Drimane-Type Sesquiterpenoids Derived from the Tropical Basidiomycetes Perenniporia centrali-africana and Cerrena sp. nov
Abstract
1. Introduction
2. Results and Discussion
2.1. Fungal Identification and Crude Extracts
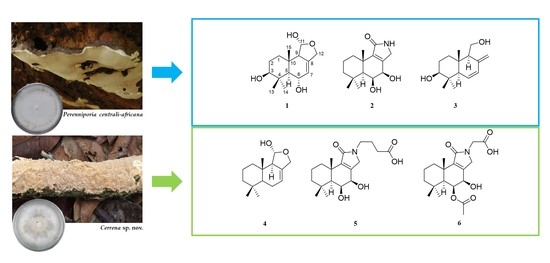
2.2. Isolation and Structure Elucidation of Metabolites from Perenniporia centrali-africana
2.3. Isolation and Structure Elucidation of Compounds 4–6 from Cerrena sp.
2.4. Biological Assays
3. Materials and Methods
3.1. General Information
3.2. Fungal Material
3.3. Seed Culture and Scale-Up of Fermentation
3.4. Harvest, Extraction, and Analytical HPLC
3.5. Isolation of the Compounds
3.6. Cytotoxicity Assay
3.7. Antimicrobial Assay
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Gressler, M.; Löhr, N.A.; Schäfer, T.; Lawrinowitz, S.; Seibold, P.S.; Hoffmeister, D. Mind the mushroom: Natural product biosynthetic genes and enzymes of Basidiomycota. Nat. Prod. Rep. 2021, 38, 702–722. [Google Scholar] [CrossRef]
- Sandargo, B.; Chepkirui, C.; Cheng, T.; Chaverra-Muñoz, L.; Thongbai, B.; Stadler, M.; Hüttel, S. Biological and chemical diversity go hand in hand: Basidomycota as source of new pharmaceuticals and agrochemicals. Biotechnol. Adv. 2019, 37, 107344. [Google Scholar] [CrossRef] [PubMed]
- Sandargo, B.; Kaysan, L.; Teponno, R.B.; Richter, C.; Thongbai, B.; Surup, F.; Stadler, M. Analogs of the carotane antibiotic fulvoferruginin from submerged cultures of a Thai Marasmius sp. Beilstein J. Org. Chem. 2021, 17, 1385–1391. [Google Scholar] [CrossRef] [PubMed]
- Richter, C.; Helaly, S.E.; Thongbai, B.; Hyde, K.D.; Stadler, M. Pyristriatins A and B: Pyridino-Cyathane antibiotics from the Basidiomycete Cyathus cf. striatus. J. Nat. Prod. 2016, 79, 1684–1688. [Google Scholar] [CrossRef]
- Cheng, T.; Chepkirui, C.; Decock, C.; Matasyoh, J.C.; Stadler, M. Skeletocutins M–Q: Biologically active compounds from the fruiting bodies of the basidiomycete Skeletocutis sp. Beilstein J. Org. Chem. 2019, 15, 2782–2789. [Google Scholar] [CrossRef]
- Cheng, T.; Chepkirui, C.; Decock, C.; Matasyoh, J.C.; Stadler, M. Sesquiterpenes from an Eastern African medicinal mushroom belonging to the genus Sanghuangporus. J. Nat. Prod. 2019, 82, 1283–1291. [Google Scholar] [CrossRef]
- Zhao, C.L.; Cui, B.K.; Dai, Y.C. New species and phylogeny of Perenniporia based on morphological and molecular characters. Fungal Divers. 2013, 58, 47–60. [Google Scholar] [CrossRef]
- Lee, J.S.; Lim, Y.W. Cerrena aurantiopora sp. nov. (Polyporaceae) from eastern Asia. Mycologia 2010, 102, 211–216. [Google Scholar] [CrossRef]
- Winquist, E.; Moilanen, U.; Mettälä, A.; Leisola, M.; Hatakka, A. Production of lignin modifying enzymes on industrial waste material by solid-state cultivation of fungi. Biochem. Eng. J. 2008, 42, 128–132. [Google Scholar] [CrossRef]
