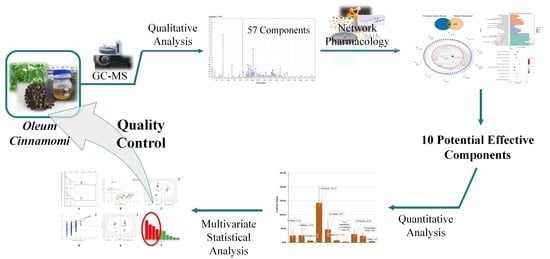
Quality Control of Oleum Cinnamomi Assisted by Network Pharmacology Strategy
Abstract
1. Introduction
2. Results and Discussion
2.1. Optimal Conditions for Sample Pretreatment and the Apparatus
2.2. GC-MS to Identify Components of Oleum Cinnamomi
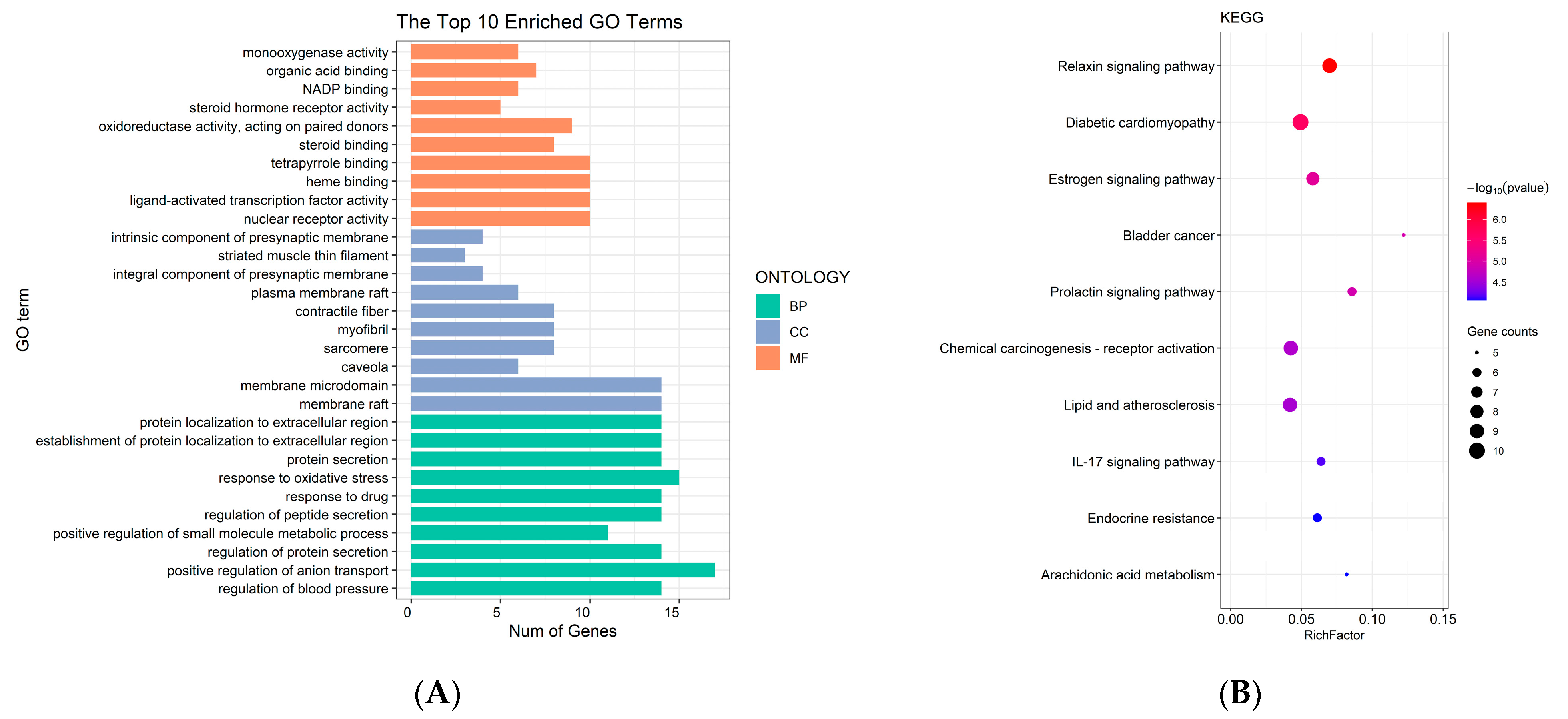
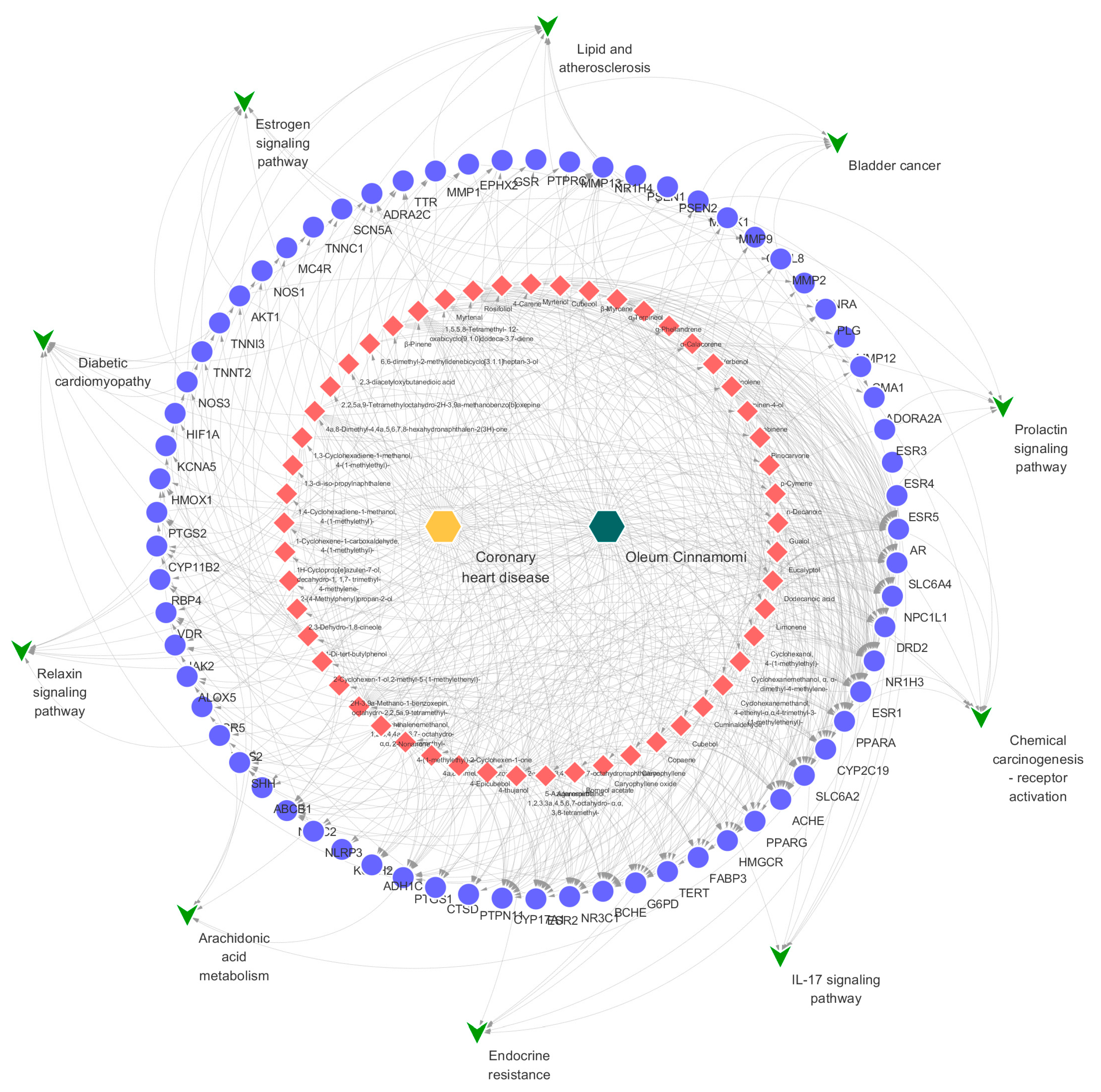
2.3. Network Pharmacological Analysis of Oleum Cinnamomi
2.4. Selection of Index Components for the Quantitative Analysis of Oleum Cinnamomi
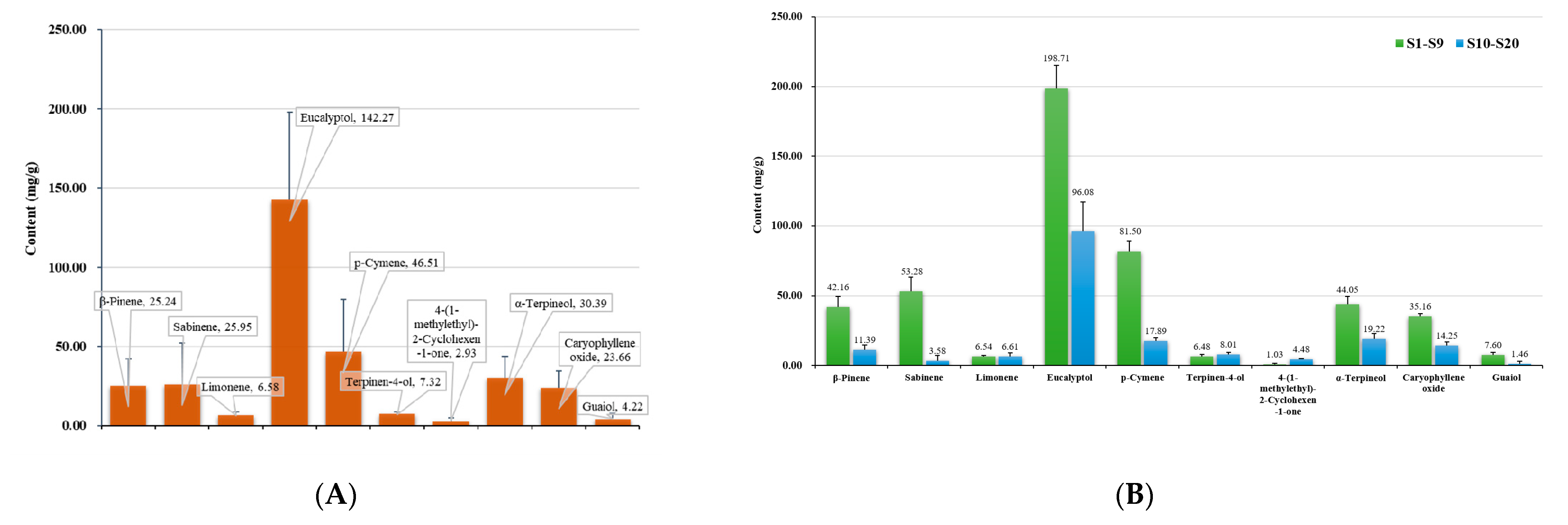
2.5. Quantitative Analysis of Multiple Batches of Oleum Cinnamomi
2.5.1. Method Validation
2.5.2. Assay Result
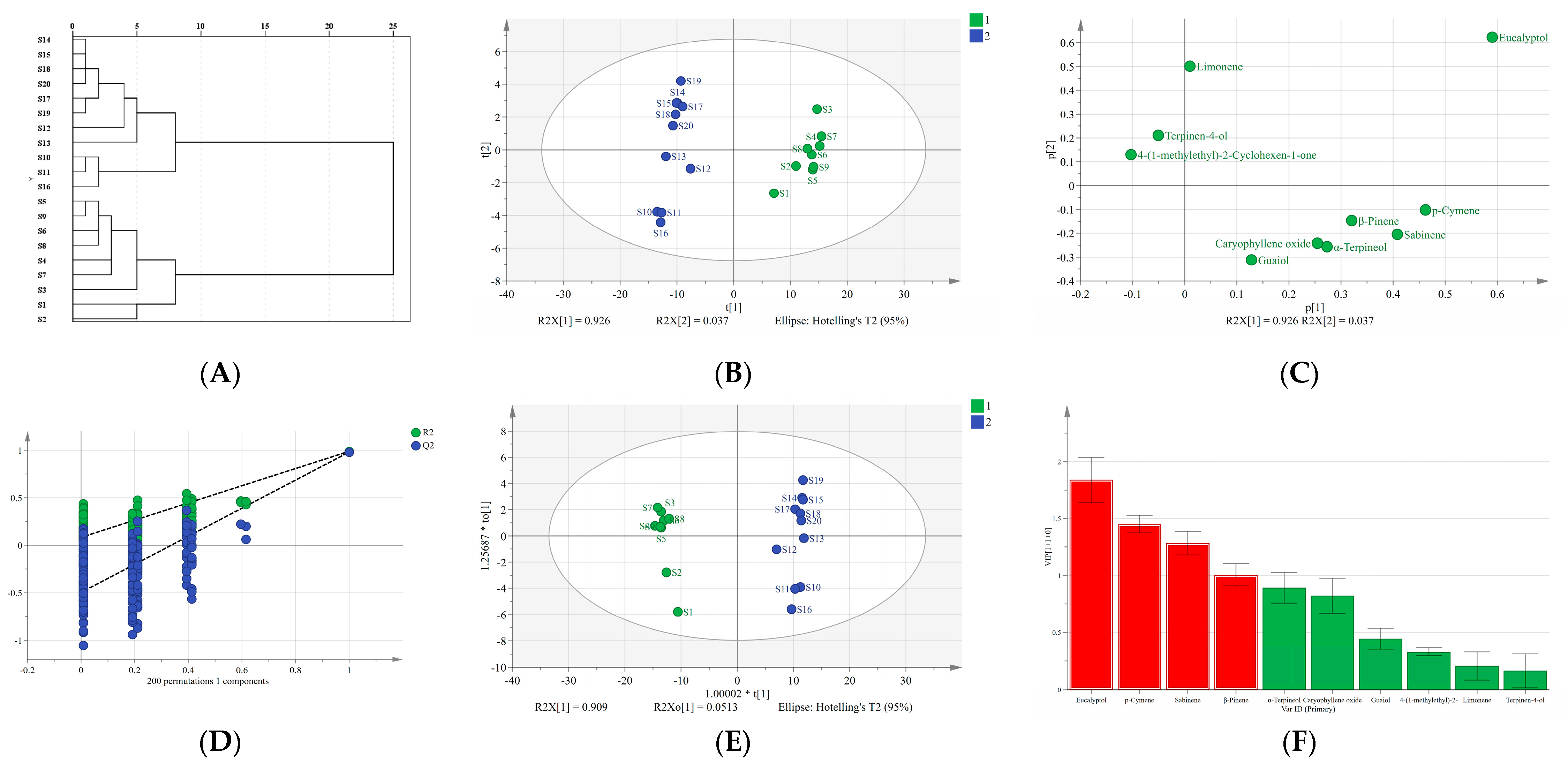
2.6. Multivariate Statistical Analysis
2.6.1. Hierarchical Clustering Analysis (HCA)
2.6.2. Principal Component Analysis (PCA)
2.6.3. Orthogonal Partial Least Squares Discriminant Analysis (OPLS-DA)
3. Materials and Methods
3.1. Reagents and Materials
3.2. Extraction of Oleum Cinnamomi
3.3. Preparation of the Standard Solution and Sample Solution
3.4. Chromatographic and Mass Spectrometric Conditions for Qualitative and Quantitative Analysis
3.5. Network Pharmacological Analysis of Oleum Cinnamomi
3.6. Statistical Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Administration, G.M.P. Quality Standard of Traditional Chinese Medicinal Materials and Ethnic Medicinal Materials in Guizhou Province; China Medical Science Press: Beijing, China, 2019; pp. 8–9. [Google Scholar]
- Liu, J.; Guo, J. Research on volatile oil of Litsealancilimba Merr. Chem. Eng. 2019, 33, 48–50. [Google Scholar]
- Muhammad, I.; Luo, W.; Shoaib, R.M.; Li, G.L.; Shams Ul Hassan, S.; Yang, Z.H.; Xiao, X.; Tu, G.L.; Yan, S.K.; Ma, X.P.; et al. Guaiane-type sesquiterpenoids from Cinnamomum migao H. W. Li: And their anti-inflammatory activities. Phytochemistry 2021, 190, 112850. Phytochemistry 2021, 190, 112850. [Google Scholar] [CrossRef] [PubMed]
