M.; Buschow, S.I.; Anderton, S.M.; Stoorvogel, W.; Wauben, M.H.M. Activated T cells recruit exosomes secreted by dendritic cells via LFA-1. Blood 2009, 113, 1977–1981. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shi, Y.; Du, L.Y.; Lv, D.D.; Li, Y.; Zhang, Z.L.; Huang, X.L.; Tang, H. Emerging role and therapeutic application of exosome in hepatitis virus infection and associated diseases. J. Gastroenterol. 2021, 56, 336–349. [Google Scholar] [CrossRef] [PubMed]
- Srikanthan, S.; Li, W.; Silverstein, R.L.; McIntyre, T.M. Exosome poly-ubiquitin inhibits platelet activation, downregulates CD36 and inhibits pro-atherothombotic cellular functions. J. Thromb. Haemost. 2014, 12, 1906–1917. [Google Scholar] [CrossRef] [Green Version]
- Yan, S.S.; Dang, G.W.; Zhang, X.Y.; Jin, C.W.; Qin, L.; Wang, Y.G.; Shi, M.; Huang, H.J.; Duan, Q.H. Downregulation of circulating exosomal miR-638 predicts poor prognosis in colon cancer patients. Oncotarget 2017, 8, 72220–72226. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Bi, J.Y.; Huang, J.Y.; Tang, Y.N.; Du, S.Y.; Li, P.Y. Exosome: A Review of Its Classification, Isolation Techniques, Storage, Diagnostic and Targeted Therapy Applications. Int. J. Nanomed. 2020, 15, 6917–6934. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.Y.; Du, S.L.; Liang, A.L.; Liu, Y.J. Exosomes and breast cancer: A comprehensive review of novel therapeutic strategies from diagnosis to treatment. Cancer Gene Therapy. 2017, 24, 6–12. [Google Scholar] [CrossRef]
- Zhang, C.; Ji, Q.; Yang, Y.; Li, Q.; Wang, Z. Exosome: Function and Role in Cancer Metastasis and Drug Resistance. Technol. Cancer Res. Treat. 2018, 17. [Google Scholar] [CrossRef] [PubMed]
- Yan, W.; Wu, X.W.; Zhou, W.Y.; Fong, M.Y.; Cao, M.H.; Liu, J.; Liu, X.J.; Chen, C.H.; Fadare, O.; Pizzo, D.P.; et al. Cancer-cell-secreted exosomal miR-105 promotes tumour growth through the MYC-dependent metabolic reprogramming of stromal cells. Nat. Cell Biol. 2018, 20, 597–609. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, W.Y.; Fong, M.Y.; Min, Y.F.; Somlo, G.; Liu, L.; Palomares, M.R.; Yu, Y.; Chow, A.; O’Connor, S.T.F.; Chin, A.R.; et al. Cancer-Secreted miR-105 Destroys Vascular Endothelial Barriers to Promote Metastasis. Cancer Cell 2014, 25, 501–515. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, T.T.; Hooda, J.; Atkinson, J.M.; Whiteside, T.L.; Oesterreich, S.; Lee, A.V. Exosomes in Breast Cancer—Mechanisms of Action and Clinical Potential. Mol. Cancer Res. 2021, 19, 935–945. [Google Scholar] [CrossRef] [PubMed]
- Luga, V.; Zhang, L.; Viloria-Petit, A.M.; Ogunjimi, A.A.; Inanlou, M.R.; Chiu, E.; Buchanan, M.; Hosein, A.N.; Basik, M.; Wrana, J.L. Exosomes Mediate Stromal Mobilization of Autocrine Wnt-PCP Signaling in Breast Cancer Cell Migration. Cell 2012, 151, 1542–1556. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hench, I.B.; Hench, J.; Tolnay, M. Liquid Biopsy in Clinical Management of Breast, Lung, and Colorectal Cancer. Front. Med. 2018, 5, 9. [Google Scholar] [CrossRef]
- Vinik, Y.; Ortega, F.G.; Mills, G.B.; Lu, Y.; Jurkowicz, M.; Halperin, S.; Aharoni, M.; Gutman, M.; Lev, S. Proteomic analysis of circulating extracellular vesicles identifies potential markers of breast cancer progression, recurrence, and response. Sci. Adv. 2020, 6, eaba5714. [Google Scholar] [CrossRef] [PubMed]
- Qin, W.Y.; Tsukasaki, Y.; Dasgupta, S.; Mukhopadhyay, N.; Ikebe, M.; Sauter, E.R. Exosomes in Human Breast Milk Promote EMT. Clin. Cancer Res. 2016, 22, 4517–4524. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, D.D.; Wu, Y.; Shen, H.Y.; Lv, M.M.; Chen, W.X.; Zhang, X.H.; Zhong, S.L.; Tang, J.H.; Zhao, J.H. Exosomes in development, metastasis and drug resistance of breast cancer. Cancer Sci. 2015, 106, 959–964. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hannafon, B.N.; Trigoso, Y.D.; Calloway, C.L.; Zhao, Y.D.; Lum, D.H.; Welm, A.L.; Zhao, Z.Z.J.; Blick, K.E.; Dooley, W.C.; Ding, W.Q. Plasma exosome microRNAs are indicative of breast cancer. Breast Cancer Res. 2016, 18, 90. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eichelser, C.; Stuckrath, I.; Muller, V.; Milde-Langosch, K.; Wikman, H.; Pantel, K.; Schwarzenbach, H. Increased serum levels of circulating exosomal microRNA-373 in receptor-negative breast cancer patients. Oncotarget 2014, 5, 9650–9663. [Google Scholar] [CrossRef] [PubMed]
- Galindo-Hernandez, O.; Villegas-Comonfort, S.; Candanedo, F.; Gonzalez-Vazquez, M.-C.; Chavez-Ocana, S.; Jimenez-Villanueva, X.; Sierra-Martinez, M.; Perez Salazar, E. Elevated Concentration of Microvesicles Isolated from Peripheral Blood in Breast Cancer Patients. Arch. Med. Res. 2013, 44, 208–214. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Ning, K.; Lu, T.-x.; Sun, X.; Jin, L.; Qi, X.; Jin, J.; Hua, D. Increasing circulating exosomes-carrying TRPC5 predicts chemoresistance in metastatic breast cancer patients. Cancer Sci. 2017, 108, 448–454. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tang, S.; Zheng, K.; Tang, Y.; Li, Z.; Zou, T.; Liu, D. Overexpression of serum exosomal HOTAIR is correlated with poor survival and poor response to chemotherapy in breast cancer patients. J. Biosci. 2019, 44, 37. [Google Scholar] [CrossRef]
- Li, X.J.; Ren, Z.J.; Tang, J.H.; Yu, Q. Exosomal MicroRNA MiR-1246 Promotes Cell Proliferation, Invasion and Drug Resistance by Targeting CCNG2 in Breast Cancer. Cell. Physiol. Biochem. 2017, 44, 1741–1748. [Google Scholar] [CrossRef] [PubMed]
- Sueta, A.; Yamamoto, Y.; Tomiguchi, M.; Takeshita, T.; Yamamoto-Ibusuki, M.; Iwase, H. Differential expression of exosomal miRNAs between breast cancer patients with and without recurrence. Oncotarget 2017, 8, 69934–69944. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hood, J.L.; Wickline, S.A. A systematic approach to exosome-based translational nanomedicine. Wiley Interdiscip. Rev.—Nanomed. Nanobiotechnology 2012, 4, 458–467. [Google Scholar] [CrossRef]
