Moco Carrier and Binding Proteins
Abstract
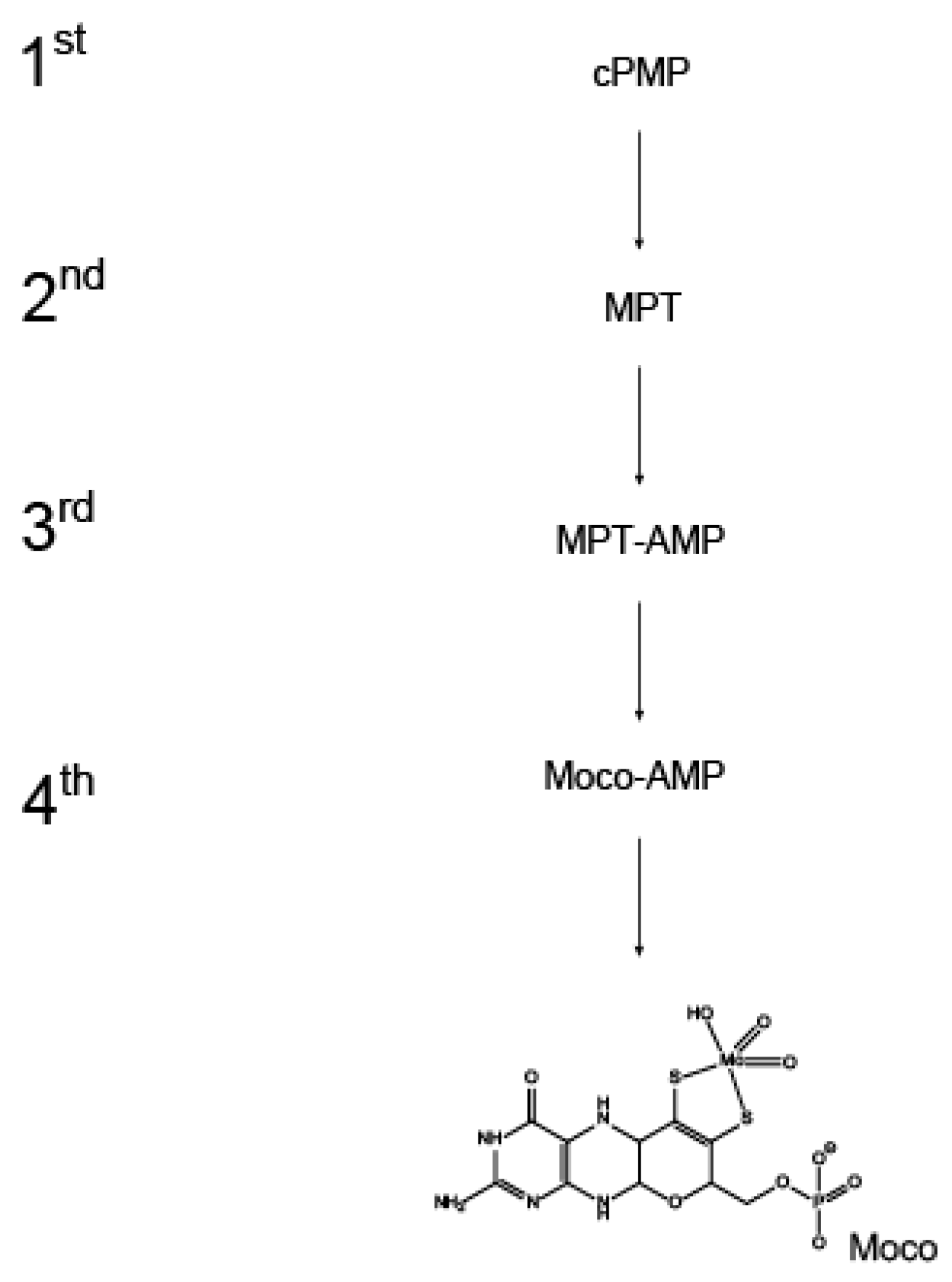
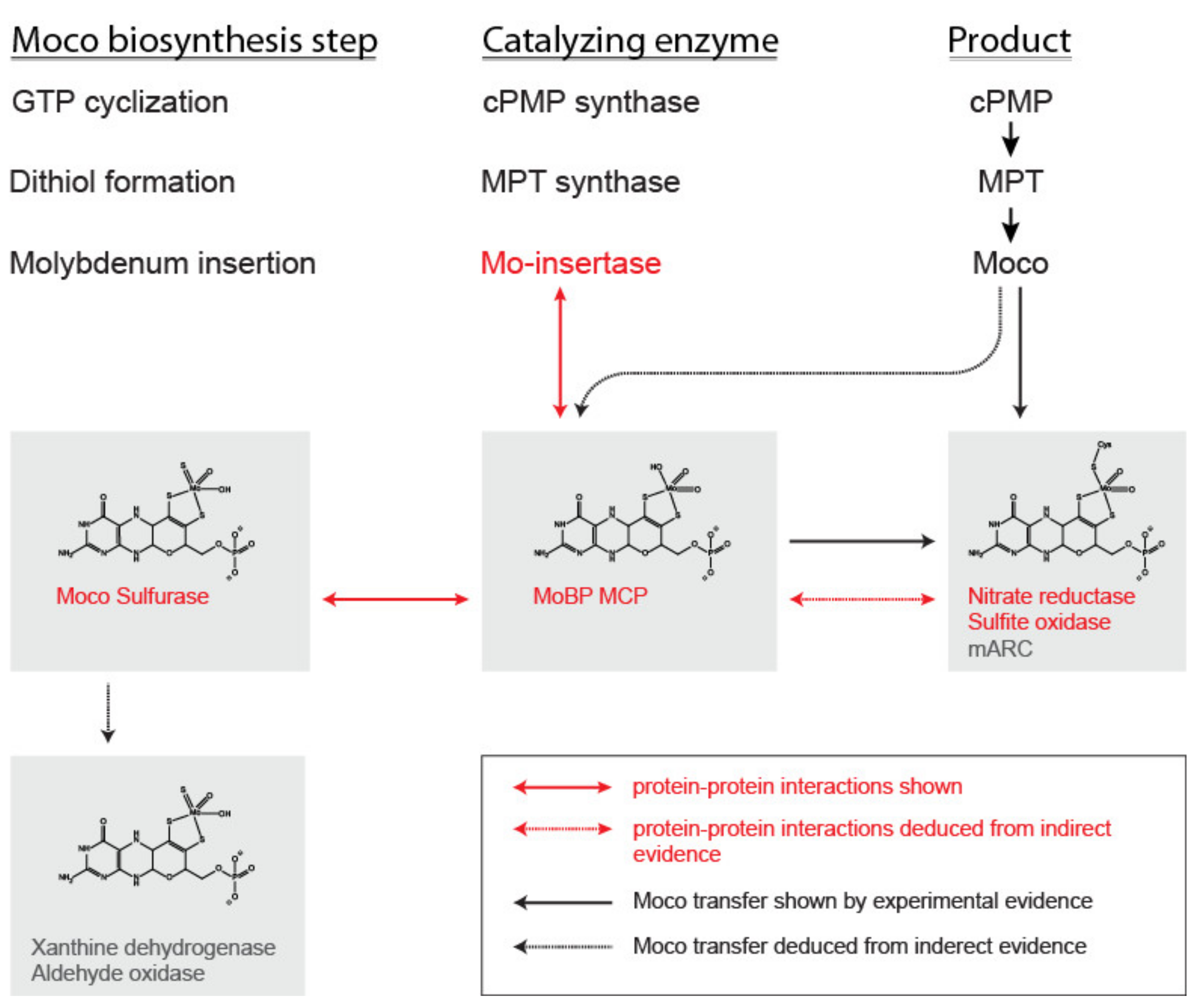
1. Introduction
2. Identification and Occurance of Moco Carrier and Binding Proteins
3. Biochemical and Structural Characterization of Moco Carrier and Binding Proteins
4. Conclusions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wuebbens, M.M.; Rajagopalan, K.V. Structural characterization of a molybdopterin precursor. J. Biol. Chem. 1993, 268, 13493–13498. [Google Scholar] [CrossRef]
- Santamaria-Araujo, J.A.; Fischer, B.; Otte, T.; Nimtz, M.; Mendel, R.R.; Wray, V.; Schwarz, G. The tetrahydropyranopterin structure of the sulfur-free and metal-free molybdenum cofactor precursor. J. Biol. Chem. 2004, 279, 15994–15999. [Google Scholar] [CrossRef] [PubMed]
- Hover, B.M.; Loksztejn, A.; Ribeiro, A.A.; Yokoyama, K. Identification of a cyclic nucleotide as a cryptic intermediate in molybdenum cofactor biosynthesis. J. Am. Chem. Soc. 2013, 135, 7019–7032. [Google Scholar] [CrossRef] [PubMed]
- Hover, B.M.; Tonthat, N.K.; Schumacher, M.A.; Yokoyama, K. Mechanism of pyranopterin ring formation in molybdenum cofactor biosynthesis. Proc. Natl. Acad. Sci. USA 2015, 112, 6347–6352. [Google Scholar] [CrossRef]
- Wajmann, S.; Hercher, T.W.; Buchmeier, S.; Hansch, R.; Mendel, R.R.; Kruse, T. The First Step of Neurospora crassa Molybdenum Cofactor Biosynthesis: Regulatory Aspects under N-Derepressing and Nitrate-Inducing Conditions. Microorganisms 2020, 8, 534. [Google Scholar] [CrossRef]
- Pitterle, D.M.; Johnson, J.L.; Rajagopalan, K.V. In vitro synthesis of molybdopterin from precursor Z using purified converting factor. Role of protein-bound sulfur in formation of the dithiolene. J. Biol. Chem. 1993, 268, 13506–13509. [Google Scholar] [CrossRef]
- Wuebbens, M.M.; Rajagopalan, K.V. Mechanistic and Mutational Studies of Escherichia coli Molybdopterin Synthase Clarify the Final Step of Molybdopterin Biosynthesis. J. Biol. Chem. 2003, 278, 14523–14532. [Google Scholar] [CrossRef]
