Anti-Inflammatory Effects of Auranamide and Patriscabratine—Mechanisms and In Silico Studies
Abstract
1. Introduction
2. Results and Discussions
2.1. Cytotoxicity of Auranamide and Patriscabratine
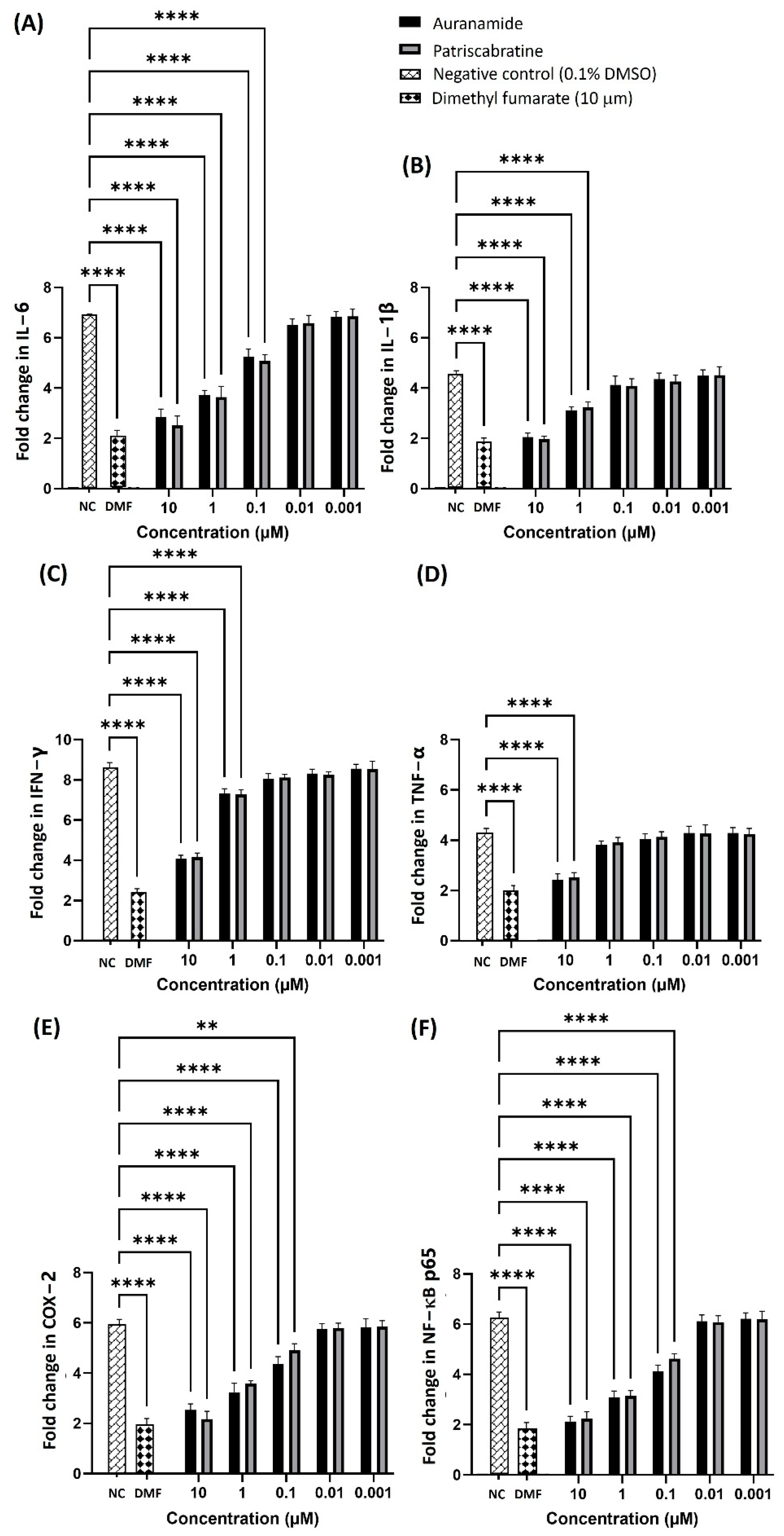
2.2. Anti-Inflammatory Effect of Auranamide and Patriscabratine
2.3. Anti-Inflammatory Mechanisms of Auranamide and Patriscabratine
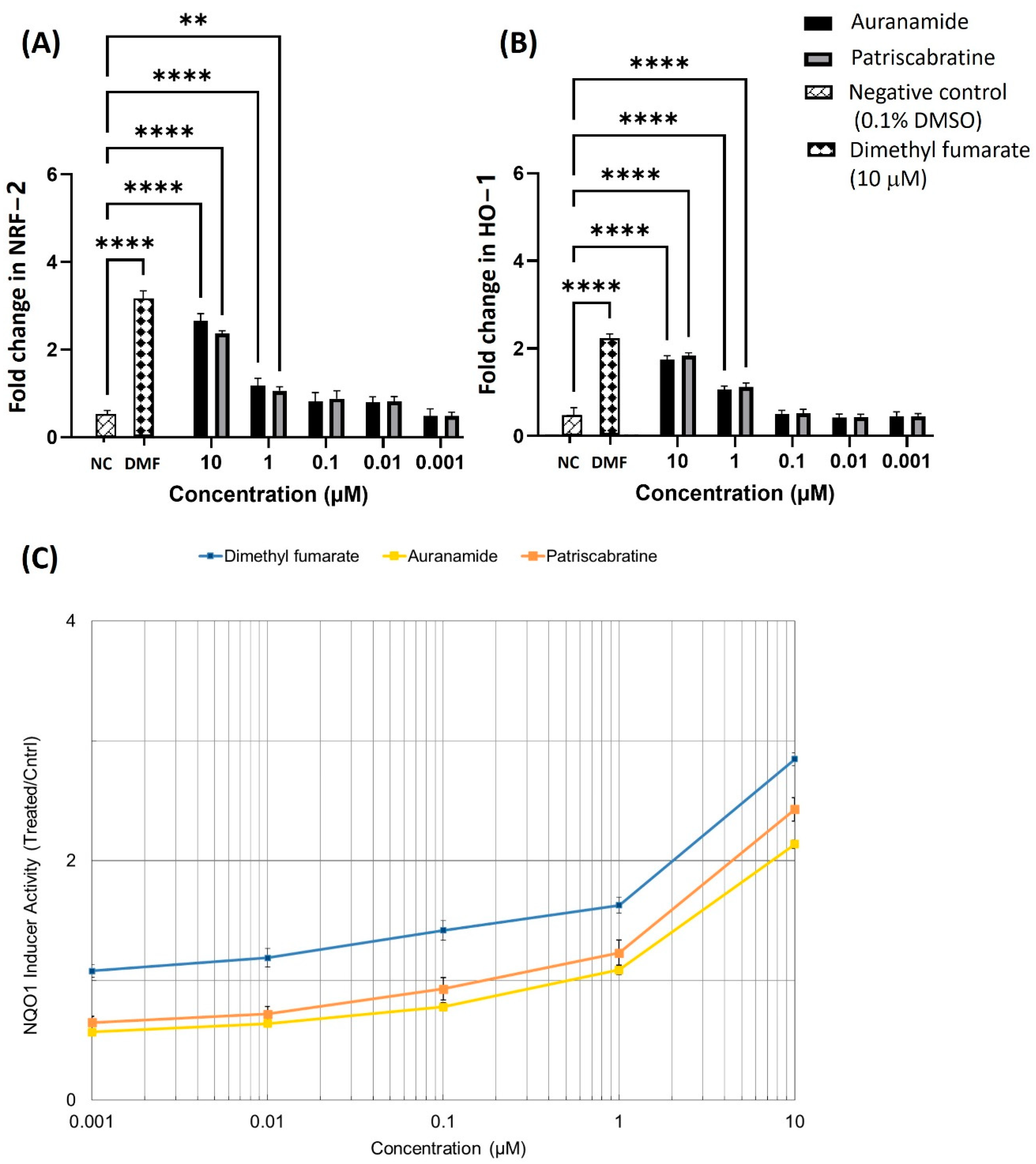
2.4. The Effect of Auranamide and Patriscabratine on NRF2 Activation
2.5. Metabolic Stability of Auranamide and Patriscabratine in Liver Microsomes
2.6. In Silico Studies
3. Materials and Methods
3.1. Preparation of Auranamide and Patriscabratine Solutions
3.2. Cytotoxicity of Auranamide and Patriscabratine
3.3. In Vitro Anti-Inflammatory Activity of Auranamide and Patriscabratine
3.4. Effect of Auranamide and Patriscabratine on NO Production in LPSEc Challenged RAW 264.7 Cells
3.5. Effect of Auranamide and Patriscabratine on Pro-Inflammatory Cytokines Expression in LPSEc Challenged RAW 264.7 Cells
3.6. Effect of Auranamide and Patriscabratine on COX-2, NF-κB p65, NRF-2 and HO-1 Expression in LPSEc-Stimulated RAW 264.7 Cells
3.7. Effect of Auranamide and Patriscabratine on NQO1 Activity in Hepa-1c1c7 Cells
3.8. Metabolic Stability of Auranamide and Patriscabratine in Liver Microsomes
3.9. In Silico Studies
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
Correction Statement
References
- Cuadrado, A.; Rojo, A.I.; Wells, G.; Hayes, J.D.; Cousin, S.P.; Rumsey, W.L.; Attucks, O.C.; Franklin, S.; Levonen, A.L.; Kensler, T.W.; et al. Therapeutic targeting of the NRF2 and KEAP1 partnership in chronic diseases. Nat. Rev. Drug Discov. 2019, 18, 295–317. [Google Scholar] [CrossRef] [PubMed]
- Arslanbaeva, L.; Bisaglia, M. Activation of the Nrf2 Pathway as a Therapeutic Strategy for ALS Treatment. Molecules 2022, 27, 1471. [Google Scholar] [CrossRef]