- Wu, B.; Wang, F.; Tang, Y.; Hu, Y.; Liu, Y.; Liu, W. Therapeutic effect of Oleum Cinnamomi on Atrial Fibrillation in Rats. Lishizhen Med. Mater. Med. Res. 2019, 30, 2128–2130. [Google Scholar]
- An, Y.; He, Y. Pharmacological action and clinical application of the constituent drugs of Xinnaoning Capsule. Beijing Med. J. 2015, 37, 103–104. [Google Scholar]
- Chen, X.; Tang, X.; Li, J.; Huang, Z.; Yang, T.; Yang, C.; Li, Y. Study on the Mechanism of oil of Cinnamomum Migao H. W. Li in the Treatment of Heart Failure Based on Network Pharmacology. Asia-Pacific Tradit. Med. 2022, 18, 143–151. [Google Scholar]
- Qiu, D.; Xie, B.; Meng, X.; Chai, L.; Yao, C.; Liao, S. Experimental Study on Anti-Influenza Virus of Dalong Mixture. J. Guizhou Univ. Tradit. Chin. Med. 1991, 2, 53–56. [Google Scholar]
- Qiu, D.; Du, M. Research and industrialization of Guizhou Miao medicine Fructus Cinnamomi. J. Guizhou Univ. Tradit. 2003, 1, 48–51. [Google Scholar]
- Chen, D.; Yang, D. In vitro antibacterial test of extracts from different formulations of Oleum Cinnamomi. Chin. J. Ethnomed Ethnopharm. 2010, 19, 42–44. [Google Scholar]
- Lee, A.Y.; Park, W.; Kang, T.W.; Cha, M.H.; Chun, J.M. Network pharmacology-based prediction of active compounds and molecular targets in Yijin-Tang acting on hyperlipidaemia and atherosclerosis. J. Ethnopharmacol. 2018, 221, 151–159. [Google Scholar]
- Zhang, R.; Zhu, X.; Bai, H.; Ning, K. Network Pharmacology Databases for Traditional Chinese Medicine: Review and Assessment. Front. Pharmacol. 2019, 10, 123. [Google Scholar]
- Wang, M.L.; Yang, Q.Q.; Ying, X.H.; Li, Y.Y.; Wu, Y.S.; Shou, Q.Y.; Ma, Q.X.; Zhu, Z.W.; Chen, M.L. Network Pharmacology-Based Approach Uncovers the Mechanism of GuanXinNing Tablet for Treating Thrombus by MAPKs Signal Pathway. Front. Pharmacol. 2020, 11, 652. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Wei, S.; Niu, S.; Ma, X.; Li, H.; Jing, M.; Zhao, Y. Network pharmacology prediction and molecular docking-based strategy to explore the potential mechanism of Huanglian Jiedu Decoction against sepsis. Comput. Biol. Med. 2022, 144, 105389. [Google Scholar] [CrossRef] [PubMed]