- Lopez-Verrilli, M.A.; Court, F.A. Exosomes: Mediators of communication in eukaryotes. Biol. Res. 2013, 46, 5–11. [Google Scholar] [CrossRef] [Green Version]
- Momen-Heravi, F.; Balaj, L.; Alian, S.; Mantel, P.Y.; Halleck, A.E.; Trachtenberg, A.J.; Soria, C.E.; Oquin, S.; Bonebreak, C.M.; Saracoglu, E.; et al. Current methods for the isolation of extracellular vesicles. Biol. Chem. 2013, 394, 1253–1262. [Google Scholar] [CrossRef]
- Baranyai, T.; Herczeg, K.; Onodi, Z.; Voszka, I.; Modos, K.; Marton, N.; Nagy, G.; Maeger, I.; Wood, M.J.; El Andaloussi, S.; et al. Isolation of Exosomes from Blood Plasma: Qualitative and Quantitative Comparison of Ultracentrifugation and Size Exclusion Chromatography Methods. PLoS ONE 2015, 10, e0145686. [Google Scholar]
- Linares, R.; Tan, S.; Gounou, C.; Arraud, N.; Brisson, A.R. High-speed centrifugation induces aggregation of extracellular vesicles. J. Extracell. Vesicles 2015, 4, 29509. [Google Scholar] [CrossRef]
- Guan, S.; Yu, H.; Yan, G.; Gao, M.; Sun, W.; Zhang, X. Characterization of Urinary Exosomes Purified with Size Exclusion Chromatography and Ultracentrifugation. J. Proteome Res. 2020, 19, 2217–2225. [Google Scholar] [CrossRef] [PubMed]
- Tauro, B.J.; Greening, D.W.; Mathias, R.A.; Ji, H.; Mathivanan, S.; Scott, A.M.; Simpson, R.J. Comparison of ultracentrifugation, density gradient separation, and immunoaffinity capture methods for isolating human colon cancer cell line LIM1863-derived exosomes. Methods 2012, 56, 293–304. [Google Scholar] [CrossRef] [PubMed]
- Welton, J.L.; Webber, J.P.; Botos, L.-A.; Jones, M.; Clayton, A. Ready-made chromatography columns for extracellular vesicle isolation from plasma. J. Extracell. Vesicles 2015, 4, 27269. [Google Scholar] [CrossRef] [PubMed]
- Musante, L.; Tataruch, D.; Gu, D.; Benito-Martin, A.; Calzaferri, G.; Aherne, S.; Holthofer, H. A Simplified Method to Recover Urinary Vesicles for Clinical Applications, and Sample Banking. Sci. Rep. 2014, 4, 7532. [Google Scholar] [CrossRef] [Green Version]
- Zhao, H.; Yang, L.; Baddour, J.; Achreja, A.; Bernard, V.; Moss, T.; Marini, J.C.; Tudawe, T.; Seviour, E.G.; San Lucas, F.A.; et al. Tumor microenvironment derived exosomes pleiotropically modulate cancer cell metabolism. Elife 2016, 5, e10250. [Google Scholar] [CrossRef] [PubMed]
- Contreras-Naranjo, J.C.; Wu, H.-J.; Ugaz, V.M. Microfluidics for exosome isolation and analysis: Enabling liquid biopsy for personalized medicine. Lab Chip 2017, 17, 3558–3577. [Google Scholar] [CrossRef] [Green Version]
- Wang, Z.; Zong, S.; Wang, Y.; Li, N.; Li, L.; Lu, J.; Wang, Z.; Chen, B.; Cui, Y. Screening and multiple detection of cancer exosomes using an SERS-based method. Nanoscale 2018, 10, 9053–9062. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Pan, B.; Zeng, F.; He, B.; Gao, Y.; Liu, X.; Song, Y. Magnetic Colloid Antibodies Accelerate Small Extracellular Vesicles Isolation for Point-of-Care Diagnostics. Nano Lett. 2021, 21, 2001–2009. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Yang, K.; Dai, Z.; Liang, Z.; Zhang, L.; Peng, X.; Zhang, Y. Cell-imprinted polydimethylsiloxane for the selective cell adhesion. Chin. Chem. Lett. 2019, 30, 672–675. [Google Scholar] [CrossRef]
- Kowal, E.J.K.; Ter-Ovanesyan, D.; Regev, A.; Church, G.M. Extracellular Vesicle Isolation and Analysis by Western Blotting. In Extracellular Vesicles: Methods and Protocols; Kuo, W.P., Jia, S., Eds.; Springer: Berlin/Heidelberg, Germany, 2017; Volume 1660, pp. 143–152. [Google Scholar]
- Coughlan, C.; Bruce, K.D.; Burgy, O.; Boyd, T.D.; Michel, C.R.; Garcia-Perez, J.E.; Adame, V.; Anton, P.; Bettcher, B.M.; Chial, H.J.; et al. Exosome Isolation by Ultracentrifugation and Precipitation and Techniques for Downstream Analyses. Curr. Protoc. Cell Biol. 2020, 88, e110. [Google Scholar] [CrossRef]
- Nolan, J.P.; Jones, J.C. Detection of platelet vesicles by flow cytometry. Platelets 2017, 28, 256–262. [Google Scholar] [CrossRef]
- Goetzl, E.J.; Schwartz, J.B.; Abner, E.L.; Jicha, G.A.; Kapogiannis, D. High complement levels in astrocyte-derived exosomes of Alzheimer disease. Ann. Neurol. 2018, 83, 544–552. [Google Scholar] [CrossRef]
- Wang, H.; Zeng, J.; Huang, J.; Cheng, H.; Chen, B.; Hu, X.; He, X.; Zhou, Y.; Wang, K. A Self-Serviced-Track 3D DNA Walker for Ultrasensitive Detection of Tumor Exosomes by Glycoprotein Profiling. Angew. Chem.—Int. Ed. 2022, 61, e202116932. [Google Scholar]
- Huang, Z.; Lin, Q.; Yang, B.; Ye, X.; Chen, H.; Weng, W.; Kong, J. Cascade signal amplification for sensitive detection of exosomes by integrating tyramide and surface-initiated enzymatic polymerization. Chem. Commun. 2020, 56, 12793–12796. [Google Scholar] [CrossRef]
- Tang, Z.; Huang, J.; He, H.; Ma, C.; Wang, K. Contributing to liquid biopsy: Optical and electrochemical methods in cancer biomarker analysis. Coord. Chem. Rev. 2020, 415, 213317. [Google Scholar] [CrossRef]
- Xu, L.; Chopdat, R.; Li, D.; Al-Jamal, K.T. Development of a simple, sensitive and selective colorimetric aptasensor for the detection of cancer-derived exosomes. Biosens. Bioelectron. 2020, 169, 112576. [Google Scholar] [CrossRef]
- Li, S.; Wang, L.; Hao, Y.; Zhang, L.; Zhou, B.; Deng, L.; Liu, Y.-N. An ultrasensitive colorimetric aptasensor for ATP based on peptide/Au nanocomposites and hemin-G-quadruplex DNAzyme. RSC Adv. 2014, 4, 23185–23190. [Google Scholar] [CrossRef]
- Yuan, Y.; Chai, Y.; Yuan, R.; Zhuo, Y.; Gan, X. An ultrasensitive electrochemical aptasensor with autonomous assembly of hemin-G-quadruplex DNAzyme nanowires for pseudo triple-enzyme cascade electrocatalytic amplification. Chem. Commun. 2013, 49, 7328–7330. [Google Scholar] [CrossRef]
- Huang, R.; He, L.; Xia, Y.; Xu, H.; Liu, C.; Xie, H.; Wang, S.; Peng, L.; Liu, Y.; Liu, Y.; et al. A Sensitive Aptasensor Based on a Hemin/G-Quadruplex-Assisted Signal Amplification Strategy for Electrochemical Detection of Gastric Cancer Exosomes. Small 2019, 15, 1900735. [Google Scholar] [CrossRef]
