CRISPR/Cas12a Coupling with Magnetic Nanoparticles and Cascaded Strand Displacement Reaction for Ultrasensitive Fluorescence Determination of Exosomal miR-21
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Extraction and Purification of Exosomes
2.3. Characterization of Exosomes
2.3.1. Transmission Electron Microscopy
2.3.2. Nanoparticle Tracking Analysis
2.4. Extraction and Purification of Total RNAs
2.5. Determination of Exosomal miR-21
2.6. Fluorescence Measurements
3. Results
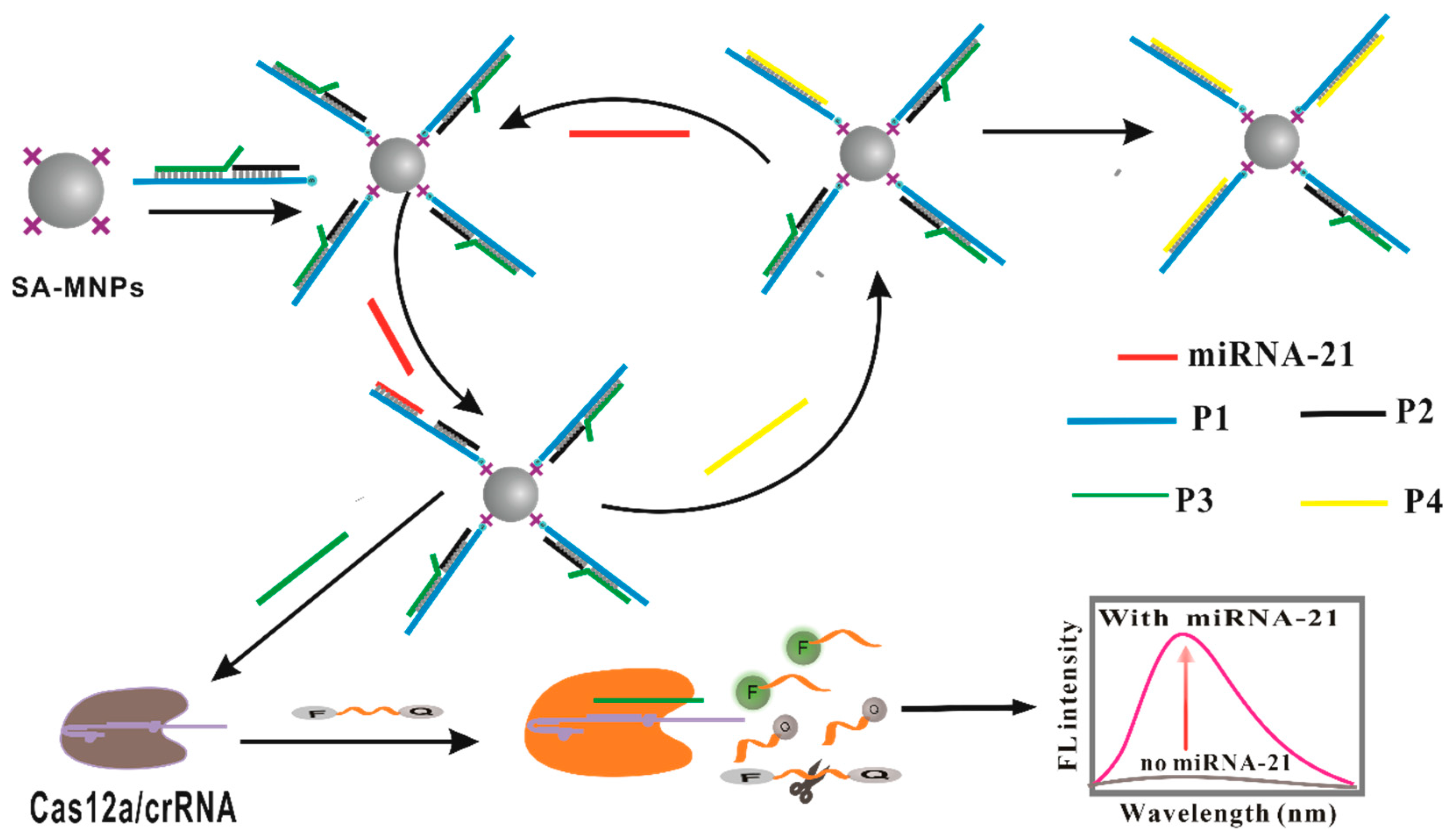
3.1. The Design and Mechanism
3.2. Characterization of Exosomes
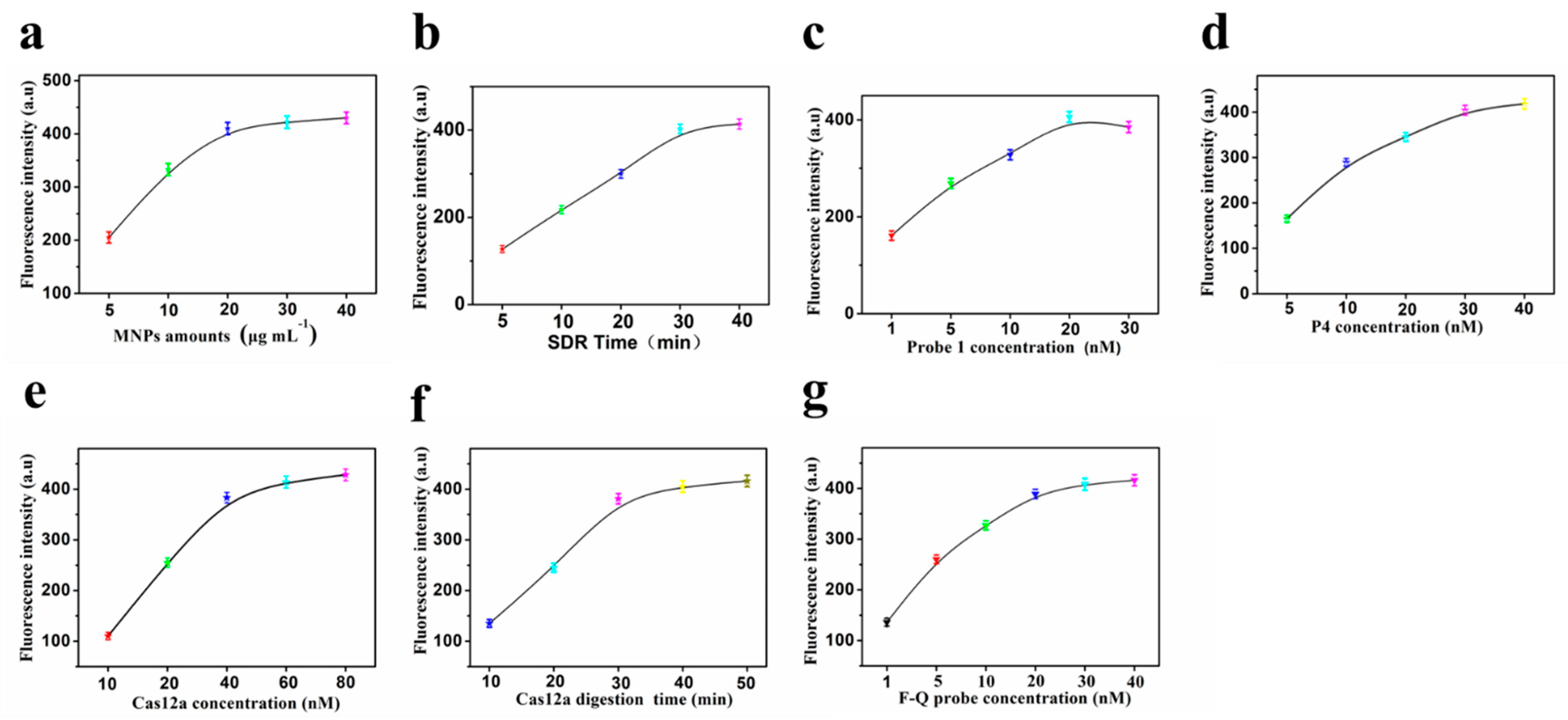
3.3. Optimization of the Assay Conditions
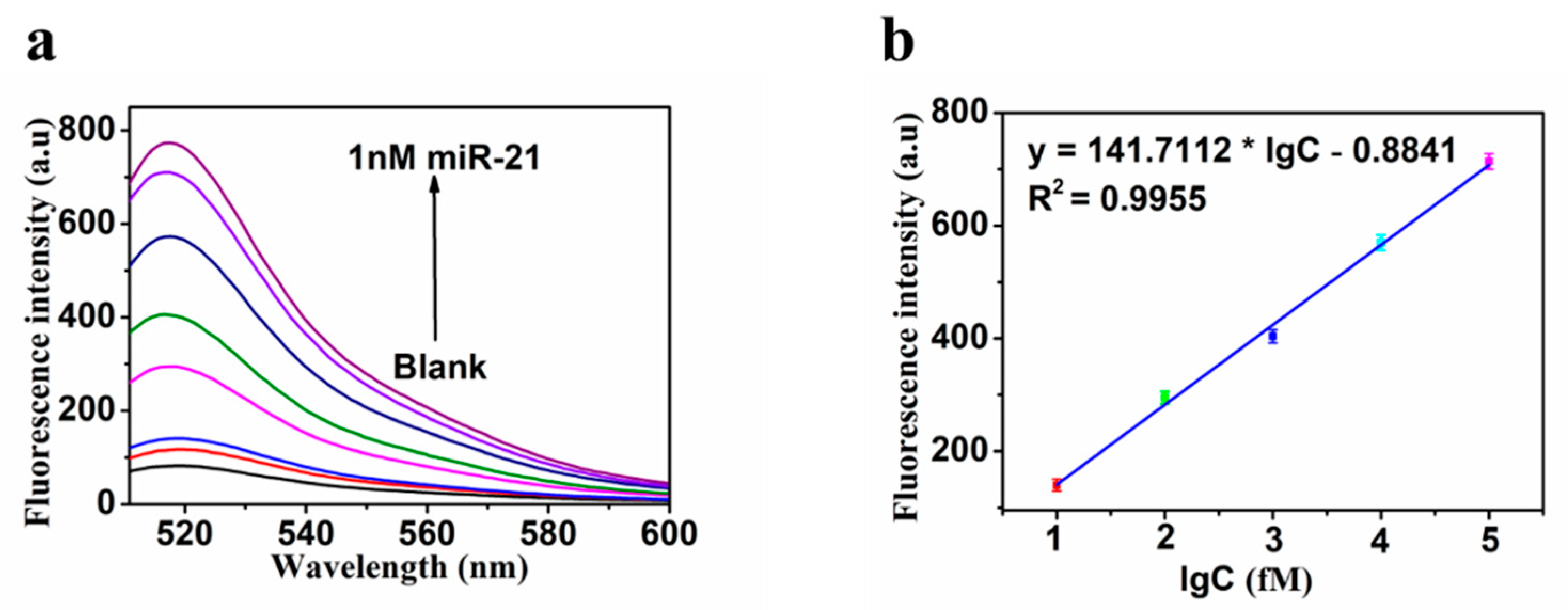
3.4. Sensitivity
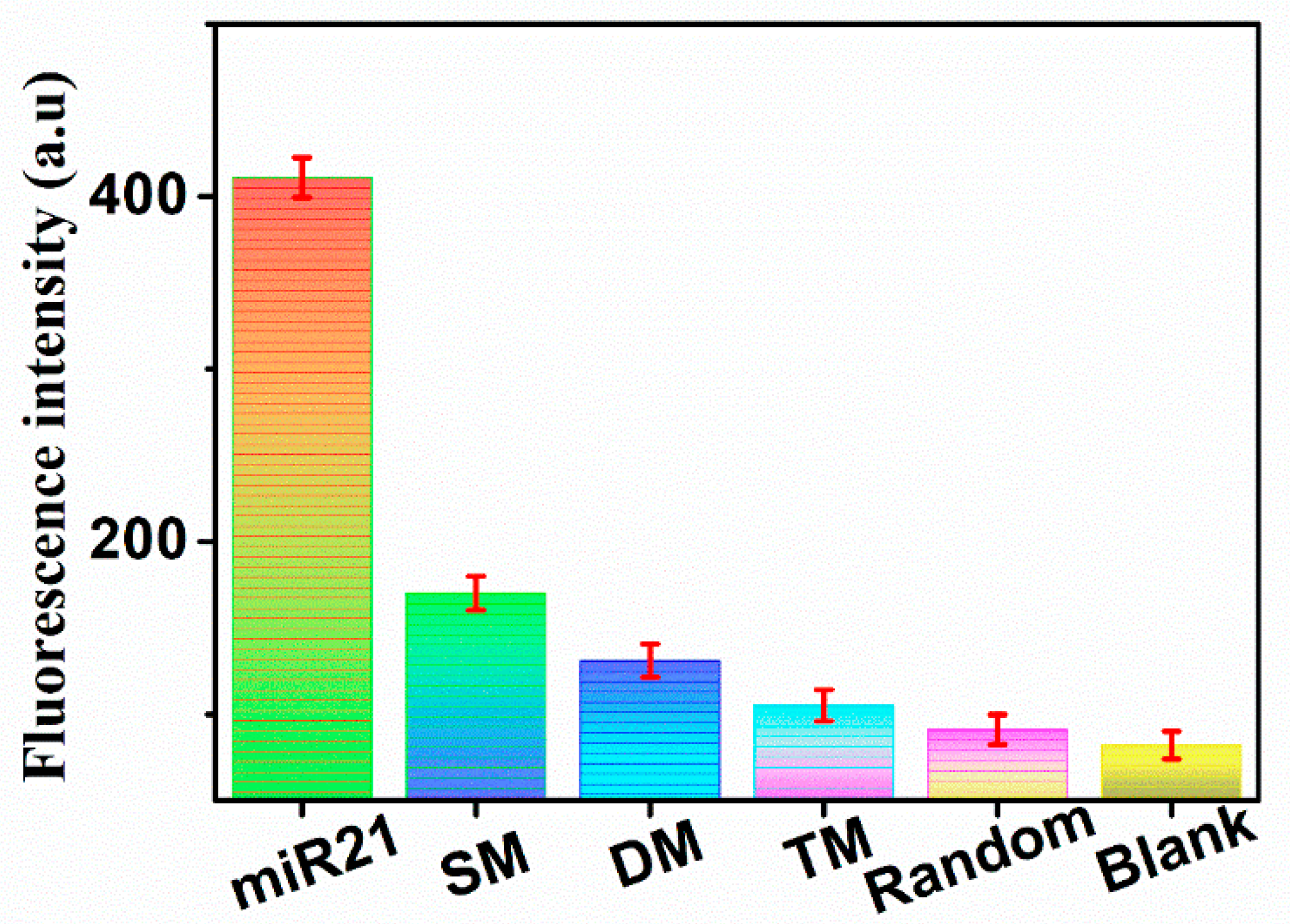
3.5. Specificity
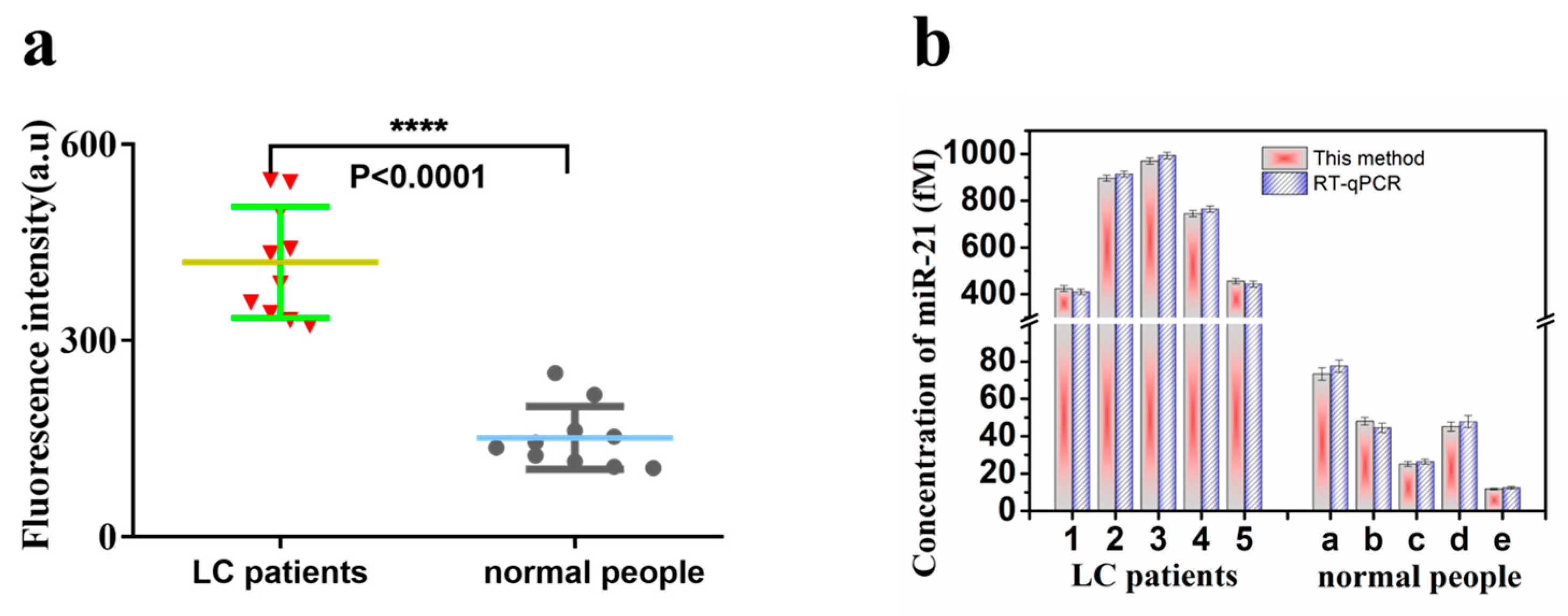
3.6. Application
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Bray, F.; Laversanne, M.; Weiderpass, E.; Soerjomataram, I. The ever-increasing importance of cancer as a leading cause of premature death worldwide. Cancer 2021, 127, 3029–3030. [Google Scholar] [CrossRef] [PubMed]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Oudkerk, M.; Liu, S.; Heuvelmans, M.A.; Walter, J.E.; Field, J.K. Lung cancer LDCT screening and mortality reduction—evidence, pitfalls and future perspectives. Nat. Rev. Clin. Oncol. 2021, 18, 135–151. [Google Scholar] [CrossRef]