- Ye, M.; Luo, G.; Ye, D.; She, M.; Sun, N.; Lu, Y.J.; Zheng, J. Network pharmacology, molecular docking integrated surface plasmon resonance technology reveals the mechanism of Toujie Quwen Granules against coronavirus disease 2019 pneumonia. Phytomedicine 2021, 85, 153401. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Wang, J. Supercritical Carbon Dioxide Extraction of Essential Oil from Cinnamomum migao H. W. Li. Chin. Med. Mat. 2003, 26, 178–180. [Google Scholar]
- Luo, J.; Zhu, D.; Liao, X.; Huang, J.; Tang, J.; Liu, W.; Bao, J.; Tan, J. Comparative Research on the Components of Volatile Oils in Litsea Lam Based on the Theory of Using Fresh Materials in Miao Medicine. Lishizhen Med. Mater. Med. Res. 2019, 30, 574–576. [Google Scholar]
- Zhou, T.; Yang, Z.; Jiang, W.; Ai, Q.; Guo, P. Variation and regularity of volatile oil constituents in fruits of national medicine Cinnamomum migao. Chin. J. Chin. Mater. Med. 2010, 35, 852–856. [Google Scholar]
- Chen, S.; Yang, X.; Wei, Z.; Zhang, Y.; Huang, Y.; Shi, Z.; Zhang, Z.; Wang, J.; Zhang, H.; Ma, J.; et al. Establishment of an anti-inflammation-based bioassay for the quality control of the 13-component TCM formula (Lianhua Qingwen). Pharm. Biol. 2021, 59, 537–545. [Google Scholar] [CrossRef]
- Tan, S.; Niu, Y.; Liu, L.; Su, A.; Hu, C.; Meng, Y. Development of a GC-MS/SIM method for the determination of phytosteryl esters. Food Chem. 2019, 281, 236–241. [Google Scholar] [CrossRef]
- Chen, S.; Tang, Z.; Yang, X.; Tu, X.; Yuan, L.; Pan, S.; Deng, W.; Zhao, T. Study on the Molecular Mechanism of Main Volatile Components of Litsea lancilimba Merr. in the Control of Coronary Heart Disease. Chin. J. Ethnomedicine Ethnopharmacy 2022, 31, 33–38. [Google Scholar]
- Zhao, J.; Ma, S.C.; Li, S.P. Advanced strategies for quality control of Chinese medicines. J. Pharm. Biomed. Anal. 2018, 147, 473–478. [Google Scholar] [CrossRef]