- Rudolph, M.J.; Wuebbens, M.M.; Turque, O.; Rajagopalan, K.V.; Schindelin, H. Structural studies of molybdopterin synthase provide insights into its catalytic mechanism. J. Biol. Chem. 2003, 278, 14514–14522. [Google Scholar] [CrossRef]
- Kruse, T. Function of Molybdenum Insertases. Molecules 2022, 27, 5372. [Google Scholar] [CrossRef]
- Kuper, J.; Llamas, A.; Hecht, H.J.; Mendel, R.R.; Schwarz, G. Structure of the molybdopterin-bound Cnx1G domain links molybdenum and copper metabolism. Nature 2004, 430, 803–806. [Google Scholar] [CrossRef]
- Probst, C.; Yang, J.; Krausze, J.; Hercher, T.W.; Richers, C.P.; Spatzal, T.; Kc, K.; Giles, L.J.; Rees, D.C.; Mendel, R.R.; et al. Mechanism of molybdate insertion into pterin-based molybdenum cofactors. Nat. Chem. 2021, 13, 758–765. [Google Scholar] [CrossRef]
- Mendel, R.R. The molybdenum cofactor. J. Biol. Chem. 2013, 288, 13165–13172. [Google Scholar] [CrossRef]
- Mendel, R.R. The History of the Molybdenum Cofactor—A Personal View. Molecules 2022, 27, 4934. [Google Scholar] [CrossRef]
- Aguilar, M.; Kalakoutskii, K.; Cardenas, J.; Fernandez, E. Direct transfer of molybdopterin cofactor to apo nitrate reductase from a carrier protein in Chlamydomonas reinhardtii. FEBS Lett. 1992, 307, 162–163. [Google Scholar] [CrossRef]
- Ataya, F.S.; Witte, C.P.; Galvan, A.; Igeno, M.I.; Fernandez, E. Mcp1 Encodes the Molybdenum Cofactor Carrier Protein in Chlamydomonas reinhardtii and Participates in Protection, Binding, and Storage Functions of the Cofactor. J. Biol. Chem. 2003, 278, 10885–10890. [Google Scholar] [CrossRef]
- Fischer, K.; Llamas, A.; Tejada-Jimenez, M.; Schrader, N.; Kuper, J.; Ataya, F.S.; Galvan, A.; Mendel, R.R.; Fernandez, E.; Schwarz, G. Function and structure of the molybdenum cofactor carrier protein from Chlamydomonas reinhardtii. J. Biol. Chem. 2006, 281, 30186–30194. [Google Scholar] [CrossRef]
- Hercher, T.W.; Krausze, J.; Yang, J.; Kirk, M.L.; Kruse, T. Identification and characterisation of the Volvox carteri Moco carrier protein. Biosci. Rep. 2020, 40, BSR20202351. [Google Scholar] [CrossRef]