- Zhou, Y.; Xu, H.; Wang, H.; Ye, B.-C. Detection of breast cancer-derived exosomes using the horseradish peroxidase-mimicking DNAzyme as an aptasensor. Analyst 2020, 145, 107–114. [Google Scholar] [CrossRef]
- Yang, Y.; Li, C.; Shi, H.; Chen, T.; Wang, Z.; Li, G. A pH-responsive bioassay for paper-based diagnosis of exosomes via mussel-inspired surface chemistry. Talanta 2019, 192, 325–330. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Zhang, X.; Liu, B.; Liu, J. Molecular Imprinting on Inorganic Nanozymes for Hundred-fold Enzyme Specificity. J. Am. Chem. Soc. 2017, 139, 5412–5419. [Google Scholar] [CrossRef]
- Gao, L.; Zhuang, J.; Nie, L.; Zhang, J.; Zhang, Y.; Gu, N.; Wang, T.; Feng, J.; Yang, D.; Perrett, S.; et al. Intrinsic peroxidase-like activity of ferromagnetic nanoparticles. Nat. Nanotechnol. 2007, 2, 577–583. [Google Scholar] [CrossRef]
- Hizir, M.S.; Top, M.; Balcioglu, M.; Rana, M.; Robertson, N.M.; Shen, F.; Sheng, J.; Yigit, M.V. Multiplexed Activity of perAuxidase: DNA-Capped AuNPs Act as Adjustable Peroxidase. Anal. Chem. 2016, 88, 600–605. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Asati, A.; Santra, S.; Kaittanis, C.; Nath, S.; Perez, J.M. Oxidase-like activity of polymer-coated cerium oxide nanoparticles. Angew. Chem. 2009, 48, 2308–2312. [Google Scholar] [CrossRef]
- Cui, R.; Han, Z.; Zhu, J.-J. Helical Carbon Nanotubes: Intrinsic Peroxidase Catalytic Activity and Its Application for Biocatalysis and Biosensing. Chem.—Eur. J. 2011, 17, 9377–9384. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.; Qu, K.; Zhao, C.; Ren, J.; Qu, X. Graphene Oxide: Intrinsic Peroxidase Catalytic Activity and Its Application to Glucose Detection. Adv. Mater. 2010, 22, 2206–2210. [Google Scholar] [CrossRef] [PubMed]
- Wei, H.; Wang, E. Nanomaterials with enzyme-like characteristics (nanozymes): Next-generation artificial enzymes. Chem. Soc. Rev. 2013, 42, 6060–6093. [Google Scholar] [CrossRef]
- Zhang, H.; Huang, Q.; Huang, Y.; Li, F.; Zhang, W.; Wei, C.; Chen, J.; Dai, P.; Huang, L.; Huang, Z.; et al. Graphitic carbon nitride nanosheets doped graphene oxide for electrochemical simultaneous determination of ascorbic acid, dopamine and uric acid. Electrochim. Acta 2014, 142, 125–131. [Google Scholar] [CrossRef]
- Lu, Q.; Deng, J.; Hou, Y.; Wang, H.; Li, H.; Zhang, Y. One-step electrochemical synthesis of ultrathin graphitic carbon nitride nanosheets and their application to the detection of uric acid. Chem. Commun. 2015, 51, 12251–12253. [Google Scholar] [CrossRef]
- Tian, J.; Liu, Q.; Ge, C.; Xing, Z.; Asiri, A.M.; Al-Youbi, A.O.; Sun, X. Ultrathin graphitic carbon nitride nanosheets: A low-cost, green, and highly efficient electrocatalyst toward the reduction of hydrogen peroxide and its glucose biosensing application. Nanoscale 2013, 5, 8921–8924. [Google Scholar] [CrossRef]
- Wang, Y.-M.; Liu, J.-W.; Adkins, G.B.; Shen, W.; Trinh, M.P.; Duan, L.-Y.; Jiang, J.-H.; Zhong, W. Enhancement of the Intrinsic Peroxidase-Like Activity of Graphitic Carbon Nitride Nanosheets by ssDNAs and Its Application for Detection of Exosomes. Anal. Chem. 2017, 89, 12327–12333. [Google Scholar] [CrossRef]
- Xia, Y.; Liu, M.; Wang, L.; Yan, A.; He, W.; Chen, M.; Lan, J.; Xu, J.; Guan, L.; Chen, J. A visible and colorimetric aptasensor based on DNA-capped single-walled carbon nanotubes for detection of exosomes. Biosens. Bioelectron. 2017, 92, 8–15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Y.; Su, Q.; Song, D.; Fan, J.; Xu, Z. Label-free detection of exosomes based on ssDNA-modulated oxidase-mimicking activity of CuCo2O4 nanorods. Anal. Chim. Acta 2021, 1145, 9–16. [Google Scholar] [CrossRef]
- Wang, C.; Irudayaraj, J. Gold Nanorod Probes for the Detection of Multiple Pathogens. Small 2008, 4, 2204–2208. [Google Scholar] [CrossRef]
- Wang, H.; Han, L.; Zheng, D.; Yang, M.; Andaloussi, Y.H.; Cheng, P.; Zhang, Z.; Ma, S.; Zaworotko, M.J.; Feng, Y.; et al. Protein-Structure-Directed Metal-Organic Zeolite-like Networks as Biomacromolecule Carriers. Angew. Chem. Int. Ed. 2020, 59, 6263–6267. [Google Scholar] [CrossRef]
- Shen, B.; Zhang, X.; Dai, J.; Ji, Y.; Huang, H. Lysosome targeting metal-organic framework probe LysFP@ZIF-8 for highly sensitive quantification of carboxylesterase 1 and organophosphates in living cells. J. Hazard. Mater. 2021, 407, 124342. [Google Scholar] [CrossRef] [PubMed]
- Ding, Z.; Lu, Y.; Wei, Y.; Song, D.; Xu, Z.; Fang, J. DNA-Engineered iron-based metal-organic framework bio-interface for rapid visual determination of exosomes. J. Colloid Interface Sci. 2022, 612, 424–433. [Google Scholar] [CrossRef] [PubMed]
- Jana, N.R.; Gearheart, L.; Obare, S.O.; Murphy, C.J. Anisotropic chemical reactivity of gold spheroids and nanorods. Langmuir 2002, 18, 922–927. [Google Scholar] [CrossRef]
- Zhang, Y.; Wang, D.; Yue, S.; Lu, Y.; Yang, C.; Fang, J.; Xu, Z. Sensitive Multicolor Visual Detection of Exosomes via Dual Signal Amplification Strategy of Enzyme-Catalyzed Metallization of Au Nanorods and Hybridization Chain Reaction. ACS Sens. 2019, 4, 3210–3218. [Google Scholar] [CrossRef]
- Lee, S.; Mayer, K.M.; Hafner, J.H. Improved Localized Surface Plasmon Resonance Immunoassay with Gold Bipyramid Substrates. Anal. Chem. 2009, 81, 4450–4455. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Zhu, X.; Li, N.; Yang, J.; Yang, Z.; Wang, J.; Yang, B. Molecular Sensitivities of Substrate-Supported Gold Nanocrystals. J. Phys. Chem. C 2019, 123, 7336–7346. [Google Scholar] [CrossRef]
- Xu, S.; Jiang, L.; Nie, Y.; Wang, J.; Li, H.; Liu, Y.; Wang, W.; Xu, G.; Luo, X. Gold Nanobipyramids as Dual-Functional Substrates for in Situ "Turn On" Analyzing Intracellular Telomerase Activity Based on Target-Triggered Plasmon-Enhanced Fluorescence. ACS Appl. Mater. Interfaces 2018, 10, 26851–26858. [Google Scholar] [CrossRef]