- Goldstraw, P.; Chansky, K.; Crowley, J.; Rami-Porta, R.; Asamura, H.; Eberhardt, W.E.; Nicholson, A.G.; Groome, P.; Mitchell, A.; Bolejack, V. The IASLC Lung Cancer Staging Project: Proposals for Revision of the TNM Stage Groupings in the Forthcoming (Eighth) Edition of the TNM Classification for Lung Cancer. J. Thorac. Oncol. 2016, 11, 39–51. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qiu, J.; Xu, J.; Zhang, K.; Gu, W.; Nie, L.; Wang, G.; Luo, Y. Refining Cancer Management Using Integrated Liquid Biopsy. Theranostics 2020, 10, 2374–2384. [Google Scholar] [CrossRef]
- Ignatiadis, M.; Sledge, G.W.; Jeffrey, S.S. Liquid biopsy enters the clinic—Implementation issues and future challenges. Nat. Rev. Clin. Oncol. 2021, 18, 297–312. [Google Scholar] [CrossRef]
- Denzer, K.; Kleijmeer, M.J.; Heijnen, H.F.; Stoorvogel, W.; Geuze, H.J. Exosome: From internal vesicle of the multivesicular body to intercellular signaling device. J. Cell. Sci. 2000, 113 Pt 19, 3365–3374. [Google Scholar] [CrossRef]
- Kalluri, R.; LeBleu, V.S. The biology, function, and biomedical applications of exosomes. Science 2020, 367, eaau6977. [Google Scholar] [CrossRef]
- Jin, X.; Chen, Y.; Chen, H.; Fei, S.; Chen, D.; Cai, X.; Liu, L.; Lin, B.; Su, H.; Zhao, L.; et al. Evaluation of Tumor-Derived Exosomal miRNA as Potential Diagnostic Biomarkers for Early-Stage Non-Small Cell Lung Cancer Using Next-Generation Sequencing. Clin. Cancer Res. 2017, 23, 5311–5319. [Google Scholar] [CrossRef] [Green Version]