- Jaganjac, M.; Matijevi, T.; Saso, L. Oxidative Stress and Cancer Heterogeneity Orchestrate NRF2 Roles Relevant for Therapy Response. Molecules 2022, 27, 1468. [Google Scholar]
- Hayes, J.D.; Dinkova-Kostova, A.T. The Nrf2 regulatory network provides an interface between redox and intermediary metabolism. Trends Biochem. Sci. 2014, 39, 199–218. [Google Scholar] [CrossRef]
- Kansanen, E.; Kuosmanen, S.M.; Leinonen, H.; Levonen, A.-L. The Keap1-Nrf2 pathway: Mechanisms of activation and dysregulation in cancer. Redox Biol. 2013, 1, 45–49. [Google Scholar] [CrossRef]
- Villeneuve, N.F.; Lau, A.; Zhang, D.D. Regulation of the Nrf2-Keap1 antioxidant response by the ubiquitin proteasome system: An insight into cullin-ring ubiquitin ligases. Antioxid. Redox Signal. 2010, 13, 1699–1712. [Google Scholar] [CrossRef] [PubMed]
- Prawan, A.; Keum, Y.S.; Khor, T.O.; Yu, S.; Nair, S.; Li, W.; Hu, L.; Kong, A.N.T. Structural Influence of Isothiocyanates on the Antioxidant Response Element (ARE)-Mediated Heme Oxygenase-1 (HO-1) Expression. Pharm. Res. 2008, 25, 836–844. [Google Scholar] [CrossRef] [PubMed]
- Mak, K.K.; Shiming, Z.; Balijepalli, M.K.; Dinkova-Kostova, A.T.; Epemolu, O.; Mohd, Z.; Pichika, M.R. Studies on the mechanism of anti-inflammatory action of swietenine, a tetranortriterpenoid isolated from Swietenia macrophylla seeds. Phytomed. Plus 2021, 1, 100018. [Google Scholar] [CrossRef]
- Baird, L.; Yamamoto, M. The Molecular Mechanisms Regulating the KEAP1-NRF2 Pathway. Mol. Cell. Biol. 2020, 40, e00099-20. [Google Scholar] [CrossRef]
- Liby, K.T.; Sporn, M.B. Synthetic oleanane triterpenoids: Multifunctional drugs with a broad range of applications for prevention and treatment of chronic disease. Pharmacol. Rev. 2012, 64, 972–1003. [Google Scholar] [CrossRef]
- Li, Y.R.; Li, G.H.; Zhou, M.X.; Xiang, L.; Ren, D.M.; Lou, H.X.; Wang, X.N.; Shen, T. Discovery of natural flavonoids as activators of Nrf2-mediated defense system: Structure-activity relationship and inhibition of intracellular oxidative insults. Bioorg. Med. Chem. 2018, 26, 5140–5150. [Google Scholar] [CrossRef]