- Zhang, Y.; Jiao, J.; Wei, Y.; Wang, D.; Yang, C.; Xu, Z. Plasmonic Colorimetric Biosensor for Sensitive Exosome Detection via Enzyme-Induced Etching of Gold Nanobipyramid@MnO2 Nanosheet Nanostructures. Anal. Chem. 2020, 92, 15244–15252. [Google Scholar] [CrossRef] [PubMed]
- Kim, C.-H.; Lee, L.-P.; Min, J.-R.; Lim, M.-W.; Jeong, S.-H. An indirect competitive assay-based aptasensor for detection of oxytetracycline in milk. Biosens. Bioelectron. 2014, 51, 426–430. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Shi, J.; Liu, W.; Zhang, K.; Zhao, H.; Zhang, H.; Zhang, Z. A simple, specific and "on-off" type MUC1 fluorescence aptasensor based on exosomes for detection of breast cancer. Sens. Actuators B-Chem. 2018, 276, 552–559. [Google Scholar] [CrossRef]
- Zhao, X.; Luo, C.; Mei, Q.; Zhang, H.; Zhang, W.; Su, D.; Fu, W.; Luo, Y. Aptamer-Cholesterol-Mediated Proximity Ligation Assay for Accurate Identification of Exosomes. Anal. Chem. 2020, 92, 5411–5418. [Google Scholar] [CrossRef]
- Wang, X.; Shang, H.; Ma, C.; Chen, L. A Fluorescence Assay for Exosome Detection Based on Bivalent Cholesterol Anchor Triggered Target Conversion and Enzyme-Free Signal Amplification. Anal. Chem. 2021, 93, 8493–8500. [Google Scholar] [CrossRef]
- Ishraq Bari, S.M.; Hossain, F.B.; Nestorova, G.G. Advances in Biosensors Technology for Detection and Characterization of Extracellular Vesicles. Sensors 2021, 21, 7645. [Google Scholar]
- Liu, J.; Zhang, Y.; Zhao, Q.; Bo, S.; Zhao, J.; Luo, S.; Li, B.; Yan, X.; Vadgama, P.; Su, L.; et al. Bifunctional aptamer-mediated catalytic hairpin assembly for the sensitive and homogenous detection of rare cancer cells. Anal. Chim. Acta 2018, 1029, 58–64. [Google Scholar] [CrossRef]
- Gao, X.H.; Li, S.; Ding, F.; Fan, H.J.; Shi, L.L.; Zhu, L.J.; Li, J.; Feng, J.; Zhu, X.Y.; Zhang, C. Rapid Detection of Exosomal MicroRNAs Using Virus-Mimicking Fusogenic Vesicles. Angew. Chem. Int. Ed. 2019, 58, 8719–8723. [Google Scholar] [CrossRef] [PubMed]
- Tayebi, M.; Zhou, Y.; Tripathi, P.; Chandramohanadas, R.; Ai, Y. Exosome Purification and Analysis Using a Facile Microfluidic Hydrodynamic Trapping Device. Anal. Chem. 2020, 92, 10733–10742. [Google Scholar] [CrossRef] [PubMed]
- Thanthri, S.H.P.; Ward, C.L.; Cornejo, M.A.; Linz, T.H. Simultaneous Preconcentration and Separation of Native Protein Variants Using Thermal Gel Electrophoresis. Anal. Chem. 2020, 92, 6741–6747. [Google Scholar] [CrossRef] [PubMed]
- Duhr, S.; Braun, D. Why molecules move along a temperature gradient. Proc. Natl. Acad. Sci. USA 2006, 103, 19678–19682. [Google Scholar] [CrossRef] [Green Version]
- Liu, C.; Zhao, J.X.; Tian, F.; Cai, L.L.; Zhang, W.; Feng, Q.; Chang, J.Q.; Wan, F.N.; Yang, Y.J.; Dai, B.; et al. Low-cost thermophoretic profiling of extracellular-vesicle surface proteins for the early detection and classification of cancers. Nat. Biomed. Eng. 2019, 3, 183–193. [Google Scholar] [CrossRef]
- Tian, F.; Zhang, S.H.; Liu, C.; Han, Z.W.; Liu, Y.; Deng, J.Q.; Li, Y.K.; Wu, X.; Cai, L.L.; Qin, L.L.; et al. Protein analysis of extracellular vesicles to monitor and predict therapeutic response in metastatic breast cancer. Nat. Commun. 2021, 12, 2536. [Google Scholar] [CrossRef]
- Li, Y.K.; Deng, J.Q.; Han, Z.W.; Liu, C.; Tian, F.; Xu, R.; Han, D.; Zhang, S.H.; Sun, J.S. Molecular Identification of Tumor-Derived Extracellular Vesicles Using Thermophoresis-Mediated DNA Computation. J. Am. Chem. Soc. 2021, 143, 1290–1295. [Google Scholar] [CrossRef]
- Huang, M.J.; Yang, J.J.; Wang, T.; Song, J.; Xia, J.L.; Wu, L.L.; Wang, W.; Wu, Q.Y.; Zhu, Z.; Song, Y.L.; et al. Homogeneous, Low-volume, Efficient, and Sensitive Quantitation of Circulating Exosomal PD-L1 for Cancer Diagnosis and Immunotherapy Response Prediction. Angew. Chem. Int. Ed. 2020, 59, 4800–4805. [Google Scholar] [CrossRef]
- Nevidalova, H.; Michalcova, L.; Glatz, Z. Capillary electrophoresis-based immunoassay and aptamer assay: A review. Electrophoresis 2020, 41, 414–433. [Google Scholar] [CrossRef]
- Khan, N.U.; Muhammad, Z.; Liu, X.; Lin, J.; Zheng, Q.; Zhang, H.; Malik, S.; He, H.; Shen, L. Ultrasensitive Detection of Exosome Using Biofunctionalized Gold Nanorods on a Silver-Island Film. Nano Lett. 2021, 21, 5532–5539. [Google Scholar] [CrossRef]
- Whittington, N.C.; Wray, S. Suppression of Red Blood Cell Autofluorescence for Immunocytochemistry on Fixed Embryonic Mouse Tissue. Curr. Protoc. Neurosci. 2017, 81, 2.28.1–2.28.12. [Google Scholar] [CrossRef] [PubMed]
- Lyu, Y.; Cui, D.; Huang, J.; Fan, W.; Miao, Y.; Pu, K. Near-Infrared Afterglow Semiconducting Nano-Polycomplexes for the Multiplex Differentiation of Cancer Exosomes. Angew. Chem. Int. Ed. 2019, 58, 4983–4987. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Xie, M.; Shi, M.; Yuan, K.; Wu, Y.; Meng, H.-M.; Qu, L.; Li, Z. Spatial Confinement-Derived Double-Accelerated DNA Cascade Reaction for Ultrafast and Highly Sensitive In Situ Monitoring of Exosomal miRNA and Exosome Tracing. Anal. Chem. 2022, 94, 2227–2235. [Google Scholar] [CrossRef] [PubMed]
- Medintz, I.L.; Uyeda, H.T.; Goldman, E.R.; Mattoussi, H. Quantum dot bioconjugates for imaging, labelling and sensing. Nat. Mater. 2005, 4, 435–446. [Google Scholar] [CrossRef] [PubMed]
- Goldman, E.R.; Balighian, E.D.; Mattoussi, H.; Kuno, M.K.; Mauro, J.M.; Tran, P.T.; Anderson, G.P. Avidin: A natural bridge for quantum dot-antibody conjugates. J. Am. Chem. Soc. 2002, 124, 6378–6382. [Google Scholar] [CrossRef]
- Wu, X.Y.; Liu, H.J.; Liu, J.Q.; Haley, K.N.; Treadway, J.A.; Larson, J.P.; Ge, N.F.; Peale, F.; Bruchez, M.P. Immunofluorescent labeling of cancer marker Her2 and other cellular targets with semiconductor quantum dots. Nat. Biotechnol. 2003, 21, 41–46. [Google Scholar] [CrossRef]
- Vinduska, V.; Gallops, C.E.; O’Connor, R.; Wang, Y.M.; Huang, X.H. Exosomal Surface Protein Detection with Quantum Dots and Immunomagnetic Capture for Cancer Detection. Nanomaterials 2021, 11, 1853. [Google Scholar] [CrossRef]