- Salehi, B.; Upadhyay, S.; Erdogan Orhan, I.; Kumar Jugran, A.; Jayaweera, S.L.D.; Dias, D.A.; Sharopov, F.; Taheri, Y.; Martins, N.; Baghalpour, N.; et al. Therapeutic Potential of α- and β-Pinene: A Miracle Gift of Nature. Biomolecules 2019, 9, 738. [Google Scholar] [CrossRef] [PubMed]
- Boskabady, M.H.; Alitaneh, S.; Alavinezhad, A. Carum copticum L.: A herbal medicine with various pharmacological effects. BioMed Res. Int. 2014, 2014, 569087. [Google Scholar] [CrossRef] [PubMed]
- Murad, H.A.S.; Alqurashi, T.M.A.; Hussien, M.A. Interactions of selected cardiovascular active natural compounds with CXCR4 and CXCR7 receptors: A molecular docking, molecular dynamics, and pharmacokinetic/toxicity prediction study. BMC Complement. Med. Ther. 2022, 22, 35. [Google Scholar] [CrossRef] [PubMed]
- Zerrouki, M.; Benkaci-Ali, F. DFT study of the mechanisms of nonenzymatic DNA repair by phytophenolic antioxidants. J. Mol. Model. 2018, 24, 78. [Google Scholar] [CrossRef] [PubMed]
- Sharma, S.; Gupta, J.; Prabhakar, P.K.; Gupta, P.; Solanki, P.; Rajput, A. Phytochemical Repurposing of Natural Molecule: Sabinene for Identification of Novel Therapeutic Benefits Using In Silico and In Vitro Approaches. Assay Drug Dev. Technol. 2019, 17, 339–351. [Google Scholar]
- Anandakumar, P.; Kamaraj, S.; Vanitha, M.K. D-limonene: A multifunctional compound with potent therapeutic effects. J. Food Biochem. 2021, 45, e13566. [Google Scholar]
- Gondim, A.N.S.; Lara, A.; Santos-Miranda, A.; Roman-Campos, D.; Lauton-Santos, S.; Menezes-Filho, J.E.R.; de Vasconcelos, C.M.L.; Conde-Garcia, E.A.; Guatimosim, S.; Cruz, J.S. (-)-Terpinen-4-ol changes intracellular Ca(2+) handling and induces pacing disturbance in rat hearts. Eur. J. Pharmacol. 2017, 807, 56–63. [Google Scholar] [CrossRef]
- Thulasiram, H.V.; Madyastha, K.M. Transformation of a monoterpene ketone, piperitenone, and related terpenoids using Mucor piriformis. Can. J. Microbiol. 2005, 51, 447–454. [Google Scholar] [CrossRef]