- Belova, O.V.; Lisov, A.V.; Vinokurova, N.G.; Kostenevich, A.A.; Sapunova, L.I.; Lobanok, A.G.; Leontievsky, A.A. Xylanase and cellulase of fungus Cerrena unicolor VKM F-3196: Production, properties, and applications for the saccharification of plant material. Appl. Biochem. Microbiol. 2014, 50, 148–153. [Google Scholar] [CrossRef]
- Sulej, J.; Janusz, G.; Osińska-Jaroszuk, M.; Rachubik, P.; Mazur, A.; Komaniecka, I.; Choma, A.; Rogalski, J. Characterization of cellobiose dehydrogenase from a biotechnologically important Cerrena unicolor strain. Appl. Biochem. Biotechnol. 2015, 176, 1638–1658. [Google Scholar] [CrossRef] [PubMed]
- Kachlishvili, E.; Jokharidze, T.; Kobakhidze, A.; Elisashvili, V. Enhancement of laccase production by Cerrena unicolor through fungal interspecies interaction and optimum conditions determination. Arch. Microbiol. 2021, 203, 3905–3917. [Google Scholar] [CrossRef] [PubMed]
- Kwon, J.; Lee, H.; Seo, Y.H.; Yun, J.; Lee, J.; Kwon, H.C.; Guo, Y.; Kang, J.S.; Kim, J.J.; Lee, D. Cytotoxic drimane sesquiterpenoids isolated from Perenniporia maackiae. J. Nat. Prod. 2018, 81, 1444–1450. [Google Scholar] [CrossRef]
- He, J.B.; Tao, J.; Miao, X.S.; Bu, W.; Zhang, S.; Dong, Z.J.; Li, Z.H.; Feng, T.; Liu, J.K. Seven new drimane-type sesquiterpenoids from cultures of fungus Laetiporus sulphureus. Fitoterapia 2015, 102, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Clarkson, C.; Madikane, E.V.; Hansen, S.H.; Smith, P.J.; Jaroszewski, J.W. HPLC-SPE-NMR characterization of sesquiterpenes in an antimycobacterial fraction from Warburgia salutaris. Planta Med. 2007, 73, 578–584. [Google Scholar] [CrossRef]
- Madikane, V.E.; Bhakta, S.; Russell, A.J.; Campbell, W.E.; Claridge, T.D.W.; Elisha, B.G.; Davies, S.G.; Smith, P.; Sim, E. Inhibition of mycobacterial arylamine N-acetyltransferase contributes to anti-mycobacterial activity of Warburgia salutaris. Bioorg. Med. Chem. 2007, 15, 3579–3586. [Google Scholar] [CrossRef]
- Kida, T.; Shibai, H.; Seto, H. Structure of new antibiotics, pereniporins A and B, from a Basidiomycete. J. Antibiot. 1986, 39, 5–24. [Google Scholar] [CrossRef][Green Version]
- Toshihiro, H.; Motoo, T.; Yasuo, M.; Yoshinori, A. Cryptoporic acids A and B, novel bitter drimane sesquiterpenoid ethers of isocitric acid, from the fungus Crptoporous volvatus. Tetrahedron Lett. 1987, 28, 6303–6304. [Google Scholar]
- Hirotani, M.; Furuya, T.; Shiro, M. Cryptoporic acids H and I, drimane sesquiterpenes from Ganoderma neo-japonicum and Cryptoporus volvatus. Phytochemistry 1991, 30, 1555–1559. [Google Scholar] [CrossRef]
- Kioy, D.; Gray, A.I.; Waterman, P.G. A comparative study of the stem-bark drimane sesquiterpenes and leaf volatile oils of Warburgia ugandensis and W. Stuhlmannii. Phytochemistry 1990, 29, 3535–3538. [Google Scholar] [CrossRef]
