Abstract
It was recently revealed that naturally occurring myricetin can inhibit ectonucleotidase ectonucleotide pyrophosphatase/phosphodiesterase 1 (ENPP1), which, in turn, can treat ischemic cardiac injury. However, due to myricetin’s poor druggability, its further developments are relatively limited, which necessitates the discovery of novel ENPP1-inhibiting myricetin analogs as alternatives. In this study, the binding model of myricetin with ENPP1 was elucidated by molecular docking and molecular dynamics studies. Subsequently, virtual screening on the self-developed flavonoid natural product database (FNPD), led to the identification of two flavonoid glycosides (Cas No: 1397173-50-0 and 1169835-58-8), as potential ENPP1 inhibitors. Docking scores and MM/GBSA binding energies predicted that they might have higher inhibitory effects than myricetin. This study provides a strong foundation for the future development of ischemic cardiac injury drugs.
1. Introduction
The ectonucleotide pyrophosphatase/phosphodiesterase (ENPP) family consists of single-pass transmembrane ectoenzymes [1]. Based on their common phosphodiesterase (PDE) domain, all ENPP members are able to hydrolyze extracellular nucleotides [1]. Each enzyme has clearly established substrate selectivity and catalytic domains that share structural similarities. Angiogenesis [2]; bone mineralization [3,4]; cell proliferation [5] and motility [6]; digestion [7]; and other activities are all impacted by ENPP catalysis. Evidence also supports that non-catalytic functions of ENPPs exist in cell signaling [8].
Among the ENPP family, ENPP1 is a type II membrane-bound glycoprotein that is essential for several physiological functions, such as insulin signaling and bone mineralization [9,10,11]. ENPP1 hydrolyzes pyrophosphate or phosphodiester linkages in a variety of extracellular substances, including nucleotides and lysophospholipids. It is conserved in vertebrates and slows the mineralization process by converting ATP into AMP, and inorganic pyrophosphate by hydrolysis [12]. Additionally, ENPP1 blocks insulin signaling, and insulin resistance is linked to an ENPP1 polymorphism [1,7].
ENPP1 has been found to be crucial for immune cell penetration into tumors [13]. It was discovered that ENPP1 regulates the cyclic GMP-AMP synthase (cGAS)-stimulator of interferon genes (STING) pathway by hydrolyzing 2’,3’-cyclic GMP-AMP (cGAMP). In the tumor microenvironment, it also helps to produce the immunosuppressive signal, adenosine, in lymphoid tissue. The cGAS/STING-driven type I IFN response is attenuated by ENPP1’s hydrolysis of cGAMP [14]. By increasing STING activation and lengthening the half-life of cGAMP in mice that have ENPP1 knocked out, inhibiting ENPP1 has emerged as a promising strategy for cancer immunotherapy [15,16].
Only very few non-nucleotide ENPP1 inhibitors (Figure 1) with different scaffolds have been discovered, including thioacetamide inhibitor 1 [17], and quinazoline–piperidine–sulfamide inhibitor 2 (QS1) [18]. Following optimization of compound 2 led to the discovery of a highly potent phosphonate ENPP1 inhibitor (STF-1084, 3) [15]. Recently Tan et al. reported a novel ENPP1 inhibitor 4, which demonstrated an IC50 value of 0.188 μM against human ENPP1 at the molecular level. Compound 4 increased the expression levels of IFN-β in vivo, which can be applied to boost antitumor immunity for cancer immunotherapy [19]. Kulkarni et al. recently reported their work on the discovery of a series of promising ENPP1 inhibitors based on a thioguanine scaffold. Their work led to the discovery of compound 5, which demonstrated to have low sub nM potency and also good efficacy in the LLC1 lung cancer syngeneic mouse model [16].
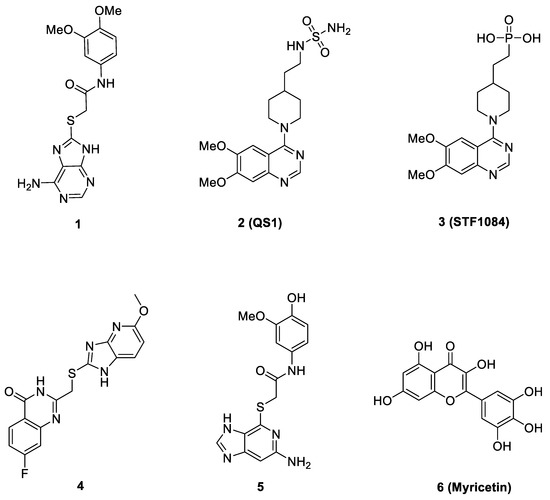
Figure 1.
Molecular structures of several representative reported ENPP1 inhibitors.
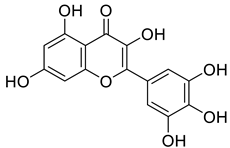
A more recent study by Li et al. reported that cardiac injury induced the expression of ENPP1 and administration of uridine, or genetic targeting of the ENPP1/AMP pathway enhanced repair, after cardiac injury [20]. In particular, their high-throughput screening (HTS) work led to the identification of myricetin, a small-molecule natural product (6, Figure 1), as a potent ENPP1 inhibitor. The administration of myricetin after heart injury rescued pyrimidine biosynthesis in nonmyocyte cells, augmented cardiac repair, and postinfarct heart function [20]. Since some other flavonoid compounds were also reported to elicit anti-myocardial ischemic effects [21,22,23,24,25,26,27,28,29], some other mechanisms including antioxidant and anti-inflammatory activities might also be involved with myricetin [30,31,32]. Novel flavonoid ENPP1 inhibitors might have similar therapeutic potential to rescue cardiac injury and thus are greatly needed. In addition, the binding model of myricetin with its target remains unknown. Thus, to address the demand for rescuing cardiac injury, it is imperative to initiate a further medicinal and chemistry effort. This effort should focus on the discovery of better myricetin-like natural product molecules that are not only potential ENPP1 inhibitors but also have some other common cardiovascular protective benefits, such as in flavonoids.