- Paulino, E.T.; Rodrigues, A.; Machado, M.; de Oliveira, K.R.V.; Bernardino, A.C.; Quintans-Júnior, L.J.; Oliveira, A.P.; Ribeiro, Ê.A.N. Alpha-terpineol prevents myocardial damage against isoproterenol-MI induced in Wistar-Kyoto rats: New possible to promote cardiovascular integrity. Life Sci. 2022, 290, 120087. [Google Scholar]
- Xiu, Z.; Zhu, Y.; Han, J.; Li, Y.; Yang, X.; Yang, G.; Song, G.; Li, S.; Li, Y.; Cheng, C.; et al. Caryophyllene Oxide Induces Ferritinophagy by Regulating the NCOA4/FTH1/LC3 Pathway in Hepatocellular Carcinoma. Front. Pharmacol. 2022, 13, 930958. [Google Scholar] [CrossRef]
- Commission, C.P. Pharmacopoeia of the People’s Republic of China: Part Four; China Medical Science Press: Beijing, China, 2020. [Google Scholar]
- Liu, Y.; Wang, Q.; Wang, F.; Xu, B.; Zhang, J.; Lu, J.; Qiao, Y. HPLC fingerprint and chemical pattern recognization of kudiezi injection. Chin. Pharm. 2013, 48, 2097–2101. [Google Scholar]
- Han, Q.; Zhou, B.; Li, Y.; Gu, Z. Quality evaluation of Shenlian capsules by HPLC fingerprint and pattern recognition. Chin. J. Pharm. Anal. 2020, 40, 1300–1308. [Google Scholar]
- Yang, Q.; Tian, G.L.; Qin, J.W.; Wu, B.Q.; Tan, L.; Xu, L.; Wu, S.Z.; Yang, J.T.; Jiang, J.H.; Yu, R.Q. Coupling bootstrap with synergy self-organizing map-based orthogonal partial least squares discriminant analysis: Stable metabolic biomarker selection for inherited metabolic diseases. Talanta 2020, 219, 121370. [Google Scholar] [CrossRef] [PubMed]
- Triba, M.N.; Le Moyec, L.; Amathieu, R.; Goossens, C.; Bouchemal, N.; Nahon, P.; Rutledge, D.N.; Savarin, P. PLS/OPLS models in metabolomics: The impact of permutation of dataset rows on the K-fold cross-validation quality parameters. Mol. Biosyst. 2015, 11, 13–19. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Huang, X.; Tong, B.; Wang, D.; Liu, J.; Liao, X.; Sun, Q. Effects of rhizosphere fungi on the chemical composition of fruits of the medicinal plant Cinnamomum migao endemic to southwestern China. BMC Microbiol. 2021, 21, 206. [Google Scholar] [CrossRef] [PubMed]
