NMR-Based Metabolomic Analysis of Cardiac Tissues Clarifies Molecular Mechanisms of CVB3-Induced Viral Myocarditis and Dilated Cardiomyopathy
Abstract
1. Introduction
2. Results
2.1. Viral Myocarditis and Dilated Cardiomyopathy Induced by CVB3 in Mice
2.2. Metabolic Alterations in CVB3-Infected Groups Compared with Normal Controls
2.3. Amino Acid Metabolism
2.4. Carbohydrate Metabolism
2.5. Lipid Metabolism
2.6. Choline Phosphorylation Metabolism
2.7. Others
2.8. Potential Biomarkers in the Progression from Acute VMC to DCM
2.9. Significantly Altered Metabolic Pathways in the Progression from Acute VMC to DCM
3. Discussion
3.1. Alanine, Aspartate and Glutamate Metabolism Is Disordered during the Progression from Acute VMC to DCM
3.2. Nicotinate and Nicotinamide Metabolism Is Impaired during the Progression from Acute VMC to DCM
3.3. D-Glutamine and D-Glutamate Metabolism Is Altered during the Progression from Acute VMC to DCM
3.4. Glycine, Serine and Threonine Metabolism Is Disturbed in the Acute VMC Stage
3.5. Taurine and Hypotaurine Metabolism Is Disordered in the aVMC Stage
4. Materials and Methods
4.1. Experimental Animal and Ethical Approval
4.2. Histological Analysis
4.3. Sample Preparation and NMR Analysis
4.4. Multivariate Statistical Analysis
4.5. Quantitative Comparison of Metabolite Levels and Identification of Characteristic Metabolites
4.6. Metabolic Pathway Analysis
4.7. Multivariate Receiver Operating Characteristic Curve Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Sample Availability
Abbreviations
| VMC | Viral myocarditis |
| DCM | Dilated cardiomyopathy |
| CVB3 | Coxsackievirus B3 |
| aVMC | Acute viral myocarditis |
| cVMC | Chronic viral myocarditis |
| CON | Normal controls |
| HF | Heart failure |
| PBS | Phosphate buffered saline |
| HCA | Hierarchical clustering analysis |
| PCA | Principal component analysis |
| RPTs | Response permutation tests |
| ROC | Receiver operating characteristic |
| PIV | Pathway impact values |
| LVEF | Left ventricle ejection fraction |
| BCAAs | Branch chain amino acids |
| PLS-DA | Partial least squares discriminant analysis |
| OPLS-DA | Orthogonal signal correction partial least-squares discriminant analysis |
References
- Van Linthout, S.; Tschope, C.; Schultheiss, H.P. Lack in treatment options for virus-induced inflammatory cardiomyopathy: Can iPS-derived cardiomyocytes close the gap? Circ. Res. 2014, 115, 540–541. [Google Scholar] [CrossRef]
- Feldman AM, M.D. myocarditis. N. Engl. J. Med. 2000, 343, 1388–1398. [Google Scholar] [CrossRef] [PubMed]