- Li, Q.; Wang, Y.; Ling, L.; Qiao, L.; Chen, H.; Ding, C.; Yu, S. Rapid and specific detection nanoplatform of serum exosomes for prostate cancer diagnosis. Microchim. Acta 2021, 188, 283. [Google Scholar] [CrossRef]
- Zhu, C.; Li, L.S.; Wang, Z.J.; Irfan, M.; Qu, F. Recent advances of aptasensors for exosomes detection. Biosens. Bioelectron. 2020, 160, 112213. [Google Scholar] [CrossRef] [PubMed]
- Sun, Z.W.; Yang, J.J.; Li, H.; Wang, C.X.; Fletcher, C.; Li, J.; Zhan, Y.; Du, L.T.; Wang, F.L.; Jiang, Y.N. Progress in the research of nanomaterial-based exosome bioanalysis and exosome-based nanomaterials tumor therapy. Biomaterials 2021, 274, 120873. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Zou, L.Y.; Yang, X.H.; Liu, X.F.; Nie, W.Y.; Zheng, Y.; Cheng, Q.; Wang, K.M. Direct quantification of cancerous exosomes via surface plasmon resonance with dual gold nanoparticle-assisted signal amplification. Biosens. Bioelectron. 2019, 135, 129–136. [Google Scholar] [CrossRef] [PubMed]
- Reiner, A.T.; Ferrer, N.G.; Venugopalan, P.; Lai, R.C.; Lim, S.K.; Dostalek, J. Magnetic nanoparticle-enhanced surface plasmon resonance biosensor for extracellular vesicle analysis. Analyst 2017, 142, 3913–3921. [Google Scholar] [CrossRef]
- Rojalin, T.; Phong, B.; Koster, H.J.; Carney, R.P. Nanoplasmonic Approaches for Sensitive Detection and Molecular Characterization of Extracellular Vesicles. Front. Chem. 2019, 7, 279. [Google Scholar] [CrossRef] [PubMed]
- Lavialle, F.; Deshayes, S.; Gonnet, F.; Larquet, E.; Kruglik, S.G.; Boisset, N.; Daniel, R.; Alfsen, A.; Tatischeff, I. Nanovesicles released by Dictyostelium cells: A potential carrier for drug delivery. Int. J. Pharm. 2009, 380, 206–215. [Google Scholar] [CrossRef] [PubMed]
- Tatischeff, I.; Larquet, E.; Falcon-Perez, J.M.; Turpin, P.-Y.; Kruglik, S.G. Fast characterisation of cell-derived extracellular vesicles by nanoparticles tracking analysis, cryo-electron microscopy, and Raman tweezers microspectroscopy. J. Extracell. Vesicles 2012, 1, 19179. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.; Carney, R.P.; Hazari, S.; Smith, Z.J.; Knudson, A.; Robertson, C.S.; Lam, K.S.; Wachsmann-Hogiu, S. 3D plasmonic nanobowl platform for the study of exosomes in solution. Nanoscale 2015, 7, 9290–9297. [Google Scholar] [CrossRef] [PubMed]
- Shin, H.; Jeong, H.; Park, J.; Hong, S.; Choi, Y. Correlation between Cancerous Exosomes and Protein Markers Based on Surface-Enhanced Raman Spectroscopy (SERS) and Principal Component Analysis (PCA). ACS Sens. 2018, 3, 2637–2643. [Google Scholar] [CrossRef]
- Li, G.; Zhu, N.; Zhou, J.; Kang, K.; Zhou, X.; Ying, B.; Yi, Q.; Wu, Y. A magnetic surface-enhanced Raman scattering platform for performing successive breast cancer exosome isolation and analysis. J. Mater. Chem. B 2021, 9, 2709–2716. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Jia, J.; Li, S. Advances in the Application of Exosomes Identification Using Surface-Enhanced Raman Spectroscopy for the Early Detection of Cancers. Front. Bioeng. Biotechnol. 2022, 9, 808933. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, N.; Marques, A.; Aguas, H.; Bandarenka, H.; Martins, R.; Bodo, C.; Costa-Silva, B.; Fortunato, E. Label-Free Nanosensing Platform for Breast Cancer Exosome Profiling. ACS Sens. 2019, 4, 2073–2083. [Google Scholar] [CrossRef] [PubMed]
- Jokerst, J.V.; Gambhir, S.S. Molecular Imaging with Theranostic Nanoparticles. Acc. Chem. Res. 2011, 44, 1050–1060. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Steinberg, I.; Huland, D.M.; Vermesh, O.; Frostig, H.E.; Tummers, W.S.; Gambhir, S.S. Photoacoustic clinical imaging. Photoacoustics 2019, 14, 77–98. [Google Scholar] [CrossRef] [PubMed]
- Ntziachristos, V.; Razansky, D. Molecular Imaging by Means of Multispectral Optoacoustic Tomography (MSOT). Chem. Rev. 2010, 110, 2783–2794. [Google Scholar] [CrossRef]
- Weber, J.; Beard, P.C.; Bohndiek, S.E. Contrast agents for molecular photoacoustic imaging. Nat. Methods 2016, 13, 639–650. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Attia, A.B.E.; Balasundaram, G.; Moothanchery, M.; Dinish, U.S.; Bi, R.; Ntziachristos, V.; Olivo, M. A review of clinical photoacoustic imaging: Current and future trends. Photoacoustics 2019, 16, 100144. [Google Scholar] [CrossRef] [PubMed]
- Nolan, J.; Sarimollaoglu, M.; Nedosekin, D.A.; Jamshidi-Parsian, A.; Galanzha, E.I.; Kore, R.A.; Griffin, R.J.; Zharov, V.P. In Vivo Flow Cytometry of Circulating Tumor-Associated Exosomes. Anal. Cell. Pathol. 2016, 2016, 1628057. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Piao, Y.J.; Kim, H.S.; Moon, W.K. Noninvasive Photoacoustic Imaging of Dendritic Cell Stimulated with Tumor Cell-Derived Exosome. Mol. Imaging Biol. 2020, 22, 612–622. [Google Scholar] [CrossRef] [PubMed]
- Dong, H.; Chen, H.; Jiang, J.; Zhang, H.; Cai, C.; Shen, Q. Highly Sensitive Electrochemical Detection of Tumor Exosomes Based on Aptamer Recognition-Induced Multi-DNA Release and Cyclic Enzymatic Amplification. Anal. Chem. 2018, 90, 4507–4513. [Google Scholar] [CrossRef] [PubMed]
- Jing, L.; Xie, C.; Li, Q.; Yang, M.; Li, S.; Li, H.; Xia, F. Electrochemical Biosensors for the Analysis of Breast Cancer Biomarkers: From Design to Application. Anal. Chem. 2022, 94, 269–296. [Google Scholar] [CrossRef]
- Wongkaew, N.; Simsek, M.; Griesche, C.; Baeumner, A.J. Functional Nanomaterials and Nanostructures Enhancing Electrochemical Biosensors and Lab-on-a-Chip Performances: Recent Progress, Applications, and Future Perspective. Chem Rev. 2019, 119, 120–194. [Google Scholar] [CrossRef] [PubMed]
- Rotariu, L.; Lagarde, F.; Jaffrezic-Renault, N.; Bala, C. Electrochemical biosensors for fast detection of food contaminants trends and perspective. Trac-Trends Anal. Chem. 2016, 79, 80–87. [Google Scholar] [CrossRef]