- Cabrera, G.M.; Julia Roberti, M.; Wright, J.E.; Seldes, A.M. Cryptoporic and isocryptoporic acids from the fungal cultures of Polyporus arcularius and P. ciliatus. Phytochemistry 2002, 61, 189–193. [Google Scholar] [CrossRef]
- Rodríguez, B.; Zapata, N.; Medina, P.; Viñuela, E. A complete 1H and 13C NMR data assignment for four drimane sesquiterpenoids isolated from Drimys winterii. Magn. Reson. Chem. 2005, 43, 82–84. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Tan, H.; Chen, K.; Chen, Y.; Li, S.; Li, H.; Zhang, W. Cerrenins A-C, cerapicane and isohirsutane sesquiterpenoids from the endophytic fungus Cerrena sp. Fitoterapia 2018, 129, 173–178. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.X.; Tan, H.B.; Chen, Y.C.; Li, S.N.; Li, H.H.; Zhang, W.M. Cytotoxic triquinane-type sesquiterpenoids from the endophytic fungus Cerrena sp. A593. Nat. Prod. Res. 2020, 34, 2430–2436. [Google Scholar] [CrossRef] [PubMed]
- Khushi, S.; Salim, A.A.; Elbanna, A.H.; Nahar, L.; Bernhardt, P.V.; Capon, R.J. Dysidealactams and dysidealactones: Sesquiterpene glycinyl-lactams, imides, and lactones from a Dysidea sp. marine sponge collected in southern Australia. J. Nat. Prod. 2020, 83, 1577–1584. [Google Scholar] [CrossRef]
- Lin, Y.C.; Abd El-Razek, M.H.; Shen, Y.C. Verticillane-type diterpenoids and an eudesmanolide-type sesquiterpene from the formosan soft coral Cespitularia hypotentaculata. Helv. Chim. Acta 2010, 93, 281–289. [Google Scholar] [CrossRef]
- Lou, Y.; Zhao, F.; Wu, Z.; Peng, K.F.; Wei, X.C.; Chen, L.X.; Qiu, F. Germacrane-type sesquiterpenes from Curcuma wenyujin. Helv. Chim. Acta 2009, 92, 1665–1672. [Google Scholar] [CrossRef]
- Nomoto, Y.; Harinantenaina, L.; Sugimoto, S.; Matsunami, K.; Otsuka, H. 3,4-seco-24-homo-28-nor-Cycloartane and drimane-type sesquiterpenes and their lactams from the EtOAc-soluble fraction of a leaf extract of Cinnamosma fragrans and their biological activity. J. Nat. Med. 2014, 68, 513–520. [Google Scholar] [CrossRef]
- Justo, A.; Miettinen, O.; Floudas, D.; Ortiz-Santana, B.; Sjökvist, E.; Lindner, D.; Nakasone, K.; Niemelä, T.; Larsson, K.H.; Ryvarden, L.; et al. A revised family-level classification of the Polyporales (Basidiomycota). Fungal Biol. 2017, 121, 798–824. [Google Scholar] [CrossRef]
- Huang, Y.; Hoefgen, S.; Valiante, V. Biosynthesis of fungal drimane-type sesquiterpene esters. Angew. Chem. Int. Ed. 2021, 60, 23763–23770. [Google Scholar] [CrossRef]
- Matio Kemkuignou, B.; Treiber, L.; Zeng, H.; Schrey, H.; Schobert, R.; Stadler, M. Macrooxazoles a–d, new 2,5-disubstituted oxazole-4-carboxylic acid derivatives from the plant pathogenic fungus Phoma macrostoma. Molecules 2020, 25, 5497. [Google Scholar] [CrossRef] [PubMed]