HTS has become a routine drug discovery process for the identification of novel hit compounds against interesting targets. This process can screen huge chemical libraries rapidly to find the most promising compounds [33,34]. However, it also requires considerable cost and time. As a powerful computational approach to screening large chemical libraries, virtual screening (VS) can save time and costs in the drug development process, which complements HTS effectively in recent years [35,36,37].
In this study, the binding model of myricetin with ENPP1 was firstly elucidated by molecular docking studies. This was further confirmed by molecular dynamics simulations and experimental determination via SAR studies of its structural analogs. Furthermore, based on the model of myricetin binding with ENPP1 and our in-depth understanding of the essential intermolecular interactions from molecular dynamics studies, virtual screening of novel myricetin-like human ENPP1 inhibitors was also performed. This was achieved via an integrated VS workflow to screen the self-developed flavonoid natural product database (FNPD); two flavonoid glycosides were identified as potential ENPP1 inhibitors, and their inhibitory effects were predicted to be superior to that of myricetin when referring to docking scores and MM/GBSA binding energies. This strongly supports future development of new ENPP1 inhibitors for treating ischemic cardiac injury.
2. Results
2.1. In Vitro ENPP1 Inhibition by Myricetin and pH Does Not Influence Its Activity
Previous studies found that the activity of ENPP1 showed metal ion dependency and pH dependency [38]. ENPP1 showed higher activity at higher pH with the addition of Ca2+ and Zn2+. Myricetin was reported to inhibit human ENPP1 activity with an IC50 of 4.8 μM in an HTS study [20]. However, whether the pH condition influences the inhibitory activity of myricetin, on ENPP1, is still uncertain. We found myricetin showed close IC50s with 414.6 nM and 432.7 nM assayed in buffer A (pH 9.5), and buffer B (pH 7.4) (Figure 2), suggesting that pH does not influence ENPP1 inhibition by myricetin. Hence, myricetin may show unique binding behavior compared to others of the reported ENPP1 inhibitors that demonstrated potent inhibition only under basic conditions [38].
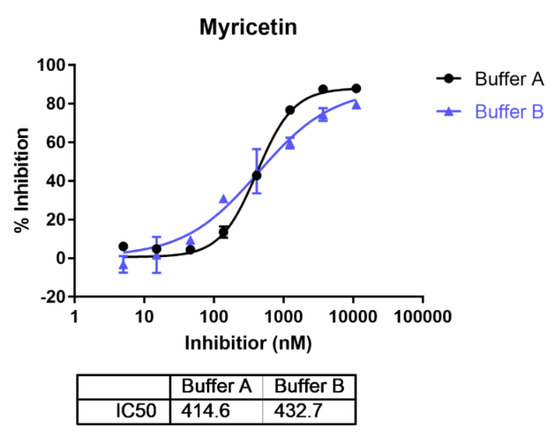
Figure 2.
The inhibitory activities for myricetin on ENPP1 assayed under different pH conditions. (Buffer A: pH 9.5; Buffer B: pH 7.4).
2.2. Molecular Docking of Myricetin with ENPP1 and Further Validations
The binding region in the entire ENPP1 is shown in Figure 3a. As a positive control molecule, the original ligand in the crystal structure (PDB ID: 6wew) was extracted and docked into the active site of ENPP1. The binding pose was superimposed considerably well with that of the crystal structure complex, and the RMSD value was 0.4945 based on the heavy atoms of the ligand. This methodological investigation indicates that the Glide docking method is applicable to this system.
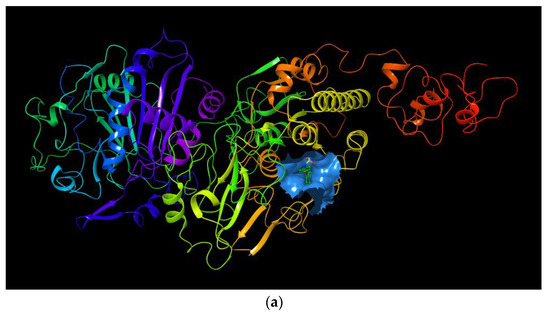
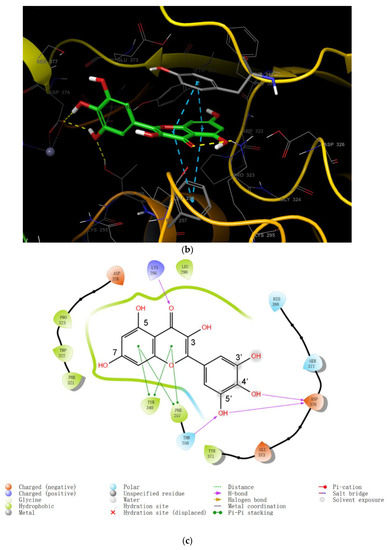
Figure 3.
(a) The binding region in the entire ENPP1; (b) the binding model of myricetin with the ENPP1 from docking study; (c) 2D summary of interaction analysis results of ENPP1-myricetin complexes.