- Li, L.; Xu, S.; Yan, H.; Li, X.; Yazd, H.S.; Li, X.; Huang, T.; Cui, C.; Jiang, J.; Tan, W. Nucleic Acid Aptamers for Molecular Diagnostics and Therapeutics: Advances and Perspectives. Angew. Chem. Int. Ed. Engl. 2021, 60, 2221–2231. [Google Scholar] [CrossRef] [PubMed]
- Chai, H.; Miao, P. Bipedal DNA Walker Based Electrochemical Genosensing Strategy. Anal. Chem. 2019, 91, 4953–4957. [Google Scholar] [CrossRef] [PubMed]
- Xu, M.; Tang, D. Recent advances in DNA walker machines and their applications coupled with signal amplification strategies: A critical review. Anal. Chim. Acta 2021, 1171, 338523. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Vietz, C.; Schröder, T.; Acuna, G.; Lalkens, B.; Tinnefeld, P. A DNA Walker as a Fluorescence Signal Amplifier. Nano Lett. 2017, 17, 5368–5374. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Sun, R.; He, P.; Zhang, X. Ultrasensitive Detection of Exosomes by Target-Triggered Three-Dimensional DNA Walking Machine and Exonuclease III-Assisted Electrochemical Ratiometric Biosensing. Anal. Chem. 2019, 91, 14773–14779. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Q.; Rahimian, A.; Son, K.; Shin, D.S.; Patel, T.; Revzin, A. Development of an aptasensor for electrochemical detection of exosomes. Methods 2016, 97, 88–93. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xuan, F.; Fan, T.W.; Hsing, I.M. Electrochemical Interrogation of Kinetically-Controlled Dendritic DNA/PNA Assembly for Immobilization-Free and Enzyme-Free Nucleic Acids Sensing. ACS Nano 2015, 9, 5027–5033. [Google Scholar] [CrossRef] [PubMed]
- Gai, P.; Gu, C.; Li, H.; Sun, X.; Li, F. Ultrasensitive Ratiometric Homogeneous Electrochemical MicroRNA Biosensing via Target-Triggered Ru(III) Release and Redox Recycling. Anal. Chem. 2017, 89, 12293–12298. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Yang, L.; Yin, X.; Liu, X.; Ge, L.; Li, F. A facile homogeneous electrochemical biosensing strategy based on displacement reaction for intracellular and extracellular hydrogen peroxide detection. Biosens Bioelectron 2019, 141, 111446. [Google Scholar] [CrossRef] [PubMed]
- Yin, X.; Hou, T.; Huang, B.; Yang, L.; Li, F. Aptamer recognition-trigged label-free homogeneous electrochemical strategy for an ultrasensitive cancer-derived exosome assay. Chem. Commun. 2019, 55, 13705–13708. [Google Scholar] [CrossRef]
- Maduraiveeran, G.; Sasidharan, M.; Ganesan, V. Electrochemical sensor and biosensor platforms based on advanced nanomaterials for biological and biomedical applications. Biosens. Bioelectron. 2018, 103, 113–129. [Google Scholar] [CrossRef] [PubMed]
- Jia, X.; Dong, S.; Wang, E. Engineering the bioelectrochemical interface using functional nanomaterials and microchip technique toward sensitive and portable electrochemical biosensors. Biosens. Bioelectron. 2016, 76, 80–90. [Google Scholar] [CrossRef] [PubMed]
- Salahandish, R.; Ghaffarinejad, A.; Omidinia, E.; Zargartalebi, H.; Majidzadeh, A.K.; Naghib, S.M.; Sanati-Nezhad, A. Label-free ultrasensitive detection of breast cancer miRNA-21 biomarker employing electrochemical nano-genosensor based on sandwiched AgNPs in PANI and N-doped graphene. Biosens. Bioelectron. 2018, 120, 129–136. [Google Scholar] [CrossRef] [PubMed]
- Cheng, G.; Li, W.Q.; Ha, L.; Han, X.H.; Hao, S.J.; Wan, Y.; Wang, Z.G.; Dong, F.P.; Zou, X.; Mao, Y.W.; et al. Self-Assembly of Extracellular Vesicle-like Metal-Organic Framework Nanoparticles for Protection and Intracellular Delivery of Biofunctional Proteins. J. Am. Chem. Soc. 2018, 140, 7282–7291. [Google Scholar] [CrossRef]
- Wang, S.Z.; Chen, Y.J.; Wang, S.Y.; Li, P.; Mirkin, C.A.; Farha, O.K. DNA-Functionalized Metal-Organic Framework Nanoparticles for Intracellular Delivery of Proteins. J. Am. Chem. Soc. 2019, 141, 2215–2219. [Google Scholar] [CrossRef] [Green Version]
- Chen, T.T.; Yi, J.T.; Zhao, Y.Y.; Chu, X. Biomineralized Metal-Organic Framework Nanoparticles Enable Intracellular Delivery and Endo-Lysosomal Release of Native Active Proteins. J. Am. Chem. Soc. 2018, 140, 9912–9920. [Google Scholar] [CrossRef]
- Yang, X.T.; Tang, Q.; Jiang, Y.; Zhang, M.N.; Wang, M.; Mao, L.Q. Nanoscale ATP-Responsive Zeolitic Imidazole Framework-90 as a General Platform for Cytosolic Protein Delivery and Genome Editing. J. Am. Chem. Soc. 2019, 141, 3782–3786. [Google Scholar] [CrossRef]
- Zhong, M.; Yang, L.; Yang, H.; Cheng, C.; Deng, W.F.; Tan, Y.M.; Xie, Q.J.; Yao, S.Z. An electrochemical immunobiosensor for ultrasensitive detection of Escherichia coil O157:H7 using CdS quantum dots-encapsulated metal-organic frameworks as signal-amplifying tags. Biosens. Bioelectron. 2019, 126, 493–500. [Google Scholar] [CrossRef]
- Zhou, Y.; Zhang, D.A.; Zeng, J.; Gan, N.; Cuan, J. A luminescent Lanthanide-free MOF nanohybrid for highly sensitive ratiometric temperature sensing in physiological range. Talanta 2018, 181, 410–415. [Google Scholar] [CrossRef]
- Cao, Y.; Wang, Y.; Yu, X.; Jiang, X.; Li, G.; Zhao, J. Identification of programmed death ligand-1 positive exosomes in breast cancer based on DNA amplification-responsive metal-organic frameworks. Biosens. Bioelectron. 2020, 166, 112452. [Google Scholar] [CrossRef] [PubMed]
- Menck, K.; Bleckmann, A.; Wachter, A.; Hennies, B.; Ries, L.; Schulz, M.; Balkenhol, M.; Pukrop, T.; Schatlo, B.; Rost, U.; et al. Characterisation of tumour-derived microvesicles in cancer patients’ blood and correlation with clinical outcome. J. Extracell. Vesicles 2017, 6, 1–16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- An, Y.; Li, R.; Zhang, F.; He, P.G. Magneto-Mediated Electrochemical Sensor for Simultaneous Analysis of Breast Cancer Exosomal Proteins. Anal. Chem. 2020, 92, 5404–5410. [Google Scholar] [CrossRef] [PubMed]