- Wendt, L.; Sir, E.B.; Kuhnert, E.; Heitkämper, S.; Lambert, C.; Hladki, A.I.; Romero, A.I.; Luangsa-ard, J.J.; Srikitikulchai, P.; Peršoh, D.; et al. Resurrection and emendation of the Hypoxylaceae, recognised from a multigene phylogeny of the Xylariales. Mycol. Prog. 2018, 17, 115–154. [Google Scholar] [CrossRef]
- Otto, A.; Laub, A.; Wendt, L.; Porzel, A.; Schmidt, J.; Palfner, G.; Becerra, J.; Krüger, D.; Stadler, M.; Wessjohann, L.; et al. Chilenopeptins A and B, peptaibols from the Chilean Sepedonium aff. chalcipori KSH 883. J. Nat. Prod. 2016, 79, 929–938. [Google Scholar] [CrossRef] [PubMed]
- Hassan, K.; Matio Kemkuignou, B.; Stadler, M. Two new triterpenes from basidiomata of the medicinal and edible mushroom, Laetiporus sulphureus. Molecules 2021, 26, 7090. [Google Scholar] [CrossRef]

| Pos | 1 | 2 | 3 | |||
|---|---|---|---|---|---|---|
| δC, Type | δH (J in HZ) | δC, Type | δH (J in HZ) | δC, Type | δH (J in HZ) | |
| 1 | 37.2, CH2 | α: 1.44, m | 37.7, CH2 | α: 1.06, m | 37.1, CH2 | α: 1.40, m |
| β: 1.70, m | β: 2.67, m | β: 1.85, dt (13.0, 3.4) | ||||
| 2 | 26.5, CH2 | 1.67, m | 19.6, CH2 | α: 1.46, m | 28.3, CH2 | 1.70, m |
| β: 1.80, m | ||||||
| 3 | 78.2, CH | α: 3.22, dd (11.4, 4.5) | 44.3, CH2 | α: 1.44, m | 79.4, CH | α: 3.26, m |
| β: 1.25, m | ||||||
| 4 | 39.0, C | - | 34.2, C | - | 39.8, C | - |
| 5 | 57.0, CH | α: 1.31, d (8.9) | 50.8, CH | α: 1.48, d (1.3) | 56.2, CH | α: 1.98, br s |
| 6 | 67.4, CH | β: 4.34, br d (8.9) | 72.7, CH | α: 4.35, br s | 133.3, CH | 6.20, dd (10.1, 3.1) |
| 7 | 120.3, CH | 5.53, m | 70.9, CH | α: 4.10, br s | 129.3, CH | 5.74, br d (10.1) |
| 8 | 139.1, C | - | 149.1, C | - | 146.6, C | - |
| 9 | 60.6, CH | α: 2.25, m | 143.0, C | - | 56.9, CH | α: 2.09, br d (4.7) |
| 10 | 38.1, C | - | 36.7, C | - | 38.8, C | - |
| 11 | 98.7, CH | β: 5.17, d (4.5) | 173.4, C | - | 60.6, CH2 | α: 3.72, dd (10.8, 7.3) |
| β: 3.924, dd (10.8, 7.3) | ||||||
| 12 | 67.7, CH2 | α: 4.46, m | 46.7, CH2 | α: 3.73, dd (18.7, 2.6) | 112.6, CH2 | 4.96, s |
| β: 4.18, m | β: 4.00, m | 5.15, s | ||||
| 13 | 15.1, CH3 | 0.99, s | 24.1, CH3 | 1.26, s | 16.6, CH3 | 0.81, s |
| 14 | 29.5, CH3 | 1.23, s | 34.1, CH3 | 1.00, s | 28.5, CH3 | 1.06, s |
| 15 | 14.3, CH3 | 0.85, s | 21.3, CH3 | 1.52, s | 15.2, CH3 | 0.74, s |
| Pos | 4 | 5 | 6 | |||
|---|---|---|---|---|---|---|
| δC, Type | δH (J in HZ) | δC, Type | δH (J in HZ) | δC, Type | δH (J in HZ) | |
| 1 | 40.0, CH2 | α: 1.24, m | 38.2, CH2 | α: 2.56, br d (12.7) | 37.7, CH2 | α: 2.68, br d (13.2) |
| β: 1.80, dq (13.7, 2.7) | β: 1.13, m | β: 1.12, m | ||||
| 2 | 18.7, CH2 | α: 1.62, qt (13.7, 3.5) | 19.9, CH2 | α: 1.85, m | 19.4, CH2 | α: 1.81, m |
| β: 1.45, m | β: 1.52, m | β: 1.54, m | ||||