- Rinschen, M.M.; Ivanisevic, J.; Giera, M.; Siuzdak, G. Identification of bioactive metabolites using activity metabolomics. Nat. Rev. Mol. Cell Biol. 2019, 20, 353–367. [Google Scholar] [CrossRef] [PubMed]
- McGarrah, R.W.; Crown, S.B.; Zhang, G.F.; Shah, S.H.; Newgard, C.B. Cardiovascular Metabolomics. Circ. Res. 2018, 122, 1238–1258. [Google Scholar] [CrossRef]
- Cui, H.; Chen, Y.; Li, K.; Zhan, R.; Zhao, M.; Xu, Y.; Lin, Z.; Fu, Y.; He, Q.; Tang, P.C.; et al. Untargeted metabolomics identifies succinate as a biomarker and therapeutic target in aortic aneurysm and dissection. Eur. Heart J. 2021, 42, 4373–4385. [Google Scholar] [CrossRef]
- Kong, Q.; Gu, J.; Lu, R.; Huang, C.; Hu, X.; Wu, W.; Lin, D. NMR-Based Metabolomic Analysis of Sera in Mouse Models of CVB3-Induced Viral Myocarditis and Dilated Cardiomyopathy. Biomolecules 2022, 12, 112. [Google Scholar] [CrossRef] [PubMed]
- Crook, A.A.; Powers, R. Quantitative NMR-Based Biomedical Metabolomics: Current Status and Applications. Molecules 2020, 25, 5128. [Google Scholar] [CrossRef]
- Ampong, I. Metabolic and Metabolomics Insights into Dilated Cardiomyopathy. Ann. Nutr. Metab. 2022, 78, 147–155. [Google Scholar] [CrossRef]
- Haas, J.; Frese, K.S.; Sedaghat-Hamedani, F.; Kayvanpour, E.; Tappu, R.; Nietsch, R.; Tugrul, O.F.; Wisdom, M.; Dietrich, C.; Amr, A.; et al. Energy Metabolites as Biomarkers in Ischemic and Dilated Cardiomyopathy. Int. J. Mol. Sci. 2021, 22, 1999. [Google Scholar] [CrossRef]
- Zhao, J.; Yang, S.; Jing, R.; Jin, H.; Hu, Y.; Wang, J.; Gu, M.; Niu, H.; Zhang, S.; Chen, L.; et al. Plasma Metabolomic Profiles Differentiate Patients With Dilated Cardiomyopathy and Ischemic Cardiomyopathy. Front. Cardiovasc. Med. 2020, 7, 597546. [Google Scholar] [CrossRef]
- Johnson, C.H.; Ivanisevic, J.; Siuzdak, G. Metabolomics: Beyond biomarkers and towards mechanisms. Nat. Rev. Mol. Cell Biol. 2016, 17, 451–459. [Google Scholar] [CrossRef] [PubMed]
- Griffin JL, N.A. Metabolomics as a functional genomic tool for understanding lipid dysfunction in diabetes, obesity and related disorders. Pharmacogenomics 2006, 7, 1095–1107. [Google Scholar] [CrossRef]
- Song, H.; Wang, L.; Liu, H.L.; Wu, X.B.; Wang, H.S.; Liu, Z.H.; Li, Y.; Diao, D.C.; Chen, H.L.; Peng, J.S. Tissue metabolomic fingerprinting reveals metabolic disorders associated with human gastric cancer morbidity. Oncol. Rep. 2011, 26, 431–438. [Google Scholar] [CrossRef] [PubMed]
- Sreekumar, A.; Poisson, L.M.; Rajendiran, T.M.; Khan, A.P.; Cao, Q.; Yu, J.; Laxman, B.; Mehra, R.; Lonigro, R.J.; Li, Y.; et al. Metabolomic profiles delineate potential role for sarcosine in prostate cancer progression. Nature 2009, 457, 910–914. [Google Scholar] [CrossRef] [PubMed]