- Satoh, T.; Lipton, S. Recent advances in understanding NRF2 as a druggable target: Development of pro-electrophilic and non-covalent NRF2 activators to overcome systemic side effects of electrophilic drugs like dimethyl fumarate. F1000Research 2017, 6, 2138. [Google Scholar] [CrossRef]
- Ma, L.; Liu, J.; Lin, Q.; Gu, Y.; Yu, W. Eugenol protects cells against oxidative stress via Nrf2. Exp. Ther. Med. 2021, 21, 107. [Google Scholar] [CrossRef] [PubMed]
- Hammer, A.; Waschbisch, A.; Kuhbandner, K.; Bayas, A.; Lee, D.H.; Duscha, A.; Haghikia, A.; Gold, R.; Linker, R.A. The NRF2 pathway as potential biomarker for dimethyl fumarate treatment in multiple sclerosis. Ann. Clin. Transl. Neurol. 2018, 5, 668–676. [Google Scholar] [CrossRef]
- Sharma, S.B.; Gupta, R. Drug development from natural resource: A systematic approach. Mini Rev. Med. Chem. 2015, 15, 52–57. [Google Scholar] [CrossRef]
- Khan, R.A. Natural products chemistry: The emerging trends and prospective goals. Saudi Pharm. J. 2018, 26, 739–753. [Google Scholar] [CrossRef]
- Janczewski, Ł. Sulforaphane and Its Bifunctional Analogs: Synthesis and Biological Activity. Molecules 2022, 27, 1750. [Google Scholar] [CrossRef]
- Taylor, E.; Kim, Y.; Zhang, K.; Chau, L.; Nguyen, B.C.; Rayalam, S.; Wang, X. Antiaging Mechanism of Natural Compounds: Effects on Autophagy and Oxidative Stress. Molecules 2022, 27, 4396. [Google Scholar] [CrossRef] [PubMed]
- Low, J.S.; Mak, K.K.; Pichika, M.R.; Marappan, P.; Zhang, S.; Jiampanichkul, A.; Chua, S.; Mohandas, K.; Balijepalli, M.K. Melastoma malabathricum L.: A review of its traditional uses, phytochemical constituents and bioactivities. J. Glob. Trends Pharm. Sci. 2021, 12, 9257–9274. [Google Scholar]
- Serna, D.M.O.; Martínez, J.H.I. Phenolics and polyphenolics from melastomataceae species. Molecules 2015, 20, 17818–17847. [Google Scholar] [CrossRef]