For myricetin, two main representative binding models were obtained, among which the biggest difference is the benzopyrone scaffold or the 2-phenyl group of myricetin, which form better π-π interactions with the target. Since π-π interaction generally can only be considered via high-level quantum chemical calculations, it is not, therefore, explicitly described in traditional molecular mechanics force fields [39]. It seems reasonable that benzopyrone can form stronger π-π interactions since it is a large, delocalized π system and the withdrawing 4-carbonyl group also helps to increase the π-π interactions with the neighboring Tyr340 or Phe257 amino acid residues. Based on these considerations, and also the satisfactory docking scores, we propose that the binding model in Figure 3b is reasonable for myricetin. In this model, the crystal structure complex was stabilized via π-π interactions and hydrogen bonding interactions. The benzopyrone skeleton of the myricetin molecule plays an important role in the interaction with ENPP1, in which the parent nucleus forms a parallel-displaced π-π stacking interaction with Tyr340 and a T-shape π-π interaction with Phe257 (Figure 3c); 4-oxo forms a hydrogen bond with Lys295; the 3′,4′-dihydroxyl of the 2-phenyl forms two hydrogen bonds with Thr256 and Asp376; and the 7-OH possibly forms a hydrogen bond with Phe321 judging from the distance.
The reasonability of this binding model of myricetin was further supported by molecular dynamics simulations. For one of the other representative models, in which the 2-phenyl group of myricetin adopts π-π interaction with ENPP1, it underwent relatively big RMS deviations during MD simulations. However, the RMSD of the ENPP1 protein Cα atoms remained stable for our proposed binding model during the whole of the MD simulations, implying the system reached equilibrium at the beginning. There are many more intermolecular forces between the ligand and the protein (Figure 4). Notably, the 5′-hydroxyl could also form a water-mediated hydrogen bond with Asp218 which would maintain for 10% of the simulation time; further, the 3′,4′-dihydroxyl formed a hydrogen bond mainly with Asp218 and Asp376, respectively. Therefore, the 3′,4′,5′-trihydroxyl group also played a very important role in the binding with the target.
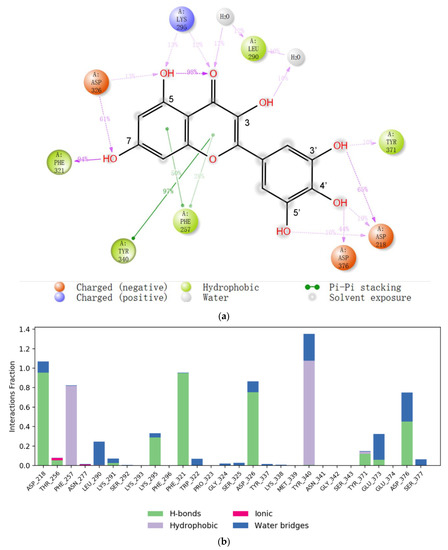
Figure 4.
(a) The 2D summary of interaction analysis results of ENPP1-myricetin complexes. The interaction pairs that occurred during more than 10% of the simulation time are included. (b) Interaction fraction summary of ENPP1–myricetin contacts. Graph is normalized for the total simulation time. Interaction-fraction values over 1.0 indicate that the residue has multiple contact routes for interacting with the ligand.
To further validate the binding model, several flavonoid compounds in Figure 5, including diosmetin, baicalein, apigenin, scutellarein, and luteolin, were purchased and determined for their enzyme inhibitory activities. The main differences between the purchased flavone compounds vary in the substitution group of the 2-phenyl group of the flavonoid scaffold. From the MD studies, the 3′,4′,5′-trihydroxyl phenyl group play an important role in binding with the target. It is interesting that from our preliminary biological investigation, luteolin was found to have a small amount of inhibitory activity (32% inhibition at 3.7 uM) while the other compounds, except myricetin, are completely inactive. Clearly, our proposed binding model can effectively explain the SARs of myricetin derivatives. Therefore, the binding model proposed by us was supported by both the theoretical computations and the SAR studies of myricetin derivatives.
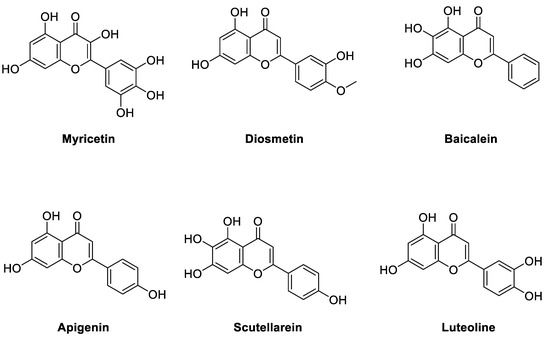
Figure 5.
Structures of the SAR studies of myricetin-like flavonoid compounds.
2.3. Virtual Screening of Novel ENPP1 Inhibitors
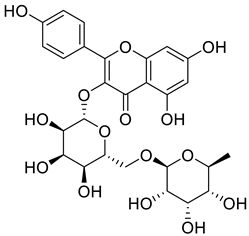
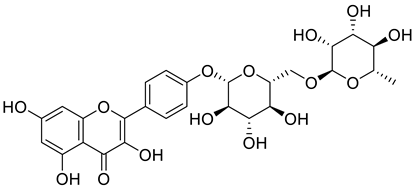
Starting from the essential scaffold of 3,5,7-trihydroxy-2-phenyl-4H-chromen-4-one of myricetin for the binding with ENPP1 (identified from computational studies), the substructure search implemented in Scifinder was then employed and 1886 flavonoid molecules were retrieved and exported to SDF file format. The 1886 molecules were further processed, and the resulting 5690 models including tautomeric, stereochemical, and ionization variations were then submitted to virtual screening studies. In the first HTVS run, 793 binding poses were obtained. In the second SP docking run, 720 poses were obtained. In the XP run, 151 binding poses were obtained. After MM/GBSA calculations, two flavonoid glycosides (Cas No: 1397173-50-0 and 1169835-58-8), Table 1) were identified as potential ENPP1 inhibitors. Based on better docking scores and MM/GBSA binding energies, their inhibitory effects were predicted to be superior to that of myricetin. These flavonoid aglycones share very similar binding orientations with myricetin, and the disaccharides formed additional hydrogen bonds with the target (Figure 6). Notably, hydrophobic interactions were also formed between the disaccharides and the protein. This strongly warrants future biological studies and might provide better medicines for ischemic cardiac injury.