- Li, B.; Cao, Y.; Sun, M.; Feng, H. Expression, regulation, and function of exosome-derived miRNAs in cancer progression and therapy. FASEB J. 2021, 35, e21916. [Google Scholar] [CrossRef]
- Bica-Pop, C.; Cojocneanu-Petric, R.; Magdo, L.; Raduly, L.; Gulei, D.; Berindan-Neagoe, I. Overview upon miR-21 in lung cancer: Focus on NSCLC. Cell. Mol. Life. Sci. 2018, 75, 3539–3551. [Google Scholar] [CrossRef] [PubMed]
- Smolarz, M.; Widlak, P. Serum Exosomes and Their miRNA Load-A Potential Biomarker of Lung Cancer. Cancers 2021, 13, 1373. [Google Scholar] [CrossRef] [PubMed]
- Forero, D.A.; González-Giraldo, Y.; Castro-Vega, L.J.; Barreto, G.E. qPCR-based methods for expression analysis of miRNAs. Biotechniques 2019, 67, 192–199. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Foiani, G.; Guelfi, G.; Mandara, M.T. MicroRNA Dysregulation in Canine Meningioma: RT-qPCR Analysis of Formalin-Fixed Paraffin-Embedded Samples. J. Neuropathol. Exp. Neurol. 2021, 80, 769–775. [Google Scholar] [CrossRef] [PubMed]
- Horny, M.C.; Dupuis, V.; Siaugue, J.M.; Gamby, J. Release and Detection of microRNA by Combining Magnetic Hyperthermia and Electrochemistry Modules on a Microfluidic Chip. Sensors 2020, 21, 185. [Google Scholar] [CrossRef]
- Ma, G.M.; Huo, L.W.; Tong, Y.X.; Wang, Y.C.; Li, C.P.; Jia, H.X. Label-free and sensitive MiRNA detection based on turn-on fluorescence of DNA-templated silver nanoclusters coupled with duplex-specific nuclease-assisted signal amplification. Mikrochim. Acta 2021, 188, 355. [Google Scholar] [CrossRef]