- Susanti, D.; Sirat, H.M.; Ahmad, F.; Ali, R.M. Bioactive Constituents from the Leaves of Melastoma melabathricum L. J. Ilm. Farm. 2008, 5, 1–8. [Google Scholar]
- Lestari, O.A.; Palupi, N.S.; Setiyono, A.; Kusnandar, F.; Yuliana, N.D. In vitro antioxidant potential and phytochemical profiling of Melastoma malabathricum leaf water extract. Food Sci. Technol. 2022, 42, e92021. [Google Scholar] [CrossRef]
- Sirat, H.M.; Susanti, D.; Ahmad, F.; Takayama, H.; Kitajima, M. Amides, triterpene and flavonoids from the leaves of Melastoma malabathricum L. J. Nat. Med. 2010, 64, 492–495. [Google Scholar] [CrossRef] [PubMed]
- Matzinger, M.; Fischhuber, K.; Heiss, E.H. Activation of Nrf2 signaling by natural products—Can it alleviate diabetes? Biotechnol. Adv. 2018, 36, 1738–1767. [Google Scholar] [CrossRef] [PubMed]
- Staurengo-Ferrari, L.; Badaro-Garcia, S.; Hohmann, M.S.; Manchope, M.F.; Zaninelli, T.H.; Casagrande, R.; Verri, W.A., Jr. Contribution of Nrf2 Modulation to the Mechanism of Action of Analgesic and Anti-inflammatory Drugs in Pre-clinical and Clinical Stages. Front. Pharmacol. 2019, 9, 1536. [Google Scholar] [CrossRef]
- Uddin, S.J.; Grice, D.; Tiralongo, E. Evaluation of cytotoxic activity of patriscabratine, tetracosane and various flavonoids isolated from the Bangladeshi medicinal plant Acrostichum aureum. Pharm. Biol. 2012, 50, 1276–1280. [Google Scholar] [CrossRef]
- Chang, Y.C.; Yang, S.F.; Huang, F.M.; Liu, C.M.; Tai, K.W.; Hsieh, Y.S. Proinflammatory cytokines induce cyclooxygenase-2 mRNA and protein expression in human pulp cell cultures. J. Endod. 2003, 29, 201–204. [Google Scholar] [CrossRef]
- Hohmann, M.S.; Zaninelli, T.H.; Staurengo-Ferrari, L.; Manchope, M.F.; Badaro-Garcia, S.; Freitas, A.D.; Casagrande, R.; Verri, W.A. Nrf2 in Immune Responses During Inflammation. In Nrf2 and Its Modulation in Inflammation; Springer: Cham, Switzerland, 2020; pp. 23–49. [Google Scholar]