Table 1.
Docking scores and MM/GBSA scores of two flavonoids predicted as bioactive ENPP1 inhibitors compared with myricetin.
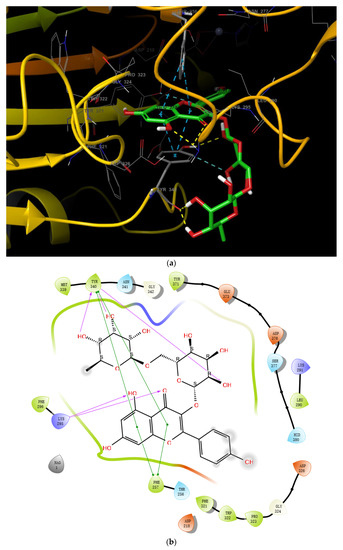
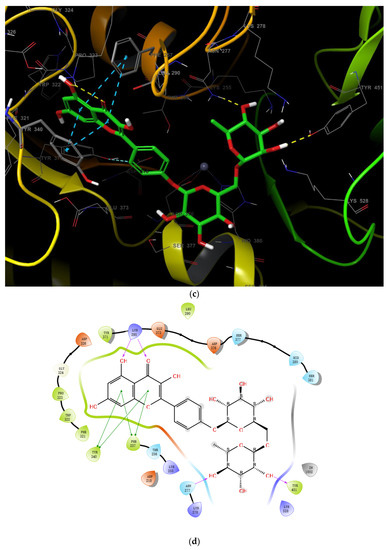
Figure 6.
The 3D and 2D binding models of the two flavonoid glycosides identified as potential ENPP1 inhibitors from virtual screening studies. (a,b) were for Compound 1 (Cas No: 1397173-50-0); (c,d) were for Compound 2 (Cas No: 1169835-58-8).
3. Discussion
Myricetin is a naturally occurring flavonoid antioxidant, which is found in vegetables, fruits, tea, and wine. This valuable dietary constituent has shown promising therapeutic potential, including anticancer, antidiabetic, osteoporosis protection, and anti-inflammatory effects [30,40]. In the case of myocardial dysfunction, myricetin can suppress the inflammatory cytokines and reduce platelet aggregation [32]. Recently it was reported to be an effective ENPP1 inhibitor. Additionally, administration of myricetin after heart injury helps to rescue pyrimidine biosynthesis in nonmyocyte cells, as well as augmented cardiac repair and postinfarct heart function [20].
Despite a wide spectrum of pharmacological effects and low toxicity, myricetin’s poor water solubility, bioavailability, and stability limit its further clinical use. Therefore, novel flavonoid compounds, which may show better therapeutic benefits via similar mechanisms, are greatly needed. In this study, two flavonoid glycosides were discovered from virtual screening studies. Generally, higher negative Glide docking score values indicate higher affinity between the receptor and the ligands. However, empirical scoring functions, such as Glide scores, cannot give a reliable prediction. MM/GBSA is much more accurate in binding free energy calculations and has been successfully applied in drug design [41,42,43,44]. The two flavonoid glycosides have shown better docking scores and MM/GBSA binding energies than those of myricetin, implying their ENPP1 inhibitory activities might outweigh that of myricetin. This suggests these two flavonoid glycosides might have considerable potential in the treatment of cardiac injury based on their probable better inhibitory activities on ENPP1.
Due to their complex chemical structures, lack of accessible commercial supply (or expensive prices), these two predicted ENPP1 inhibitors cannot be obtained at this time. Therefore, one limitation of the study is the lack of in vitro enzyme inhibition and in vivo data. Additional efforts, such as isolation from plants or chemical synthesis, are strongly recommended to obtain the two compounds and thus fully characterize their ENPP1 inhibitory activities and also to test their in vivo cardioprotective activities. Notably, the virtual screening methods used in this study will also be helpful to discover novel ENPP1 inhibitors of other chemical skeletons, which may further be used in cancer immunotherapy.
4. Materials and Methods
4.1. Molecular Docking and Structure-Based Virtual Screening
4.1.1. Receptor Preparation
The binding model studies of myricetin, and other molecules investigated here, were performed by molecular docking using the Glide software from Schrodinger (released 2021-2). The protein structures of ENPP1 (PDB ID: 6wew [45]) were downloaded from the RCSB website (http://www.rcsb.org, accessed on 20 June 2022). Routinely, protein structure preparation was achieved using the Protein Preparation Wizard, including adding hydrogen, filling in missing side chains, removing crystallographic water molecules, assigning partial charges, etc. It is noted that the phosphate anion coordinating with the essential zinc atoms of the active site, needs to be removed from the crystal structure since it is not necessary for the enzyme to elicit catalytic activity. The complex was minimized in two steps: firstly, only the hydrogen atoms were allowed to move, and secondly, all the heavy atoms were optimized until the root-mean-square deviation (RMSD) reached a maximum value of 0.3 Å.
4.1.2. Receptor Grid Generation
The Receptor Grid Generation module in Glide was applied to generate a receptor grid file for docking, in which the binding site was identified using the existing ligand in the crystal structure.