- Cutler, J.I.; Auyeung, E.; Mirkin, C.A. Spherical Nucleic Acids. J. Am. Chem. Soc. 2012, 134, 1376–1391. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Liu, J. Freezing Directed Construction of Bio/Nano Interfaces: Reagentless Conjugation, Denser Spherical Nucleic Acids, and Better Nanoflares. J. Am. Chem. Soc. 2017, 139, 9471–9474. [Google Scholar] [CrossRef] [Green Version]
- Li, J.; Fan, C.; Pei, H.; Shi, J.; Huang, Q. Smart Drug Delivery Nanocarriers with Self-Assembled DNA Nanostructures. Adv. Mater. 2013, 25, 4386–4396. [Google Scholar] [CrossRef]
- Wang, L.; Deng, Y.; Wei, J.; Huang, Y.; Wang, Z.; Li, G. Spherical nucleic acids-based cascade signal amplification for highly sensitive detection of exosomes. Biosens. Bioelectron. 2021, 191, 113465. [Google Scholar] [CrossRef]
- Yu, Y.; Jin, B.; Li, Y.; Deng, Z. Stimuli-Responsive DNA Self-Assembly: From Principles to Applications. Chemistry 2019, 25, 9785–9798. [Google Scholar] [CrossRef]
- Ke, Y.; Castro, C.; Choi, J.H. Structural DNA Nanotechnology: Artificial Nanostructures for Biomedical Research. Annu. Rev. Biomed. Eng. 2018, 20, 375–401. [Google Scholar] [CrossRef] [Green Version]
- Liu, N.; Lu, H.; Liu, L.; Ni, W.; Yao, Q.; Zhang, G.-J.; Yang, F. Ultrasensitive Exosomal MicroRNA Detection with a Supercharged DNA Framework Nanolabel. Anal. Chem. 2021, 93, 5917–5923. [Google Scholar] [CrossRef]
- Wu, M.S.; Sun, X.T.; Zhu, M.J.; Chen, H.Y.; Xu, J.J. Mesoporous silica film-assisted amplified electrochemiluminescence for cancer cell detection. Chem. Commun. 2015, 51, 14072–14075. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Chen, Y.; Zhu, J.-J. Recent Advances in Electrochemiluminescence Analysis. Anal. Chem. 2017, 89, 358–371. [Google Scholar] [CrossRef] [PubMed]
- Feng, Q.M.; Ma, P.; Cao, Q.H.; Guo, Y.H.; Xu, J.J. An aptamer-binding DNA walking machine for sensitive electrochemiluminescence detection of tumor exosomes. Chem. Commun. 2020, 56, 269–272. [Google Scholar] [CrossRef] [PubMed]
- Qiao, B.; Guo, Q.; Jiang, J.; Qi, Y.; Zhang, H.; He, B.; Cai, C.; Shen, J. An electrochemiluminescent aptasensor for amplified detection of exosomes from breast tumor cells (MCF-7 cells) based on G-quadruplex/hemin DNAzymes. Analyst 2019, 144, 3668–3675. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; An, Y.; Jin, T.Y.; Zhang, F.; He, P.G. Detection of MUC1 protein on tumor cells and their derived exosomes for breast cancer surveillance with an electrochemiluminescence aptasensor. J. Electroanal. Chem. 2021, 882, 115011. [Google Scholar] [CrossRef]
- Zhang, H.X.; Wang, Z.H.; Zhang, Q.X.; Wang, F.; Liu, Y. Ti3C2 MXenes nanosheets catalyzed highly efficient electrogenerated chemiluminescence biosensor for the detection of exosomes. Biosens. Bioelectron. 2019, 124, 184–190. [Google Scholar] [CrossRef] [PubMed]
- Cheng, N.; Song, Y.; Shi, Q.; Du, D.; Liu, D.; Luo, Y.; Xu, W.; Lin, Y. Au@Pd Nanopopcorn and Aptamer Nanoflower Assisted Lateral Flow Strip for Thermal Detection of Exosomes. Anal. Chem. 2019, 91, 13986–13993. [Google Scholar] [CrossRef]
- Vaidyanathan, R.; Naghibosadat, M.; Rauf, S.; Korbie, D.; Carrascosa, L.G.; Shiddiky, M.J.; Trau, M. Detecting exosomes specifically: A multiplexed device based on alternating current electrohydrodynamic induced nanoshearing. Anal. Chem. 2014, 86, 11125–11132. [Google Scholar] [CrossRef]

| Method | Principle | Advantages | Shortcoming |
|---|---|---|---|
| Differential centrifugation | According to the difference in density, size and shape, the separation is completed by the cooperation of low-speed centrifugation and high-speed centrifugation. | Easy operation, simple technology, large sample capacity, no additional chemicals, fewer consumables, and good reproducibility | Time consuming, low purity, small quantity, easy contamination [39,40], and may destroy the exosomes’ structure [41]. |
| Density gradient centrifugation | According to the difference in density, size, and shape, the separation is completed by ultracentrifugation in a certain concentration gradient medium. | Compared with ultracentrifugation, the purity is higher, the yield is higher, and no additional chemicals are needed. | High cost, complex technology, tedious operation, time-consuming centrifugation, and loss of a large number of samples [42], which means it is not suitable for large-scale use. |
| Ultrafiltration method | According to the molecular size or molecular weight, ultrafiltration membranes with different interception relative molecular weights are used to complete the separation. | Suitable for large volume samples, unlimited on sample size, simple technology, less time-consuming, multiple samples can be processed at the same time, and the biological activity of exosomes is not affected [43,44]. | Short service life, sample loss leads to reduced output, easy contamination by non-exosome protein, easy deformation of vesicles, and the filter pore is easily clogged. |
| Size sieving method | According to the molecular size or molecular weight, the separation is completed by a porous gel column. | Compared with ultrafiltration, it takes less time, protects exosome structure, and prevents exosome aggregation. | limited sample volume by column volume, easy contamination by particles of similar size, easy dilution and low yield. |
| Immune affinity capture method | Separation is completed based on the specific binding between antibodies and exosome membrane proteins, including enzyme-linked immunosorbent assay (ELISA) and immunomagnetic bead adsorption. | High purity and specificity. Most of the target proteins are CD63, CD9, and CD81, which are commonly found on the surface of exosomes [42,45]. | High cost, limited output, time-consuming, low extraction efficiency, the antibody can be blocked, the steps of special antibody membrane are cumbersome |
| Polymer precipitation method | Using hydrophilic polymers or other chemical reagents to change the dispersion or water solubility of the exosomes to precipitate the exosomes. | Easy operation and no need for special instruments. | Low purity, Easy mixing with impurities, need to remove impurities. |