- Luo, X.; Zhu, J.; Jia, W.; Fang, N.; Wu, P.; Cai, C.; Zhu, J.J. Boosting Long-Range Surface-Enhanced Raman Scattering on Plasmonic Nanohole Arrays for Ultrasensitive Detection of MiRNA. ACS. Appl. Mater. Interfaces 2021, 13, 18301–18313. [Google Scholar] [CrossRef]
- Huang, Y.; Sun, T.; Liu, L.; Xia, N.; Zhao, Y.; Yi, X. Surface plasmon resonance biosensor for the detection of miRNAs by combining the advantages of homogeneous reaction and heterogeneous detection. Talanta 2021, 234, 122622. [Google Scholar] [CrossRef]
- Chen, X.; Xu, K.; Li, J.; Yang, M.; Li, X.; Chen, Q.; Lu, C.; Yang, H. Switch-conversional ratiometric fluorescence biosensor for miRNA detection. Biosens. Bioelectron. 2020, 155, 112104. [Google Scholar] [CrossRef]
- Makarova, K.S.; Wolf, Y.I.; Iranzo, J.; Shmakov, S.A.; Alkhnbashi, O.S.; Brouns, S.J.J.; Charpentier, E.; Cheng, D.; Haft, D.H.; Horvath, P.; et al. Evolutionary classification of CRISPR-Cas systems: A burst of class 2 and derived variants. Nat. Rev. Microbiol. 2020, 18, 67–83. [Google Scholar] [CrossRef]
- Hanna, R.E.; Doench, J.G. Design and analysis of CRISPR-Cas experiments. Nat. Biotechnol. 2020, 38, 813–823. [Google Scholar] [CrossRef] [PubMed]
- Li, S.Y.; Cheng, Q.X.; Liu, J.K.; Nie, X.Q.; Zhao, G.P.; Wang, J. CRISPR-Cas12a has both cis- and trans-cleavage activities on single-stranded DNA. Cell. Res. 2018, 28, 491–493. [Google Scholar] [CrossRef] [PubMed]