- Tam, P.E. Coxsackievirus myocarditis: Interplay between virus and host in the pathogenesis of heart disease. Viral Immunol. 2006, 19, 133–146. [Google Scholar] [CrossRef] [PubMed]
- Masui, R.; Sasaki, M.; Funaki, Y.; Ogasawara, N.; Mizuno, M.; Iida, A.; Izawa, S.; Kondo, Y.; Ito, Y.; Tamura, Y.; et al. G protein-coupled receptor 43 moderates gut inflammation through cytokine regulation from mononuclear cells. Inflamm. Bowel Dis. 2013, 19, 2848–2856. [Google Scholar] [CrossRef]
- Pirozzi, C.; Francisco, V.; Guida, F.D.; Gomez, R.; Lago, F.; Pino, J.; Meli, R.; Gualillo, O. Butyrate Modulates Inflammation in Chondrocytes via GPR43 Receptor. Cell. Physiol. Biochem. 2018, 51, 228–243. [Google Scholar] [CrossRef]
- Nakajima, A.; Nakatani, A.; Hasegawa, S.; Irie, J.; Ozawa, K.; Tsujimoto, G.; Suganami, T.; Itoh, H.; Kimura, I. The short chain fatty acid receptor GPR43 regulates inflammatory signals in adipose tissue M2-type macrophages. PLoS ONE 2017, 12, e0179696. [Google Scholar] [CrossRef]
- Mizuta, K.; Matoba, A.; Shibata, S.; Masaki, E.; Emala, C.W., Sr. Obesity-induced asthma: Role of free fatty acid receptors. Jpn. Dent. Sci. Rev. 2019, 55, 103–107. [Google Scholar] [CrossRef]
- Zambell, K.L.; Fitch, M.D.; Fleming, S.E. Acetate and butyrate are the major substrates for de novo lipogenesis in rat colonic epithelial cells. J. Nutr. 2003, 133, 3509–3515. [Google Scholar] [CrossRef]
- McCarty, M.F.; O’Keefe, J.H.; DiNicolantonio, J.J. Dietary Glycine Is Rate-Limiting for Glutathione Synthesis and May Have Broad Potential for Health Protection. Ochsner J. 2018, 18, 81–87. [Google Scholar] [PubMed]
- Ji, Y.; Fan, X.; Zhang, Y.; Li, J.; Dai, Z.; Wu, Z. Glycine regulates mucosal immunity and the intestinal microbial composition in weaned piglets. Amino Acids 2022, 54, 385–398. [Google Scholar] [CrossRef] [PubMed]
- Cruz, M.; Maldonado-Bernal, C.; Mondragon-Gonzalez, R.; Sanchez-Barrera, R.; Wacher, N.H.; Carvajal-Sandoval, G.; Kumate, J. Glycine treatment decreases proinflammatory cytokines and increases interferon-gamma in patients with type 2 diabetes. J. Endocrinol. Investig. 2008, 31, 694–699. [Google Scholar] [CrossRef]
- Sa, O.M.S.; Lopes, N.N.F.; Alves, M.T.S.; Caran, E.M.M. Effects of Glycine on Collagen, PDGF, and EGF Expression in Model of Oral Mucositis. Nutrients 2018, 10, 1485. [Google Scholar] [CrossRef]