4.1.3. Preparation of Flavonoid Natural Products Database
Starting from the scaffold of 3,5,7-trihydroxy-2-phenyl-4H-chromen-4-one (which was identified to be essential for the binding with ENPP1 identified from docking studies), the substructure search function implemented in Scifinder was employed and 1886 flavonoid molecules with the scaffold were retrieved and exported to SDF file format. All the 2D molecules were prepared by LigPrep to produce corresponding low-energy 3D structures for further virtual screening studies. LigPrep is a powerful tool that can consider ionization states, tautomers, stereochemistries, and ring conformations. It can generate 32 stereoisomers at most for each input structure. The optimized flavonoid molecules were stored in Mae file format as our self-built Flavonoid Natural Product Database (FNPD). Such a name is used for convenience and does not mean it covers all the reported flavonoid compounds in the literature.
4.1.4. Molecular Docking Studies of Myricetin
The docking accuracy was evaluated with the existing ligand of the crystal structure and used as a positive control molecule for methodological investigation. The positive control molecule and myricetin were docked into the binding site of ENPP1 using the Glide XP mode. The ligands were treated as flexible; all the docked compounds were evaluated by the Glide scoring function, and the binding models with satisfactory docking scores were also analyzed using the MM/GBSA free energy calculations.
4.1.5. Identification of Novel Human ENPP1 Inhibitors by Structure-Based Virtual Screening of FNPD
The virtual screening workflow protocol in the Glide software was used to discover better myricetin-like compounds from our FNPD. The protocol starts from HTVS to SP then to XP, and it enriches the data at each level such that only a few compounds, satisfying some requirements, are submitted to the next highest accuracy level calculations.
4.2. Molecular Dynamics Simulation and Analysis
Molecular Dynamics (MD) simulations were performed using Desmond software. The System Builder in Maestro was used to set up the simulation systems. The ENPP1/ligand complex models were placed in the orthorhombic box at a buffer distance of 10 Å to create a hydration model. An SPC water model was used to create the hydration model [46]. The cut-off radii for van der Waals and electrostatic interactions, time step, and the initial temperature and pressure of the system were set to 9 Å, 2.0 fs, 300 K, and 1.01325 bar, respectively. Additionally, the sampling interval during the simulation was set to 100 ps. Finally, MD simulations were performed under the NPT ensemble for 40 ns using an OPLS4 force field [47]. Interaction analysis between the target and the ligands was performed with The Simulation Interactions Diagram.
4.3. In Vitro ENPP1 Enzyme Inhibition Assay
The assay was performed at 37 ℃ in a total volume of 50 μL in a clear bottom 384-well microplate. The reaction mixture contained 200 μM p-Nph-5′-TMP and the test compound (as per the concentration required) in buffer (consisting of 50 mM Tris, 250 mM NaCl, 500 μM CaCl2, 10 μM ZnCl2) with two pH conditions (Buffer A: pH9.5, Buffer B: pH7.4). The total volume of 50 μL contained 25 μL of the reaction mixture; 5 μL of tested compound (with final DMSO 1% (v/v)); and 20 μL of hENPP1 (with a final concentration of 1 nM). The reaction was then initiated by the addition of 25 μL of the p-Nph-5-TMP substrate at a final concentration of 0.2 mM, and the reaction mixture was incubated for 30–60 min at 37 °C. The amounts of released p-nitrophenolate were measured at 405 nm. The activity of each compound was compared with the reaction found in the absence of synthesized compounds. Percentage hENPP1 activity was determined, for different concentrations of the test inhibitors, by comparing the absorbance of inhibitors versus blank. The IC50 values were determined by plotting the percent inhibition versus inhibitor concentration curves using a four-parameter non-linear regression curve fit in GraphPad Prism® software.
5. Conclusions
Based on the latest report on myricetin’s ENPP1 inhibitory activity and promising in vivo activities in treating cardiac injury, this study was designed to discover novel human ENPP1 inhibitors from natural sources.
In the present study, in vitro ENPP1 inhibitory assay proved the absence of influence via pH conditions on myricetin’s inhibitory activity on ENPP1. Secondly, it was found that the binding model of myricetin with ENPP1 was elucidated through molecular docking, which was further validated by molecular dynamics simulations. In the binding model, the benzopyrone skeleton of myricetin plays an important role in the interaction with ENPP1 via π-π interaction with Phe257 and Tyr340. Furthermore, the 3′,4′,5′-trihydroxyl phenyl group also formed essential hydrogen bond interactions with the target. It is interesting that from our biological results, luteolin was found to have a small amount of inhibitory activity (32% inhibition at 3.7 uM) while the other myricetin analogs are completely inactive, which can be adequately explained by our proposed binding model of myricetin.
In addition, on the basis of the self-developed flavonoid natural products database (FNPD), virtual screening of novel human ENPP1 inhibitors was conducted. The most important and clinically relevant finding was that the two natural flavonoid glycosides were predicted to have strong ENPP1 inhibiting potential, suggesting their expected therapeutic benefits in cardiac injury treatment.
These findings have significant implications for the understanding of myricetin and similar flavonoid compounds. The proposed myricetin binding model with ENPP1 has extended our knowledge of myricetin’s model of action and will be helpful for structural optimization to find novel flavonoid compounds with better ENPP1 inhibition and druggability properties. In particular, the two flavonoid glycosides (Cas No: 1397173-50-0 and 1169835-58-8) identified from the virtual screening study could have even superior activities on ENPP1 and thus this warrants further biological evaluations. In conclusion, this work provides a strong basis for future development of new ENPP1 inhibitors for treating ischemic cardiac injury.
Author Contributions
S.S.: Investigation, data curation; formal analysis; validation, and writing—original draft; Z.S.: Conceptualization, methodology, project administration, resources, supervision, and writing—review and editing. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no funding.