- Dai, Y.; Somoza, R.A.; Wang, L.; Welter, J.F.; Li, Y.; Caplan, A.I.; Liu, C.C. Exploring the Trans-Cleavage Activity of CRISPR-Cas12a (cpf1) for the Development of a Universal Electrochemical Biosensor. Angew. Chem. Int. Ed. Engl. 2019, 58, 17399–17405. [Google Scholar] [CrossRef] [PubMed]
- Yao, Y.; Zhang, H.; Tian, T.; Liu, Y.; Zhu, R.; Ji, J.; Liu, B. Iodide-modified Ag nanoparticles coupled with DSN-Assisted cycling amplification for label-free and ultrasensitive SERS detection of MicroRNA-21. Talanta 2021, 235, 122728. [Google Scholar] [CrossRef] [PubMed]
- Fang, C.; Ouyang, P.; Yang, Y.; Qing, Y.; Han, J.; Shang, W.; Chen, Y.; Du, J. MiRNA Detection Using a Rolling Circle Amplification and RNA-Cutting Allosteric Deoxyribozyme Dual Signal Amplification Strategy. Biosensors 2021, 11, 222. [Google Scholar] [CrossRef]
- Zhu, Y.; Wang, J.; Xie, H.; Liu, H.; Liu, S.; He, D.; Mi, P.; He, S.; Wang, J.; Sun, Y. NIR-to-Vis Handheld Platforms for Detecting miRNA Level and Mutation Based on Sub-10 nm Sulfide Nanodots and HCR Amplification. ACS Appl Mater. Interfaces 2022, 14, 10212–10226. [Google Scholar] [CrossRef]
- Wang, Q.; Liu, J.; Zeng, J.; Yang, Z.; Ran, F.; Wu, L.; Yang, G.; Mei, Q.; Wang, X.; Chen, Q. Determination of miRNA derived from exosomes of prostate cancer via toehold-aided cyclic amplification combined with HRP enzyme catalysis and magnetic nanoparticles. Anal. Biochem. 2021, 630, 114336. [Google Scholar] [CrossRef]