- Sivakumar, R.; Babu, P.V.; Shyamaladevi, C.S. Aspartate and glutamate prevents isoproterenol-induced cardiac toxicity by alleviating oxidative stress in rats. Exp. Toxicol. Pathol. 2011, 63, 137–142. [Google Scholar] [CrossRef] [PubMed]
- Marrocco, A.; Frawley, K.; Pearce, L.L.; Peterson, J.; O’Brien, J.P.; Mullett, S.J.; Wendell, S.G.; St Croix, C.M.; Mischler, S.E.; Ortiz, L.A. Metabolic Adaptation of Macrophages as Mechanism of Defense against Crystalline Silica. J. Immunol. 2021, 207, 1627–1640. [Google Scholar] [CrossRef]
- Jia, X.; Gao, Y.; Liu, L.; Guo, Y.; Wang, J.; Ma, H.; Zhao, R.; Li, B.; Du, Y.; Yang, Q. Artemisinin Alleviates Intestinal Inflammation and Metabolic Disturbance in Ulcerative Colitis Rats Induced by DSS. Evid. Based Complement. Altern. Med. 2022, 2022, 6211215. [Google Scholar] [CrossRef]
- Diguet, N.; Trammell, S.A.; Tannous, C.; Deloux, R.; Piquereau, J.; Mougenot, N.; Gouge, A.; Gressette, M.; Manoury, B.; Blanc, J.; et al. Nicotinamide Riboside Preserves Cardiac Function in a Mouse Model of Dilated Cardiomyopathy. Circulation 2018, 137, 2256–2273. [Google Scholar] [CrossRef]
- Canto, C.; Menzies, K.J.; Auwerx, J. NAD+ Metabolism and the Control of Energy Homeostasis: A Balancing Act between Mitochondria and the Nucleus. Cell Metab. 2015, 22, 31–53. [Google Scholar] [CrossRef]
- Galli, U.; Travelli, C.; Massarotti, A.; Fakhfouri, G.; Rahimian, R.; Tron, G.C.; Genazzani, A.A. Medicinal chemistry of nicotinamide phosphoribosyltransferase (NAMPT) inhibitors. J. Med. Chem. 2013, 56, 6279–6296. [Google Scholar] [CrossRef]
- Hsu, C.P.; Yamamoto, T.; Oka, S.; Sadoshima, J. The function of nicotinamide phosphoribosyltransferase in the heart. DNA Repair 2014, 23, 64–68. [Google Scholar] [CrossRef] [PubMed]
- Murashige, D.; Jang, C.; Neinast, M.; Edwards, J.J.; Cowan, A.; Hyman, M.C.; Rabinowitz, J.D.; Frankel, D.S.; Arany, Z. Comprehensive quantification of fuel use by the failing and nonfailing human heart. Science 2020, 370, 364–368. [Google Scholar] [CrossRef] [PubMed]
- Neinast, M.; Murashige, D.; Arany, Z. Branched Chain Amino Acids. Annu. Rev. Physiol. 2019, 81, 139–164. [Google Scholar] [CrossRef] [PubMed]
- Sun, H.; Olson, K.C.; Gao, C.; Prosdocimo, D.A.; Zhou, M.; Wang, Z.; Jeyaraj, D.; Youn, J.Y.; Ren, S.; Liu, Y.; et al. Catabolic Defect of Branched-Chain Amino Acids Promotes Heart Failure. Circulation 2016, 133, 2038–2049. [Google Scholar] [CrossRef] [PubMed]
- Amin, A.M.; Sheau Chin, L.; Teh, C.H.; Mostafa, H.; Mohamed Noor, D.A.; Abdul Kader, M.; Kah Hay, Y.; Ibrahim, B. Pharmacometabolomics analysis of plasma to phenotype clopidogrel high on treatment platelets reactivity in coronary artery disease patients. Eur. J. Pharm. Sci. 2018, 117, 351–361. [Google Scholar] [CrossRef]