| 3 | 42.7, CH2 | α: 1.25, m | 44.6, CH2 | α: 1.28, m | 44.2, CH2 | α: 1.29, m |
| β: 1.45, m | β: 1.46, m | β: 1.49, m | ||||
| 4 | 33.0, C | - | 34.6, C | - | 34.1, C | - |
| 5 | 50.4, CH | 1.31, dd (11.6, 5.4) | 51.2, CH | 1.45, m | 49.7, CH | 1.73, d (1.0) |
| 6 | 23.9, CH2 | α: 2.14, m | 72.5, CH | 4.28, br s | 73.8, CH | 5.46, m |
| β: 1.92, m | ||||||
| 7 | 116.2, CH | 5.47, br s | 70.7, CH | 4.05, d (1.5) | 67.1, CH | 4.12, br d (1.7) |
| 8 | 138.2, C | - | 147.6, C | - | 147.0, C | - |
| 9 | 62.0, CH | 2.15, m | 144.2, C | - | 142.5, C | - |
| 10 | 33.7, C | - | 37.3, C | - | 36.8, C | - |
| 11 | 99.2, CH | 5.19, dd (4.5, 4.0) 11-OH: 5.11, br d (4.5) | 172.5, C | - | 170.4, C | - |
| 12 | 68.1, CH2 | α: 4.33, br d (11.3) | 52.3, CH2 | α: 4.09, d (19.4) | 51.6, CH2 | α: 4.18, d (18.6) |
| β: 4.04, m | β: 3.84, d (19.4) | β: 3.92, d (18.6) | ||||
| 13 | 21.3, CH3 | 0.93, s | 24.1, CH3 | 1.24, s | 23.5, CH3 | 1.04, s |
| 14 | 33.0, CH3 | 0.88, s | 34.2, CH3 | 1.03, s | 33.7, CH3 | 1.01, s |
| 15 | 13.8, CH3 | 0.81, s | 21.3, CH3 | 1.49, s | 21.3, CH3 | 1.50, s |
| 1′ | - | - | 176.8, C | - | 171.2, C | - |
| 2′ | - | - | 32.3, CH2 | 2.31, t (7.4) | 43.4, CH2 | 4.21, m |
| 3′ | - | - | 25.2, CH2 | 1.88, quin (7.4) | - | - |
| 4′ | - | - | 42.6, CH2 | 3.47, t (7.0) | - | - |
| 1″ | - | - | - | - | 170.6, C | - |
| 2″ | - | - | - | - | 21.5, CH3 | 2.01, s |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pathompong, P.; Pfütze, S.; Surup, F.; Boonpratuang, T.; Choeyklin, R.; Matasyoh, J.C.; Decock, C.; Stadler, M.; Boonchird, C. Drimane-Type Sesquiterpenoids Derived from the Tropical Basidiomycetes Perenniporia centrali-africana and Cerrena sp. nov. Molecules 2022, 27, 5968. https://doi.org/10.3390/molecules27185968
Pathompong P, Pfütze S, Surup F, Boonpratuang T, Choeyklin R, Matasyoh JC, Decock C, Stadler M, Boonchird C. Drimane-Type Sesquiterpenoids Derived from the Tropical Basidiomycetes Perenniporia centrali-africana and Cerrena sp. nov. Molecules. 2022; 27(18):5968. https://doi.org/10.3390/molecules27185968
Chicago/Turabian StylePathompong, Paomephan, Sebastian Pfütze, Frank Surup, Thitiya Boonpratuang, Rattaket Choeyklin, Josphat C. Matasyoh, Cony Decock, Marc Stadler, and Chuenchit Boonchird. 2022. "Drimane-Type Sesquiterpenoids Derived from the Tropical Basidiomycetes Perenniporia centrali-africana and Cerrena sp. nov" Molecules 27, no. 18: 5968. https://doi.org/10.3390/molecules27185968
APA StylePathompong, P., Pfütze, S., Surup, F., Boonpratuang, T., Choeyklin, R., Matasyoh, J. C., Decock, C., Stadler, M., & Boonchird, C. (2022). Drimane-Type Sesquiterpenoids Derived from the Tropical Basidiomycetes Perenniporia centrali-africana and Cerrena sp. nov. Molecules, 27(18), 5968. https://doi.org/10.3390/molecules27185968