- Han, Z.; Song, W. Spatiotemporal variations in cropland abandonment in the Guizhou-Guangxi karst mountain area, China. J. Clean. Prod. 2019, 238, 117888. [Google Scholar] [CrossRef]
- Xu, Y.; Wang, S.; Bai, X.; Shu, D.; Tian, Y. Runoff response to climate change and human activities in a typical karst watershed, SW China. PLoS ONE 2018, 13, e0193073. [Google Scholar] [CrossRef]
- Zhou, W.; Chen, Z.; Li, W.; Wang, Y.; Li, X.; Yu, H.; Ran, P.; Liu, Z. Systems pharmacology uncovers the mechanisms of anti-asthma herbal medicine intervention (ASHMI) for the prevention of asthma. J. Funct. Foods 2019, 52, 611–619. [Google Scholar] [CrossRef]
- Li, Y.; Huang, L.; Zeng, X.; Zhong, G.; Ying, M.; Huang, M.; Bi, H. Down-regulation of P-gp expression and function after Mulberroside A treatment: Potential role of protein kinase C and NF-kappa B. Chem. Biol. Interact. 2014, 213, 44–50. [Google Scholar] [CrossRef]
- Liu, X.; Zheng, X.; Wang, N.; Cao, H.; Lu, Y.; Long, Y.; Zhao, K.; Zhou, H.; Zheng, J. Kukoamine B, a novel dual inhibitor of LPS and CpG DNA, is a potential candidate for sepsis treatment. Br. J. Pharmacol. 2011, 162, 1274–1290. [Google Scholar] [CrossRef]
- PubChem. Available online: https://pubchem.ncbi.nlm.nih.gov (accessed on 18 June 2022).
- SwissTarget Prediction. Available online: https://new.swisstargetprediction.ch/ (accessed on 18 June 2022).
- Therapeutic Target Database. Available online: https://bidd.nus.edu.sg/bidd-databases/ttd/ttd.asp (accessed on 20 June 2022).
- OMIM. Available online: https://www.omim.org/ (accessed on 20 June 2022).
- DisGeNet. Available online: https://www.disgenet.org/search (accessed on 20 June 2022).
- GeneCards. Available online: https://www.genecards.org/ (accessed on 20 June 2022).
- UniProt. Available online: https://www.uniprot.org/ (accessed on 23 June 2022).
- Cytoscape. Available online: https://cytoscape.org/ (accessed on 26 June 2022).