Institutional Review Board Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Borza, R.; Salgado-Polo, F.; Moolenaar, W.H.; Perrakis, A. Structure and function of the ecto-nucleotide pyrophosphatase/phosphodiesterase (ENPP) family: Tidying up diversity. J. Biol. Chem. 2022, 298, 101526. [Google Scholar] [CrossRef] [PubMed]
- Perez-Valencia, J.A.; Prosdocimi, F.; Cesari, I.M.; da Costa, I.R.; Furtado, C.; Agostini, M.; Rumjanek, F.D. Angiogenesis and evading immune destruction are the main related transcriptomic characteristics to the invasive process of oral tongue cancer. Sci. Rep. 2018, 8, 2007. [Google Scholar] [CrossRef] [PubMed]
- Mackenzie, N.C.; Zhu, D.; Milne, E.M.; van’t Hof, R.; Martin, A.; Quarles, L.D.; Millan, J.L.; Farquharson, C.; MacRae, V.E. Altered bone development and an increase in FGF-23 expression in Enpp1(-/-) mice. PLoS ONE 2012, 7, e32177. [Google Scholar] [CrossRef]
- Korostishevsky, M.; Cohen, Z.; Malkin, I.; Ermakov, S.; Yarenchuk, O.; Livshits, G. Morphological and biochemical features of obesity are associated with mineralization genes’ polymorphisms. Int. J. Obes. 2010, 34, 1308–1318. [Google Scholar] [CrossRef]
- Wang, H.; Ye, F.; Zhou, C.; Cheng, Q.; Chen, H. High expression of ENPP1 in high-grade serous ovarian carcinoma predicts poor prognosis and as a molecular therapy target. PLoS ONE 2021, 16, e0245733. [Google Scholar] [CrossRef]
- Ledderose, C.; Hefti, M.M.; Chen, Y.; Bao, Y.; Seier, T.; Li, L.; Woehrle, T.; Zhang, J.; Junger, W.G. Adenosine arrests breast cancer cell motility by A3 receptor stimulation. Purinergic Signal. 2016, 12, 673–685. [Google Scholar] [CrossRef]
- Li, N.; Pan, H.; Cui, M.; Li, Q. Association of K121Q Polymorphism in Ectonucleotide Pyrophosphatase/Phosphodiesterase 1 with Clinical Characteristics of Metabolic Syndrom. Exp. Clin. Endocrinol. Diabetes 2016, 124, 313–317. [Google Scholar] [CrossRef] [PubMed]
- Kato, K.; Nishimasu, H.; Okudaira, S.; Mihara, E.; Ishitani, R.; Takagi, J.; Aoki, J.; Nureki, O. Crystal structure of Enpp1, an extracellular glycoprotein involved in bone mineralization and insulin signaling. Proc. Natl. Acad. Sci. USA 2012, 109, 16876–16881. [Google Scholar] [CrossRef] [PubMed]
- Di Paola, R.; Caporarello, N.; Marucci, A.; Dimatteo, C.; Iadicicco, C.; del Guerra, S.; Prudente, S.; Sudano, D.; Miele, C.; Parrino, C.; et al. ENPP1 affects insulin action and secretion: Evidences from in vitro studies. PLoS ONE 2011, 6, e19462. [Google Scholar] [CrossRef] [PubMed]
- Chin, C.N.; Dallas-Yang, Q.; Liu, F.; Ho, T.; Ellsworth, K.; Fischer, P.; Natasha, T.; Ireland, C.; Lu, P.; Li, C.; et al. Evidence that inhibition of insulin receptor signaling activity by PC-1/ENPP1 is dependent on its enzyme activity. Eur. J. Pharmacol. 2009, 606, 17–24. [Google Scholar] [CrossRef]
- Szeri, F.; Niaziorimi, F.; Donnelly, S.; Fariha, N.; Tertyshnaia, M.; Patel, D.; Lundkvist, S.; van de Wetering, K. The Mineralization Regulator ANKH Mediates Cellular Efflux of ATP, Not Pyrophosphate. J. Bone Miner. Res. 2022, 37, 1024–1031. [Google Scholar] [CrossRef] [PubMed]
- Huitema, L.F.; Apschner, A.; Logister, I.; Spoorendonk, K.M.; Bussmann, J.; Hammond, C.L.; Schulte-Merker, S. Entpd5 is essential for skeletal mineralization and regulates phosphate homeostasis in zebrafish. Proc. Natl. Acad. Sci. USA 2012, 109, 21372–21377. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Duran, M.A.; Dhanota, N.; Chatila, W.K.; Bettigole, S.E.; Kwon, J.; Sriram, R.K.; Humphries, M.P.; Salto-Tellez, M.; James, J.A.; et al. Metastasis and Immune Evasion from Extracellular cGAMP Hydrolysis. Cancer Discov. 2021, 11, 1212–1227. [Google Scholar] [CrossRef]
- Ritchie, C.; Carozza, J.A.; Li, L. Biochemistry, Cell Biology, and Pathophysiology of the Innate Immune cGAS-cGAMP-STING Pathway. Annu. Rev. Biochem. 2022, 91, 599–628. [Google Scholar] [CrossRef] [PubMed]
- Carozza, J.A.; Bohnert, V.; Nguyen, K.C.; Skariah, G.; Shaw, K.E.; Brown, J.A.; Rafat, M.; von Eyben, R.; Graves, E.E.; Glenn, J.S.; et al. Extracellular cGAMP is a cancer cell-produced immunotransmitter involved in radiation-induced anti-cancer immunity. Nat. Cancer 2020, 1, 184–196. [Google Scholar] [CrossRef] [PubMed]