- Li, P.; Ge, J.; Li, H. Lysine acetyltransferases and lysine deacetylases as targets for cardiovascular disease. Nat. Rev. Cardiol. 2020, 17, 96–115. [Google Scholar] [CrossRef]
- McMaster, W.G.; Kirabo, A.; Madhur, M.S.; Harrison, D.G. Inflammation, immunity, and hypertensive end-organ damage. Circ. Res. 2015, 116, 1022–1033. [Google Scholar] [CrossRef]
- Gizak, A.; McCubrey, J.A.; Rakus, D. Cell-to-cell lactate shuttle operates in heart and is important in age-related heart failure. Aging 2020, 12, 3388–3406. [Google Scholar] [CrossRef]
- GA, B. Lactate shuttles in nature. Biochem. Soc. Trans. 2002, 30, 258–264. [Google Scholar]
- Shimada, Y.J.; Batra, J.; Kochav, S.M.; Patel, P.; Jung, J.; Maurer, M.S.; Hasegawa, K.; Reilly, M.P.; Fifer, M.A. Difference in Metabolomic Response to Exercise between Patients with and without Hypertrophic Cardiomyopathy. J. Cardiovasc. Transl. Res. 2021, 14, 246–255. [Google Scholar] [CrossRef]
- Lai, Q.; Yuan, G.Y.; Wang, H.; Liu, Z.L.; Kou, J.P.; Yu, B.Y.; Li, F. Exploring the protective effects of schizandrol A in acute myocardial ischemia mice by comprehensive metabolomics profiling integrated with molecular mechanism studies. Acta Pharmacol. Sin. 2020, 41, 1058–1072. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.K.; Zaidi, S.H.; Liu, P.; Dawood, F.; Cheah, A.Y.L.; Wen, W.H.; Saiki, Y.; Rabinovitch, M. A serine elastase inhibitor reduces inflammation and fibrosis and preserves cardiac function after experimentally-induced murine myocarditis. Nat. Med. 1998, 4, 1383–1391. [Google Scholar] [CrossRef] [PubMed]
- Xu, M.; Xue, R.Q.; Lu, Y.; Yong, S.Y.; Wu, Q.; Cui, Y.L.; Zuo, X.T.; Yu, X.J.; Zhao, M.; Zang, W.J. Choline ameliorates cardiac hypertrophy by regulating metabolic remodelling and UPRmt through SIRT3-AMPK pathway. Cardiovasc. Res. 2019, 115, 530–545. [Google Scholar] [CrossRef] [PubMed]
- Tuboly, E.; Gaspar, R.; Ibor, M.O.; Gomori, K.; Kiss, B.; Strifler, G.; Hartmann, P.; Ferdinandy, P.; Bartekova, M.; Boros, M.; et al. L-Alpha-glycerylphosphorylcholine can be cytoprotective or cytotoxic in neonatal rat cardiac myocytes: A double-edged sword phenomenon. Mol. Cell Biochem. 2019, 460, 195–203. [Google Scholar] [CrossRef] [PubMed]
- Fajardo, V.M.; Feng, I.; Chen, B.Y.; Perez-Ramirez, C.A.; Shi, B.; Clark, P.; Tian, R.; Lien, C.L.; Pellegrini, M.; Christofk, H.; et al. GLUT1 overexpression enhances glucose metabolism and promotes neonatal heart regeneration. Sci. Rep. 2021, 11, 8669. [Google Scholar] [CrossRef]