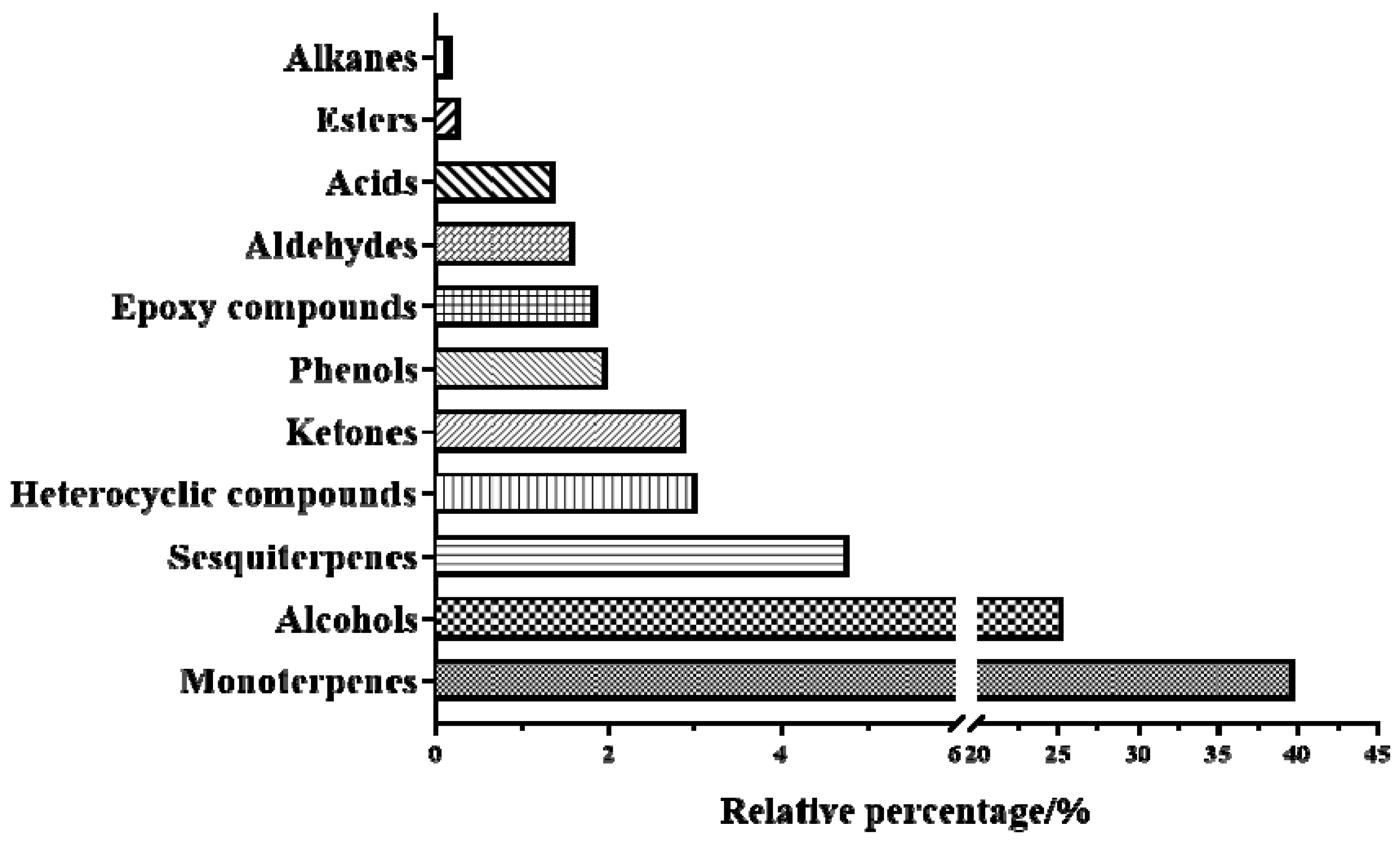
| NO. | Chemical Constituents | Molecular Formula | Molecular Weight | Relative Percentage (%) |
|---|---|---|---|---|
| 1 * | β-Pinene | C10H16 | 136 | 3.53 |
| 2 * | Sabinene | C10H16 | 136 | 2.53 |
| 3 | α-Phellandrene | C10H16 | 136 | 0.95 |
| 4 * | β-Myrcene | C10H16 | 136 | 0.49 |
| 5 | Terpinolene | C10H16 | 136 | 0.25 |
| 6 | 2,3-Dehydro-1,8-cineole | C10H16O | 152 | 0.23 |
| 7 * | Limonene | C10H16 | 136 | 1.43 |
| 8 * | Eucalyptol | C10H18O | 154 | 25.03 |
| 9 * | γ-Terpinene | C10H16 | 136 | 0.51 |
| 10 * | p-Cymene | C10H14 | 134 | 4.66 |
| 11 * | 4-Carene | C10H16 | 136 | 0.19 |
| 12 * | 2-Nonanone | C9H18O | 142 | 0.24 |
| 13 | Copaene | C15H24 | 204 | 0.83 |
| 14 * | 4-Thujanol | C10H18O | 154 | 0.41 |
| 15 | Pinocarvone | C10H14O | 150 | 0.41 |
| 16 * | Borneol acetate | C12H20O2 | 196 | 0.28 |
| 17 * | Caryophyllene | C15H24 | 204 | 0.77 |
| 18 * | Terpinen-4-ol | C10H18O | 154 | 1.70 |
| 19 | Dodecane, 2-methyl- | C13H28 | 184 | 0.18 |
| 20 * | Myrtenal | C10H14O | 150 | 0.45 |
| 21 | Butanedioic acid, 2,3-bis(acetyloxy)- | C8H10O8 | 234 | 0.22 |
| 22 | Cyclohexanol, 4-(1-methylethyl)- | C9H18O | 142 | 0.38 |
| 23 | 6,6-dimethyl-2-methylidenebicyclo[3.1.1]heptan-3-ol | C10H16O | 152 | 0.90 |
| 24 * | 4-(1-methylethyl)-2-Cyclohexen-1-one | C9H14O | 138 | 1.68 |
| 25 | Cyclohexanemethanol, α, α- dimethyl-4-methylene- | C10H18O | 154 | 0.66 |
| 26 | Verbenol | C10H16O | 152 | 0.58 |
| 27 | 4a,8-Dimethyl-2-(prop-1-en-2-yl)-1,2,3,4,4a,5,6,7-octahydronaphthalene | C15H24 | 204 | 0.33 |
| 28 * | α-Terpineol | C10H18O | 154 | 6.23 |
| 29 | 2,2,5a,9-Tetramethyloctahydro-2H-3,9a-methanobenzo[b]oxepine | C15H26O | 222 | 1.02 |
| 30 | 1-Cyclohexene-1-carboxaldehyde, 4-(1-methylethyl)- | C10H16O | 152 | 0.39 |
| 31 | β-cadinene | C15H24 | 204 | 2.60 |
| 32* | Cuminaldehyde | C10H12O | 148 | 0.76 |
| 33* | Myrtenol | C10H16O | 152 | 0.51 |
| 34 | 2H-3,9a-Methano-1-benzoxepin, octahydro-2,2,5a,9-tetramethyl- | C15H26O | 222 | 0.60 |
| 35 | 2-Cyclohexen-1-ol, 2-methyl-5-(1-methylethenyl)- | C10H16O | 152 | 0.36 |
| 36 * | 2-(4-Methylphenyl)propan-2-ol | C10H14O | 150 | 0.54 |
| 37 | 4-Epicubebol | C15H26O | 222 | 0.18 |
| 38 | α-Calacorene | C15H20 | 200 | 0.57 |
| 39 | Cubebol | C15H26O | 222 | 0.26 |
| 40 * | Caryophyllene oxide | C15H24O | 220 | 1.30 |
| 41 | 1,3-Cyclohexadiene-1-methanol, 4-(1-methylethyl)- | C10H16O | 152 | 0.3 |