| Microfluidic method | Controls the fluid behavior in the microchannel to accurately control the droplet shape and particle size. | Light equipment, low sample size, low cost, simple operation, high purity, integration of separation and analysis, process automation [46]. | Lack of standard and large-scale testing, low output, special design is needed, high cost. |
| Artificial antibody method | Based on the mutual recognition between artificial antibody and exosome, such as aptamer technique [47] and molecular imprinting technique [48], exosome separation is completed. | Easy to prepare, economical and applicable, suitable for large-scale use, and universal [49]. | Highly professional technology and the kinds of ligands that need to be developed. |
| Type of Biosensor | Exosome Source | Exosomal Biomarker | Bio- Receptor | Probe | Amplification Method | Limit of Detection | Refs. |
|---|---|---|---|---|---|---|---|
| Colorimetric biosensors | MCF-7 cells, patient’s serum | CD63 | aptamer | g-C3N4NSs nanozyme- H2O2–TMB | NA | NA | [73] |
| patient’s serum | CD63 | aptamer | s–SWCNT nanozyme- H2O2–TMB | NA | 5.2 × 105 particles/μL | [74] | |
| patient’s serum | CD63 | aptamer | CuCo2O4 nanorods | NA | 4.5 × 103 particles/μL | [75] | |
| patient’s serum | CD63 | aptamer | Au-NRs | hybridization chain reaction (HCR) | 1.6 × 102 particles/μL (spectroscopy) 9 × 103 particles/μL (naked eyes) | [81] | |
| patient’s serum | CD63 | aptamer | Au-NBP@ MnO2-NSs | competitive reaction between exosomes, inducing placeholder chains | 1.35 × 102 particles/μL | [85] | |
| MCF-7 cells | CD63 | aptamer | Fe–MOF | NA | 5.2 × 104 particles/μL | [79] | |
| MCF-7 cells, patient’s serum | CD63 | aptamer | HRP–H2O2–PDA | NA | 7.7 × 103 particles/μL | [57] | |
| MCF-7 cells | CD63 | antibody | HRP-PDA (paper based) | NA | 4.46 × 103 particles/μL | [62] | |
| Fluorescent biosensors | MCF-7 cells | MUC1 | aptamer | Tamra–Dabcyl | NA | 4.2 × 104 particles/μL | [87] |
| MCF-7 cells, patient’s serum | CD63 cholesterol | aptamer | Connector–Backbone (proximity ligation) | rolling circle amplification (RCA) | NA | [88] [89] | |
| MCF-7 cells | HER2 EpCAM | aptamer | ASPNC, BHQ-2 | TPP amplify the afterglow signal | NA | [103] | |
| BT-474 cells UACC-812 cells MCF-7 cells patient’s serum | HER2 EpCAM CD36 | aptamer | GelRed H1-H2 toehold nanostructure | hybridization chain reaction (HCR) | 2.8 × 102 particles/μL | [98] | |
| plasma | CA15-3 CA125 CEA HER2 EGFR PSMA EpCAM VEGF | aptamer | Cy-5 | localized laser heating for thermophoretic accumulation | 3.8 × 104 particles/μL | [97] | |
| Fluorescent biosensors | MCF-7 cells plasma | PD-L1 | aptamer | infrared laser | NA | <200 pg/mL | [99] |
| MCF-7 cells patient’s serum | miR-21 | Fusion protein | Vir-FVs | NA | NA | [92] | |
| SK-BR-3 cells | HER2 | IgG Streptavidin | Quantum dots | NA | NA | [107] | |
| patient’s plasma | CD81 HER2 | antibody | Quantum dots | NA | NA | [108] | |
| Surface plasmon Resonance (SPR) biosensors | MCF-7 cells | CD63 | aptamer | Au NPs | electronic coupling plasmonic nanostructures | 5 × 103 exosomes/mL | [112] |
| Surface- Enhanced Raman Scattering (SERS) biosensors | MCF-7 MDA-MB-231 patient’s serum | CD9 | antibody | MNPs@Au PEG | NA | NA | [119] |
| MCF-7 MDA-MB-231 | Rhodamine 6G | bacterial cellulose membrane | Ag NPs | NA | 10−11M | [121] | |
| Photoacoustic Imaging | breast cancer mice | folate | membrane | gold nanorods | NA | NA | [127] |
| DC cells | TEX | NA | gold nanoparticles | NA | NA | [128] | |
| Electro- chemical biosensors | MCF-7 cells | CD63 | aptamer | DOX, P1, P2, HP | Exo III auxiliary signal technology | 1.2 × 104 particles/mL | [142] |
| MCF-7 cells | CD63 EPCAM-251 | probe DNA | truncated probe A | TDT extends SNAs into PolyT chain, ExoIII-induced catalytic cleavage | 3.8 × 104 particles/μL | [158] | |
| MCF-7 cells plasma | CD63 EpCAM | aptamer | Au, P1, P2, MCH | 3D DNA walking machine and Exo III-assisted recycling | 1.3 × 104 particles/mL | [137] | |
| MCF-7 cells | PD-L1 | probe DNA | PVP@HRP@ZIF-8 | hyperbranched rolling circle amplification | 3.4 × 104 particles/mL | [152] | |
| Electro- chemical biosensors | MCF-7 SK-BR-3 MDA-MB-231 BT474 | CD63 MUC1 CEA HER2 EpCAM | aptamer | MB, SiO2 NPs | FcNHSSNH2 as signal molecule, magnetic separation | NA | [154] |
| plasma | miR-21 | probe DNA | TDN probe | base-stacking effect | 34 aM | [161] | |
| Electrochem-iluminescence biosensors | MCF-7 cells patient’s serum | CD63 | aptamer | Cy3, Ti3C2 MXenes nanocomplex | G-quadruplex/ hemin-DNAzyme, catalyze H2O2 decomposition | 7.41 × 104 particles/mL | [165] |
| patient’s serum | MUC1, CD63 | aptamer | Ru@SiO2 NPs | NA | 2.73 × 10−4 μg/mL | [166] | |
| MCF-7 cells | EpCAM | aptamer | Ti3C2 MXenes nanocomplex | Large specific surface area, electrical conductivity and catalytic performance | 5 × 103 particles/μL | [167] | |
| Thermal signal biosensors | Breast cancer cells | CD63 | aptamer nanoflowers | Au@Pd nanopopcorn ANAN-LFS | rolling circle amplification | 1.4 × 104 exosomes/μL | [168] |
| Ac electrohy- draulic dynamics | patient’s serum | HER2, PSA | antibody | Au/Biotin-BSA/Strep-tavidin/Biotin Ab | NA | 2760 exosomes/μL | [169] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tang, Q.; Xiao, X.; Li, R.; He, H.; Li, S.; Ma, C. Recent Advances in Detection for Breast-Cancer-Derived Exosomes. Molecules 2022, 27, 6673. https://doi.org/10.3390/molecules27196673
Tang Q, Xiao X, Li R, He H, Li S, Ma C. Recent Advances in Detection for Breast-Cancer-Derived Exosomes. Molecules. 2022; 27(19):6673. https://doi.org/10.3390/molecules27196673
Chicago/Turabian StyleTang, Qin, Xinying Xiao, Ranhao Li, Hailun He, Shanni Li, and Changbei Ma. 2022. "Recent Advances in Detection for Breast-Cancer-Derived Exosomes" Molecules 27, no. 19: 6673. https://doi.org/10.3390/molecules27196673