- Shi, M.; Ellingsen, O.; Bathen, T.F.; Hoydal, M.A.; Stolen, T.; Esmaeili, M. The Effect of Exercise Training on Myocardial and Skeletal Muscle Metabolism by MR Spectroscopy in Rats with Heart Failure. Metabolites 2019, 9, 53. [Google Scholar] [CrossRef]
- Soukoulis, V.; Dihu, J.B.; Sole, M.; Anker, S.D.; Cleland, J.; Fonarow, G.C.; Metra, M.; Pasini, E.; Strzelczyk, T.; Taegtmeyer, H.; et al. Micronutrient deficiencies an unmet need in heart failure. J. Am. Coll. Cardiol. 2009, 54, 1660–1673. [Google Scholar] [CrossRef]
- Doliba, N.M.; Babsky, A.M.; Doliba, N.M.; Wehrli, S.L.; Osbakken, M.D. AMP promotes oxygen consumption and ATP synthesis in heart mitochondria through the adenylate kinase reaction: An NMR spectroscopy and polarography study. Cell Biochem. Funct. 2015, 33, 67–72. [Google Scholar] [CrossRef]
- Girones, N.; Carbajosa, S.; Guerrero, N.A.; Poveda, C.; Chillon-Marinas, C.; Fresno, M. Global metabolomic profiling of acute myocarditis caused by Trypanosoma cruzi infection. PLoS Negl. Trop. Dis. 2014, 8, e3337. [Google Scholar] [CrossRef]
- Klawitter, J.; Klawitter, J.; Agardi, E.; Corby, K.; Leibfritz, D.; Lowes, B.D.; Christians, U.; Seres, T. Association of DJ-1/PTEN/AKT- and ASK1/p38-mediated cell signalling with ischaemic cardiomyopathy. Cardiovasc. Res. 2013, 97, 66–76. [Google Scholar] [CrossRef]
- Liu, C.; Li, R.; Liu, Y.; Li, Z.; Sun, Y.; Yin, P.; Huang, R. Characteristics of Blood Metabolic Profile in Coronary Heart Disease, Dilated Cardiomyopathy and Valvular Heart Disease Induced Heart Failure. Front. Cardiovasc. Med. 2020, 7, 622236. [Google Scholar] [CrossRef] [PubMed]
- Alexander, D.; Lombardi, R.; Rodriguez, G.; Mitchell, M.M.; Marian, A.J. Metabolomic distinction and insights into the pathogenesis of human primary dilated cardiomyopathy. Eur. J. Clin. Investig. 2011, 41, 527–538. [Google Scholar] [CrossRef] [PubMed]
- Fairweather, D.; Rose, N.R. Coxsackievirus-induced myocarditis in mice: A model of autoimmune disease for studying immunotoxicity. Methods 2007, 41, 118–122. [Google Scholar] [CrossRef]
- Chen, P.; Xie, Y.; Shen, E.; Li, G.G.; Yu, Y.; Zhang, C.B.; Yang, Y.; Zou, Y.; Ge, J.; Chen, R.; et al. Astragaloside IV attenuates myocardial fibrosis by inhibiting TGF-beta1 signaling in coxsackievirus B3-induced cardiomyopathy. Eur. J. Pharmacol. 2011, 658, 168–174. [Google Scholar] [CrossRef] [PubMed]
- Eriksson, U.; Ricci, R.; Hunziker, L.; Kurrer, M.O.; Oudit, G.Y.; Watts, T.H.; Sonderegger, I.; Bachmaier, K.; Kopf, M.; Penninger, J.M. Dendritic cell-induced autoimmune heart failure requires cooperation between adaptive and innate immunity. Nat. Med. 2003, 9, 1484–1490. [Google Scholar] [CrossRef]