| 42 | 1,5,5,8-Tetramethyl- 12- oxabicyclo[9.1.0]dodeca-3,7-diene | C15H24O | 220 | 0.55 |
| 43 | 1,4-Cyclohexadiene-1-methanol, 4-(1-methylethyl)- | C10H16O | 152 | 0.38 |
| 44 | Cyclohexanemethanol, 4-ethenyl-α,α,4-trimethyl-3- (1-methylethenyl)- | C15H26O | 222 | 1.26 |
| 45 * | Guaiol | C15H26O | 222 | 1.17 |
| 46 | Rosifoliol | C15H26O | 222 | 2.48 |
| 47 | 2-Naphthalenemethanol, 1,2,3,4,4a,5,6,7- octahydro- α,α, 4a,8-tetramethyl- | C15H26O | 222 | 1.96 |
| 48 | 1H-Cycloprop[e]azulen-7-ol, decahydro-1, 1,7- trimethyl- 4-methylene-, [1ar-(1aα,4aα,7β,7aβ,7bα)]- | C15H24O | 220 | 1.07 |
| 49 | 4a,8-Dimethyl-4,4a,5,6,7,8-hexahydronaphthalen-2(3H)-one | C12H18O | 178 | 0.56 |
| 50 | 1,7-di-iso-propylnaphthalene | C16H20 | 212 | 0.49 |
| 51 | 1,3-di-iso-propylnaphthalene | C16H20 | 212 | 0.35 |
| 52 | 5-Azulenemethanol, 1,2,3,3a,4,5,6,7-octahydro- α,α, 3,8-tetramethyl- | C15H26O | 222 | 0.39 |
| 53 | Agarospirol | C15H26O | 222 | 3.59 |
| 54 | 1,4-di-iso-propylnaphthalene | C16H20 | 212 | 0.23 |
| 55 | n-Decanoic acid | C10H20O2 | 172 | 0.93 |
| 56 | 2,4-Di-tert-butylphenol | C14H22O | 206 | 1.97 |
| 57 | Dodecanoic acid | C12H24O2 | 200 | 0.23 |
| Total (%) | 83.05 | |||
| Rate (°C/min) | Temperature (°C) | Retention Time (min) |
|---|---|---|
| - | 40 | 5 |
| 2 | 50 | - |
| 10 | 100 | - |
| 5 | 150 | - |
| 2 | 190 | 1 |
| 50 | 230 | 3 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zheng, L.; Zhou, Y.; Yan, T.; Gong, Z.; Li, Y.; Chen, S.; Huang, Y.; Chi, M. Quality Control of Oleum Cinnamomi Assisted by Network Pharmacology Strategy. Molecules 2022, 27, 6391. https://doi.org/10.3390/molecules27196391
Zheng L, Zhou Y, Yan T, Gong Z, Li Y, Chen S, Huang Y, Chi M. Quality Control of Oleum Cinnamomi Assisted by Network Pharmacology Strategy. Molecules. 2022; 27(19):6391. https://doi.org/10.3390/molecules27196391
Chicago/Turabian StyleZheng, Lin, Yang Zhou, Ting Yan, Zipeng Gong, Yueting Li, Siying Chen, Yong Huang, and Mingyan Chi. 2022. "Quality Control of Oleum Cinnamomi Assisted by Network Pharmacology Strategy" Molecules 27, no. 19: 6391. https://doi.org/10.3390/molecules27196391
APA StyleZheng, L., Zhou, Y., Yan, T., Gong, Z., Li, Y., Chen, S., Huang, Y., & Chi, M. (2022). Quality Control of Oleum Cinnamomi Assisted by Network Pharmacology Strategy. Molecules, 27(19), 6391. https://doi.org/10.3390/molecules27196391