- Park, H.J.; Lee, S.S. QCM sensing of miR-21 by formation of microRNA-DNA hybrid duplexes and intercalation on surface-functionalized pyrene. Analyst 2019, 144, 6936–6943. [Google Scholar] [CrossRef]
- Lim, J.Y.; Lee, S.S. Sensitive detection of microRNA using QCM biosensors: Sandwich hybridization and signal amplification by TiO2 nanoparticles. Anal. Methods 2020, 12, 5103–5109. [Google Scholar] [CrossRef]
- Hosseinzadeh, E.; Ravan, H.; Mohammadi, A.; Pourghadamyari, H. Colorimetric detection of miRNA-21 by DNAzyme-coupled branched DNA constructs. Talanta 2020, 216, 120913. [Google Scholar] [CrossRef]
- Luo, L.; Wang, L.; Zeng, L.; Wang, Y.; Weng, Y.; Liao, Y.; Chen, T.; Xia, Y.; Zhang, J.; Chen, J. A ratiometric electrochemical DNA biosensor for detection of exosomal MicroRNA. Talanta 2020, 207, 120298. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; He, C.; Wu, W.; Yang, H.; Dong, J.; Wen, L.; Hu, Z.; Yang, M.; Hou, C.; Huo, D. MXene-MoS2 heterostructure collaborated with catalyzed hairpin assembly for label-free electrochemical detection of microRNA-21. Talanta 2022, 237, 122927. [Google Scholar] [CrossRef] [PubMed]
- Sabahi, A.; Salahandish, R.; Ghaffarinejad, A.; Omidinia, E. Electrochemical nano-genosensor for highly sensitive detection of miR-21 biomarker based on SWCNT-grafted dendritic Au nanostructure for early detection of prostate cancer. Talanta 2020, 209, 120595. [Google Scholar] [CrossRef] [PubMed]