- Querejeta, R.; Varo, N.; Lopez, B.; Larman, M.; Artinano, E.; Etayo, J.C.; Martinez Ubago, J.L.; Gutierrez-Stampa, M.; Emparanza, J.I.; Gil, M.J.; et al. Serum carboxy-terminal propeptide of procollagen type I is a marker of myocardial fibrosis in hypertensive heart disease. Circulation 2000, 101, 1729–1735. [Google Scholar] [CrossRef]
- Beckonert, O.; Keun, H.C.; Ebbels, T.M.; Bundy, J.; Holmes, E.; Lindon, J.C.; Nicholson, J.K. Metabolic profiling, metabolomic and metabonomic procedures for NMR spectroscopy of urine, plasma, serum and tissue extracts. Nat. Protoc. 2007, 2, 2692–2703. [Google Scholar] [CrossRef]
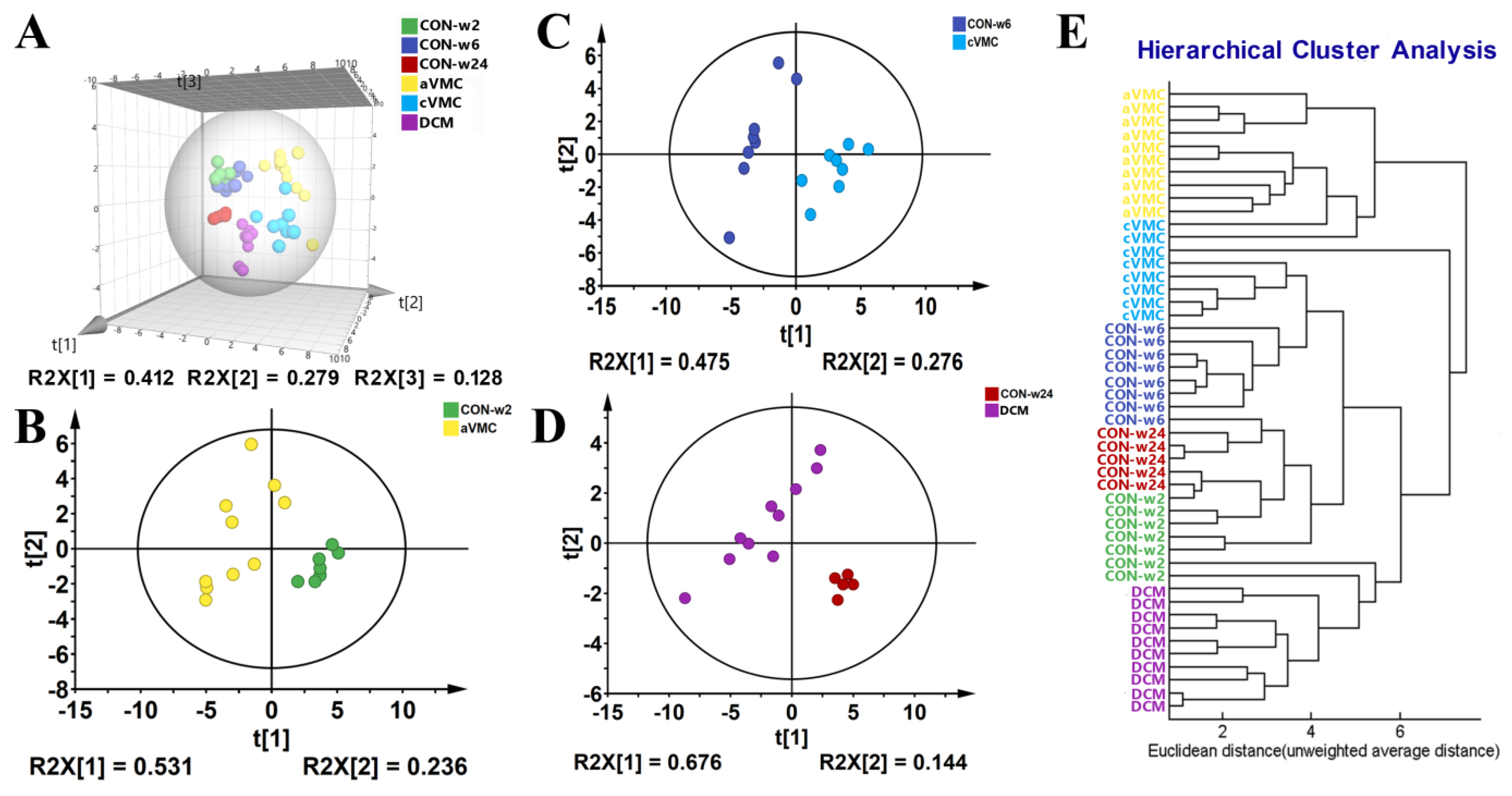

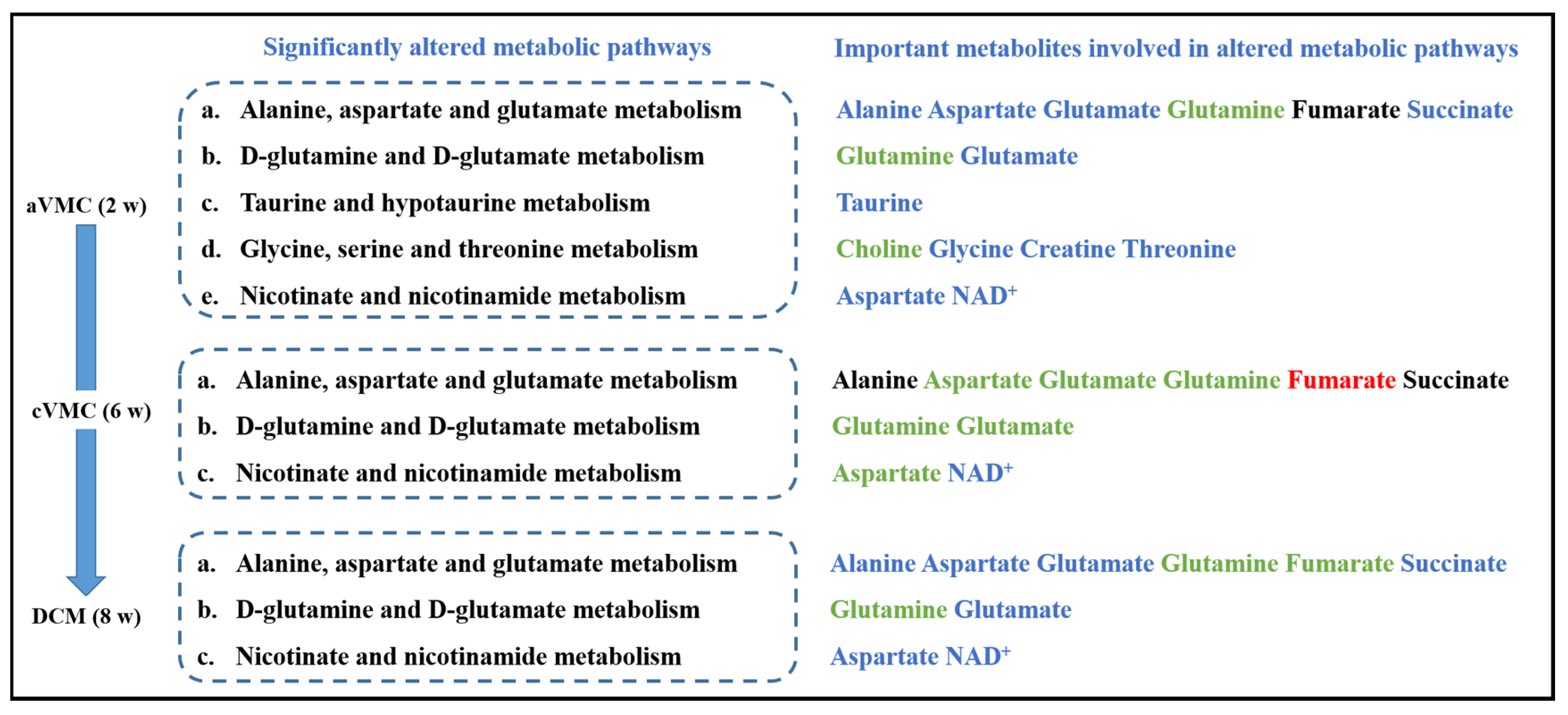

| CON-w2 | CON-w6 | CON-w24 | aVMC | cVMC | DCM | |
|---|---|---|---|---|---|---|
| Amino acid metabolism | ||||||
| leucine | 0.693 ± 0.036 | 0.651 ± 0.064 | 0.616 ± 0.006 | 0.666 ± 0.110 | 0.823 ± 0.077↑ ** | 0.552 ± 0.057 * |
| isoleucine | 0.132 ± 0.013 | 0.120 ± 0.021 | 0.124 ± 0.005 | 0.140 ± 0.024 | 0.195 ± 0.026↑ *** | 0.112 ± 0.015 |
| valine | 0.253 ± 0.019 | 0.238 ± 0.030 | 0.246 ± 0.007 | 0.226 ± 0.043 | 0.319 ± 0.032↑ ** | 0.218 ± 0.025 |
| threonine | 1.928 ± 0.090 | 1.831 ± 0.118 | 1.918 ± 0.023 | 1.468 ± 0.146↓ *** | 1.848 ± 0.056 | 1.667 ± 0.148 ** |
| glycine | 1.615 ± 0.086 | 1.567 ± 0.199 | 1.270 ± 0.042 | 1.378 ± 0.189 * | 1.284 ± 0.084↓ * | 0.813 ± 0.074↓ *** |
| lysine | 0.851 ± 0.040 | 0.793 ± 0.086 | 0.813 ± 0.039 | 0.842 ± 0.167 | 1.089 ± 0.110↑ *** | 0.747 ± 0.095 |
| alanine | 5.654 ± 0.122 | 5.242 ± 0.511 | 4.815 ± 0.260 | 3.911 ± 0.510↓ *** | 5.008 ± 0.358 | 3.892 ± 0.415↓ ** |
| taurine | 38.357 ± 1.702 | 37.347 ± 3.062 | 38.409 ± 0.900 | 30.821 ± 3.664↓ *** | 39.134 ± 2.041 | 35.727 ± 3.223 |
| glutamine | 7.763 ± 0.280 | 7.393 ± 0.614 | 7.944 ± 0.196 | 6.620 ± 1.081 | 8.164 ± 0.666 | 7.427 ± 0.680 |
| glutamate | 3.669 ± 0.193 | 3.288 ± 0.278 | 3.417 ± 0.127 | 2.768 ± 0.433↓ *** | 2.985 ± 0.121 | 2.933 ± 0.330 * |
| aspartate | 0.866 ± 0.106 | 0.848 ± 0.101 | 0.849 ± 0.052 | 0.547 ± 0.106↓ *** | 0.798 ± 0.092 | 0.625 ± 0.094↓ *** |
| Carbohydrate metabolism | ||||||
| creatine | 21.283 ± 0.634 | 21.633 ± 1.790 | 23.039 ± 0.763 | 16.821 ± 2.414↓ ** | 22.511 ± 1.253 | 21.865 ± 2.261 |
| acetate | 0.314 ± 0.016 | 0.288 ± 0.033 | 0.270 ± 0.007 | 0.374 ± 0.097 | 0.323 ± 0.025 | 0.273 ± 0.037 |
| glucose | 0.360 ± 0.094 | 0.417 ± 0.069 | 0.228 ± 0.012 | 0.381 ± 0.095 | 0.283 ± 0.076↓ * | 0.051 ± 0.040↓ *** |