- Kruse, T.; Gehl, C.; Geisler, M.; Lehrke, M.; Ringel, P.; Hallier, S.; Hansch, R.; Mendel, R.R. Identification and biochemical characterization of molybdenum cofactor-binding proteins from Arabidopsis thaliana. J. Biol. Chem. 2010, 285, 6623–6635. [Google Scholar] [CrossRef]
- Llamas, A.; Tejada-Jimenez, M.; Fernandez, E.; Galvan, A. Molybdenum metabolism in the alga Chlamydomonas stands at the crossroad of those in Arabidopsis and humans. Metallomics 2011, 3, 578–590. [Google Scholar] [CrossRef]
- Krausze, J.; Hercher, T.W.; Archna, A.; Kruse, T. The structure of the Moco carrier protein from Rippkaea orientalis. Acta Crystallogr. F Struct. Biol.Commun. 2020, 76, 453–463. [Google Scholar] [CrossRef]
- Kaufholdt, D.; Baillie, C.K.; Meyer, M.H.; Schwich, O.D.; Timmerer, U.L.; Tobias, L.; van Thiel, D.; Hansch, R.; Mendel, R.R. Identification of a protein-protein interaction network downstream of molybdenum cofactor biosynthesis in Arabidopsis thaliana. J. Plant Physiol. 2016, 207, 42–50. [Google Scholar] [CrossRef] [PubMed]
- Ringel, P.; Krausze, J.; van den Heuvel, J.; Curth, U.; Pierik, A.J.; Herzog, S.; Mendel, R.R.; Kruse, T. Biochemical characterization of molybdenum cofactor-free nitrate reductase from Neurospora crassa. J. Biol. Chem. 2013, 288, 14657–14671. [Google Scholar] [CrossRef] [PubMed]
- Llamas, A.; Otte, T.; Multhaup, G.; Mendel, R.R.; Schwarz, G. The Mechanism of nucleotide-assisted molybdenum insertion into molybdopterin. A novel route toward metal cofactor assembly. J. Biol. Chem. 2006, 281, 18343–18350. [Google Scholar] [CrossRef] [PubMed]
- Amy, N.K.; Rajagopalan, K.V. Characterization of molybdenum cofactor from Escherichia coli. J. Bacteriol. 1979, 140, 114–124. [Google Scholar] [CrossRef]
- Kalakoutskii, K.; Fernandez, E. Different forms of molybdenum cofactor in Vicia faba seeds: The presence of molybdenum cofactor carrier protein and its purification. Planta 1996, 201, 64–70. [Google Scholar] [CrossRef]
- Palmer, T.; Santini, C.-L.; Iobbi-Nivol, C.; Eaves, D.J.; Boxer, D.H.; Giordano, G. Involvement of the narJ and mob gene products in distinct steps in the biosynthesis of the molybdoenzyme nitrate reductase in Escherichia coli. Mol. Microbiol. 1996, 20, 875–884. [Google Scholar] [CrossRef]