- Meng, T.; Shang, N.; Nsabimana, A.; Ye, H.; Wang, H.; Wang, C.; Zhang, Y. An enzyme-free electrochemical biosensor based on target-catalytic hairpin assembly and Pd@UiO-66 for the ultrasensitive detection of microRNA-21. Anal. Chim. Acta 2020, 1138, 59–68. [Google Scholar] [CrossRef]
- Ai, X.; Zhao, H.; Hu, T.; Yan, Y.; He, H.; Ma, C. A signal-on fluorescence-based strategy for detection of microRNA-21 based on graphene oxide and λ exonuclease-based signal amplification. Anal. Methods 2021, 13, 2107–2113. [Google Scholar] [CrossRef]
- Gu, J.; Qiao, Z.; He, X.; Yu, Y.; Lei, Y.; Tang, J.; Shi, H.; He, D.; Wang, K. Enzyme-free amplified detection of miRNA based on target-catalyzed hairpin assembly and DNA-stabilized fluorescent silver nanoclusters. Analyst 2020, 145, 5194–5199. [Google Scholar] [CrossRef]
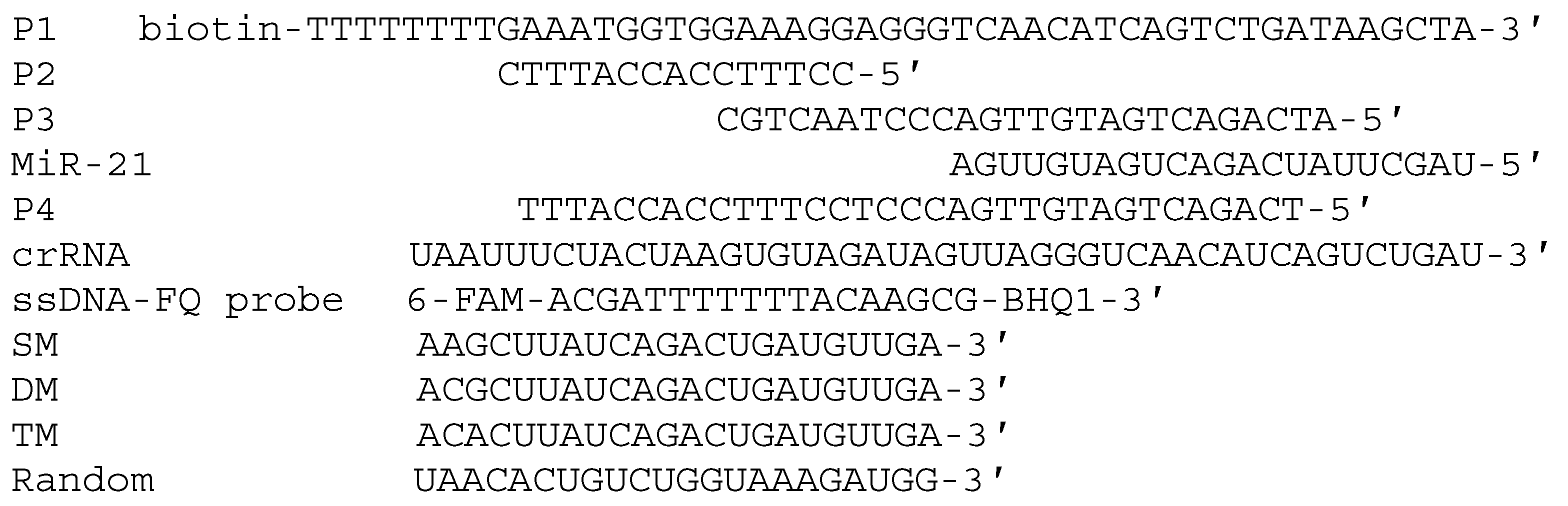

| Analytical Methods | Detection Limit | Linear Range | Ref. |
|---|---|---|---|
| Quartz crystal microbalance | 3.6 pM | 2.5 pM to 2.5 μM | [28] |
| Quartz crystal microbalance | 0.87 pM | 1.0 pM to 1.0 mM | [29] |
| Colorimetric | 1 pM | 1 pM to 1 nM | [30] |
| Electrochemistry | 2.3 fM | 10 to 70 fM | [31] |
| Electrochemistry | 26 fM | 100 fM to 100 nM | [32] |
| Electrochemistry | 0.01 fM | 0.01 fM to 1 μM | [33] |
| Electrochemistry | 0.713 fM | 20 fM to 600 pM | [34] |
| Fluorescence | 200 pM | 200 pM to 20 nM | [35] |
| Fluorescence | 0.02 nM | 0.02 to 5 nM | [36] |