| lactate | 47.617 ± 1.393 | 43.705 ± 4.052 | 41.368 ± 3.622 | 36.914 ± 4.610↓ *** | 44.133 ± 1.694 | 33.388 ± 3.968 * |
| succinate | 3.761 ± 0.244 | 3.252 ± 0.481 | 3.520 ± 0.139 | 2.015 ± 0.330↓ *** | 2.968 ± 0.316 | 2.969 ± 0.147 *** |
| dimethylamine | 0.241 ± 0.033 | 0.229 ± 0.022 | 0.236 ± 0.014 | 0.138 ± 0.042↓ *** | 0.125 ± 0.046↓ ** | 0.177 ± 0.024↓ *** |
| formate | 0.164 ± 0.020 | 0.136 ± 0.015 | 0.186 ± 0.010 | 0.054 ± 0.018↓ *** | 0.075 ± 0.023↓ *** | 0.151 ± 0.011↓ ** |
| fumarate | 0.072 ± 0.011 | 0.055 ± 0.008 | 0.071 ± 0.010 | 0.077 ± 0.013 | 0.097 ± 0.016↑ *** | 0.067 ± 0.012 |
| Lipid metabolism | ||||||
| 3-HB | 0.236 ± 0.051 | 0.142 ± 0.023 | 0.184 ± 0.027 | 0.182 ± 0.034 | 0.229 ± 0.047↑ ** | 0.164 ± 0.030 |
| Choline phosphorylation metabolism | ||||||
| GPC | 5.585 ± 0.282 | 5.803 ± 0.463 | 5.792 ± 0.174 | 5.030 ± 0.576 | 6.241 ± 0.506 | 4.983 ± 0.494 ** |
| Choline | 0.683 ± 0.036 | 0.765 ± 0.119 | 0.802 ± 0.026 | 0.892 ± 0.256 | 1.089 ± 0.185↑* | 1.015 ± 0.165↑ * |
| Others | ||||||
| myoinositol | 0.823 ± 0.057 | 0.798 ± 0.062 | 0.796 ± 0.014 | 0.789 ± 0.092 | 0.875 ± 0.058 | 0.722 ± 0.077 |
| NAD | 0.175 ± 0.021 | 0.152 ± 0.014 | 0.201 ± 0.009 | 0.078 ± 0.015↓ *** | 0.093 ± 0.024↓ *** | 0.166 ± 0.013 *** |
| AMP | 1.428 ± 0.114 | 1.058 ± 0.071 | 1.278 ± 0.067 | 0.683 ± 0.116↓ *** | 0.760 ± 0.142↓ ** | 0.933 ± 0.095↓ *** |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kong, Q.; Gu, J.; Lu, R.; Huang, C.; Chen, L.; Wu, W.; Lin, D. NMR-Based Metabolomic Analysis of Cardiac Tissues Clarifies Molecular Mechanisms of CVB3-Induced Viral Myocarditis and Dilated Cardiomyopathy. Molecules 2022, 27, 6115. https://doi.org/10.3390/molecules27186115
Kong Q, Gu J, Lu R, Huang C, Chen L, Wu W, Lin D. NMR-Based Metabolomic Analysis of Cardiac Tissues Clarifies Molecular Mechanisms of CVB3-Induced Viral Myocarditis and Dilated Cardiomyopathy. Molecules. 2022; 27(18):6115. https://doi.org/10.3390/molecules27186115
Chicago/Turabian StyleKong, Qing, Jinping Gu, Ruohan Lu, Caihua Huang, Lili Chen, Weifeng Wu, and Donghai Lin. 2022. "NMR-Based Metabolomic Analysis of Cardiac Tissues Clarifies Molecular Mechanisms of CVB3-Induced Viral Myocarditis and Dilated Cardiomyopathy" Molecules 27, no. 18: 6115. https://doi.org/10.3390/molecules27186115
APA StyleKong, Q., Gu, J., Lu, R., Huang, C., Chen, L., Wu, W., & Lin, D. (2022). NMR-Based Metabolomic Analysis of Cardiac Tissues Clarifies Molecular Mechanisms of CVB3-Induced Viral Myocarditis and Dilated Cardiomyopathy. Molecules, 27(18), 6115. https://doi.org/10.3390/molecules27186115