- Gangar, M.; Goyal, S.; Raykar, D.; Khurana, P.; Martis, A.M.; Goswami, A.; Ghoshal, I.; Patel, K.V.; Nagare, Y.; Raikar, S.; et al. Design, synthesis and biological evaluation studies of novel small molecule ENPP1 inhibitors for cancer immunotherapy. Bioorg. Chem. 2022, 119, 105549. [Google Scholar] [CrossRef]
- Chang, L.; Lee, S.Y.; Leonczak, P.; Rozenski, J.; de Jonghe, S.; Hanck, T.; Muller, C.E.; Herdewijn, P. Imidazopyridine- and purine-thioacetamide derivatives: Potent inhibitors of nucleotide pyrophosphatase/phosphodiesterase 1 (NPP1). J. Med. Chem. 2014, 57, 10080–10100. [Google Scholar] [CrossRef]
- Patel, S.D.; Habeski, W.M.; Cheng, A.C.; de la Cruz, E.; Loh, C.; Kablaoui, N.M. Quinazolin-4-piperidin-4-methyl sulfamide PC-1 inhibitors: Alleviating hERG interactions through structure based design. Bioorg. Med. Chem. Lett. 2009, 19, 3339–3343. [Google Scholar] [CrossRef]
- Wang, X.; Lu, X.; Yan, D.; Zhou, Y.; Tan, X. Development of Novel Ecto-Nucleotide Pyrophosphatase/Phosphodiesterase 1 (ENPP1) Inhibitors for Tumor Immunotherapy. Int. J. Mol. Sci. 2022, 23, 7104. [Google Scholar] [CrossRef]
- Li, S.; Yokota, T.; Wang, P.; Hoeve, J.T.; Ma, F.; Le, T.M.; Abt, E.R.; Zhou, Y.; Wu, R.; Nanthavongdouangsy, M.; et al. Cardiomyocytes disrupt pyrimidine biosynthesis in nonmyocytes to regulate heart repair. J. Clin. Investig. 2022, 132, e156296. [Google Scholar] [CrossRef]
- Prem, P.N.; Sivakumar, B.; Boovarahan, S.R.; Kurian, G.A. Recent advances in potential of Fisetin in the management of myocardial ischemia-reperfusion injury—A systematic review. Phytomedicine 2022, 101, 154123. [Google Scholar] [CrossRef] [PubMed]
- Prem, P.N.; Kurian, G.A. Fisetin attenuates renal ischemia/reperfusion injury by improving mitochondrial quality, reducing apoptosis and oxidative stress. Naunyn-Schmiedeberg’s Arch. Pharmacol. 2022, 395, 547–561. [Google Scholar] [CrossRef]
- Wu, J.; Chen, H.; Qin, J.; Chen, N.; Lu, S.; Jin, J.; Li, Y. Baicalin Improves Cardiac Outcome and Survival by Suppressing Drp1-Mediated Mitochondrial Fission after Cardiac Arrest-Induced Myocardial Damage. Oxidative Med. Cell. Longev. 2021, 2021, 8865762. [Google Scholar] [CrossRef] [PubMed]
- Ashrafizadeh, M.; Samarghandian, S.; Hushmandi, K.; Zabolian, A.; Shahinozzaman, M.; Saleki, H.; Esmaeili, H.; Raei, M.; Entezari, M.; Zarrabi, A.; et al. Quercetin in Attenuation of Ischemic/Reperfusion Injury: A Review. Curr. Mol. Pharmacol. 2021, 14, 537–558. [Google Scholar] [CrossRef] [PubMed]
- Hamid, A.A.; Aminuddin, A.; Yunus, M.H.M.; Murthy, J.K.; Hui, C.K.; Ugusman, A. Antioxidative and anti-inflammatory activities of Polygonum minus: A review of literature. Rev. Cardiovasc. Med. 2020, 21, 275–287. [Google Scholar]
- Li, J.; Gu, Z.; Liu, Y.; Wang, Y.; Zhao, M. Astilbin attenuates cerebral ischemia/reperfusion injury by inhibiting the TLR4/MyD88/NF-kappaB pathway. Toxicol. Res. 2019, 8, 1002–1008. [Google Scholar] [CrossRef]
- Zhang, Y.; Wang, X.; Wang, X.; Xu, Z.; Liu, Z.; Ni, Q.; Chu, X.; Qiu, M.; Zhao, A.; Jia, W. Protective effect of flavonoids from Scutellaria baicalensis Georgi on cerebral ischemia injury. J. Ethnopharmacol. 2006, 108, 355–360. [Google Scholar] [CrossRef] [PubMed]
- Bak, I.; Lekli, I.; Juhasz, B.; Nagy, N.; Varga, E.; Varadi, J.; Gesztelyi, R.; Szabo, G.; Szendrei, L.; Bacskay, I.; et al. Cardioprotective mechanisms of Prunus cerasus (sour cherry) seed extract against ischemia-reperfusion-induced damage in isolated rat hearts. Am. J. Physiol. Heart Circ. Physiol. 2006, 291, H1329–H1336. [Google Scholar] [CrossRef]
- Zenebe, W.; Pechanova, O.; Bernatova, I. Protective effects of red wine polyphenolic compounds on the cardiovascular system. Exp. Clin. Cardiol. 2001, 6, 153–158. [Google Scholar]
- Song, X.; Tan, L.; Wang, M.; Ren, C.; Guo, C.; Yang, B.; Ren, Y.; Cao, Z.; Li, Y.; Pei, J. Myricetin: A review of the most recent research. Biomed. Pharmacother. 2021, 134, 111017. [Google Scholar] [CrossRef]
- Salimi, A.; Jamali, Z.; Shabani, M. Antioxidant Potential and Inhibition of Mitochondrial Permeability Transition Pore by Myricetin Reduces Aluminium Phosphide-Induced Cytotoxicity and Mitochondrial Impairments. Front. Pharmacol. 2021, 12, 719081. [Google Scholar] [CrossRef] [PubMed]