| Fluorescence | 0.89 fM | 10 to 1 × 105 fM | This method |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, Q.; Liu, J.; He, N.; Zhang, M.; Wu, L.; Chen, X.; Zhu, J.; Ran, F.; Chen, Q.; Zhang, H. CRISPR/Cas12a Coupling with Magnetic Nanoparticles and Cascaded Strand Displacement Reaction for Ultrasensitive Fluorescence Determination of Exosomal miR-21. Molecules 2022, 27, 5338. https://doi.org/10.3390/molecules27165338
Liu Q, Liu J, He N, Zhang M, Wu L, Chen X, Zhu J, Ran F, Chen Q, Zhang H. CRISPR/Cas12a Coupling with Magnetic Nanoparticles and Cascaded Strand Displacement Reaction for Ultrasensitive Fluorescence Determination of Exosomal miR-21. Molecules. 2022; 27(16):5338. https://doi.org/10.3390/molecules27165338
Chicago/Turabian StyleLiu, Qing, Jingjian Liu, Na He, Moli Zhang, Lun Wu, Xiyu Chen, Jun Zhu, Fengying Ran, Qinhua Chen, and Hua Zhang. 2022. "CRISPR/Cas12a Coupling with Magnetic Nanoparticles and Cascaded Strand Displacement Reaction for Ultrasensitive Fluorescence Determination of Exosomal miR-21" Molecules 27, no. 16: 5338. https://doi.org/10.3390/molecules27165338
APA StyleLiu, Q., Liu, J., He, N., Zhang, M., Wu, L., Chen, X., Zhu, J., Ran, F., Chen, Q., & Zhang, H. (2022). CRISPR/Cas12a Coupling with Magnetic Nanoparticles and Cascaded Strand Displacement Reaction for Ultrasensitive Fluorescence Determination of Exosomal miR-21. Molecules, 27(16), 5338. https://doi.org/10.3390/molecules27165338