- Stewart, V.; MacGregor, C.H. Nitrate reductase in Escherichia coli K-12: Involvement of chlC, chlE, and chlG loci. J. Bacteriol. 1982, 151, 788–799. [Google Scholar] [CrossRef]
- Schwarz, G.; Boxer, D.H.; Mendel, R.R. Molybdenum cofactor biosynthesis. The plant protein Cnx1 binds molybdopterin with high affinity. J. Biol. Chem. 1997, 272, 26811–26814. [Google Scholar] [CrossRef]
- Kaufholdt, D.; Baillie, C.K.; Bikker, R.; Burkart, V.; Dudek, C.A.; von Pein, L.; Rothkegel, M.; Mendel, R.R.; Hansch, R. The molybdenum cofactor biosynthesis complex interacts with actin filaments via molybdenum insertase Cnx1 as anchor protein in Arabidopsis thaliana. Plant Sci. 2016, 244, 8–18. [Google Scholar] [CrossRef]
- Jeon, W.B.; Allard, S.T.; Bingman, C.A.; Bitto, E.; Han, B.W.; Wesenberg, G.E.; Phillips, G.N., Jr. X-ray crystal structures of the conserved hypothetical proteins from Arabidopsis thaliana gene loci At5g11950 and AT2g37210. Proteins 2006, 65, 1051–1054. [Google Scholar] [CrossRef]
| Accession Number | Annotation | Publication | Organism |
|---|---|---|---|
| AY039706 | CeMcp1 | [15] | Chlamydomonas reinhardtii |
| At2g28305 | MoBP1 | [18] | Arabidopsis thaliana |
| At5g11950 | MoBP2 | ||
| At2g37210 | MoBP3 | ||
| At4g35190 | MoBP4 | ||
| At3g53450 | MoBP5 | ||
| At5g03270 | MoBP6 | ||
| WP_012595913 | RoMCP | [20] | Rippkaea orientalis |
| XP_002954772.1 | VcMCP | [17] | Volvox carteri |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kruse, T. Moco Carrier and Binding Proteins. Molecules 2022, 27, 6571. https://doi.org/10.3390/molecules27196571
Kruse T. Moco Carrier and Binding Proteins. Molecules. 2022; 27(19):6571. https://doi.org/10.3390/molecules27196571
Chicago/Turabian StyleKruse, Tobias. 2022. "Moco Carrier and Binding Proteins" Molecules 27, no. 19: 6571. https://doi.org/10.3390/molecules27196571
APA StyleKruse, T. (2022). Moco Carrier and Binding Proteins. Molecules, 27(19), 6571. https://doi.org/10.3390/molecules27196571