- Lin, X.; Lin, C.H.; Liu, R.; Li, C.; Jiao, S.; Yi, X.; Walker, M.J.; Xu, X.M.; Zhao, T.; Huang, P.C.; et al. Myricetin against myocardial injury in rat heat stroke model. Biomed. Pharmacother. 2020, 127, 110194. [Google Scholar] [CrossRef] [PubMed]
- Giri, A.K.; Ianevski, A. High-throughput screening for drug discovery targeting the cancer cell-microenvironment interactions in hematological cancers. Expert Opin. Drug Discov. 2022, 17, 181–190. [Google Scholar] [CrossRef] [PubMed]
- Gavriilidou, A.F.M.; Sokratous, K.; Yen, H.Y.; de Colibus, L. High-Throughput Native Mass Spectrometry Screening in Drug Discovery. Front. Mol. Biosci. 2022, 9, 837901. [Google Scholar] [CrossRef] [PubMed]
- Vasina, M.; Velecky, J.; Planas-Iglesias, J.; Marques, S.M.; Skarupova, J.; Damborsky, J.; Bednar, D.; Mazurenko, S.; Prokop, Z. Tools for computational design and high-throughput screening of therapeutic enzymes. Adv. Drug Deliv. Rev. 2022, 183, 114143. [Google Scholar] [CrossRef]
- Cetin, Y.; Aydinlik, S.; Gungor, A.; Kan, T.; Avsar, T.; Durdagi, S. Review on in silico Methods, High-throughput Screening Techniques, and Cell Culture Based In Vitro Assays for SARS-CoV-2. In Current Medicinal Chemistry; Bentham Science Publishers: Sharjah, United Arab Emirates, 2022. [Google Scholar]
- Lu, S.; He, X.; Yang, Z.; Chai, Z.; Zhou, S.; Wang, J.; Rehman, A.U.; Ni, D.; Pu, J.; Sun, J.; et al. Activation pathway of a G protein-coupled receptor uncovers conformational intermediates as targets for allosteric drug design. Nat. Commun. 2021, 12, 4721. [Google Scholar] [CrossRef]
- Li, L.; Yin, Q.; Kuss, P.; Maliga, Z.; Millan, J.L.; Wu, H.; Mitchison, T.J. Hydrolysis of 2’3’-cGAMP by ENPP1 and design of nonhydrolyzable analogs. Nat. Chem. Biol. 2014, 10, 1043–1048. [Google Scholar] [CrossRef]
- Yuki, H.; Tanaka, Y.; Hata, M.; Ishikawa, H.; Neya, S.; Hoshino, T. Implementation of pi-pi interactions in molecular dynamics simulation. J. Comput. Chem. 2007, 28, 1091–1099. [Google Scholar] [CrossRef]
- Gupta, G.; Siddiqui, M.A.; Khan, M.M.; Ajmal, M.; Ahsan, R.; Rahaman, M.A.; Ahmad, M.A.; Arshad, M.; Khushtar, M. Current Pharmacological Trends on Myricetin. Drug Res. 2020, 70, 448–454. [Google Scholar] [CrossRef]
- Lu, S.; Chen, Y.; Wei, J.; Zhao, M.; Ni, D.; He, X.; Zhang, J. Mechanism of allosteric activation of SIRT6 revealed by the action of rationally designed activators. Acta Pharm. Sin. B 2021, 11, 1355–1361. [Google Scholar] [CrossRef]
- Lu, S.; Ni, D.; Wang, C.; He, X.; Lin, H.; Wang, Z.; Zhang, J. Deactivation Pathway of Ras GTPase Underlies Conformational Substates as Targets for Drug Design. ACS Catal. 2019, 9, 7188–7196. [Google Scholar] [CrossRef]
- Ni, D.; Wei, J.; He, X.; Rehman, A.U.; Li, X.; Qiu, Y.; Pu, J.; Lu, S.; Zhang, J. Discovery of cryptic allosteric sites using reversed allosteric communication by a combined computational and experimental strategy. Chem. Sci. 2020, 12, 464–476. [Google Scholar] [CrossRef] [PubMed]
- Wang, E.; Sun, H.; Wang, J.; Wang, Z.; Liu, H.; Zhang, J.Z.H.; Hou, T. End-Point Binding Free Energy Calculation with MM/PBSA and MM/GBSA: Strategies and Applications in Drug Design. Chem. Rev. 2019, 119, 9478–9508. [Google Scholar] [CrossRef] [PubMed]
- Dennis, M.L.; Newman, J.; Dolezal, O.; Hattarki, M.; Surjadi, R.N.; Nuttall, S.D.; Pham, T.; Nebl, T.; Camerino, M.; Khoo, P.S.; et al. Crystal structures of human ENPP1 in apo and bound forms. Acta Crystallogr. Sect. D Struct. Biol. 2020, 76, 889–898. [Google Scholar] [CrossRef]
- Berendsen, H.J.C.; Postma, J.P.M.; van Gunsteren, W.F.; Hermans, J. Interaction Models for Water in Relation to Protein Hydration. In Intermolecular Forces, Proceedings of the Fourteenth Jerusalem Symposium on Quantum Chemistry and Biochemistry, Jerusalem, Israel, 13–16 April 1981; Pullman, B., Ed.; Springer: Dordrecht, The Netherlands, 1981; pp. 331–342. [Google Scholar]
- Lu, C.; Wu, C.; Ghoreishi, D.; Chen, W.; Wang, L.; Damm, W.; Ross, G.A.; Dahlgren, M.K.; Russell, E.; von Bargen, C.D.; et al. OPLS4: Improving Force Field Accuracy on Challenging Regimes of Chemical Space. J. Chem. Theory Comput. 2021, 17, 4291–4300. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).