- Dinkova-Kostova, A.T.; Talalay, P.; Sharkey, J.; Zhang, Y.; Holtzclaw, W.D.; Wang, X.J.; David, E.; Schiavoni, K.H.; Finlayson, S.; Mierke, D.F.; et al. An exceptionally potent inducer of cytoprotective enzymes: Elucidation of the structural features that determine inducer potency and reactivity with Keap1. J. Biol. Chem. 2010, 285, 33747–33755. [Google Scholar] [CrossRef]
- Dinkova-Kostova, A.T.; Talalay, P. NAD(P)H:quinone acceptor oxidoreductase 1 (NQO1), a multifunctional antioxidant enzyme and exceptionally versatile cytoprotector. Arch. Biochem. Biophys. 2010, 501, 116. [Google Scholar] [CrossRef]
- Dinkova-Kostova, A.T.; Fahey, J.W.; Kostov, R.V.; Kensler, T.W. KEAP1 and Done? Targeting the NRF2 Pathway with Sulforaphane. Trends Food Sci. Technol. 2017, 69, 257–269. [Google Scholar] [CrossRef] [PubMed]
- Mak, K.K.; Epemolu, O.; Pichika, M.R. The role of DMPK science in improving pharmaceutical research and development efficiency. Drug Discov. Today 2022, 27, 705–729. [Google Scholar] [CrossRef] [PubMed]
- Siramshetty, V.B.; Shah, P.; Kerns, E.; Nguyen, K.; Yu, K.R.; Kabir, M.; Williams, J.; Neyra, J.; Southall, N.; Nguyễn, Ð.T.; et al. Retrospective assessment of rat liver microsomal stability at NCATS: Data and QSAR models. Sci. Rep. 2020, 10, 20713. [Google Scholar] [CrossRef] [PubMed]
- Fahey, J.W.; Dinkova-Kostova, A.T.; Stephenson, K.K.; Talalay, P. The “Prochaska” Microtiter Plate Bioassay for Inducers of NQO1. Methods Enzymol. 2004, 382, 243–258. [Google Scholar] [PubMed]
- Iwatsubo, T.; Hirota, N.; Ooie, T.; Suzuki, H.; Shimada, N.; Chiba, K.; Ishizaki, T.; Green, C.E.; Tyson, C.A.; Sugiyama, Y. Prediction of in vivo drug metabolism in the human liver from in vitro metabolism data. Pharmacol. Ther. 1997, 73, 147–171. [Google Scholar] [CrossRef]

| Compound | Human | Rat | Mouse | |
|---|---|---|---|---|
| Clint (mL/min/g liver) | Auranamide | 10.18 ± 0.35 | 17.22 ± 0.46 | 16.18 ± 0.43 |
| Patriscabratine | 13.27 ± 0.42 | 18.53 ± 0.56 | 18.64 ± 0.51 | |
| Half-life (T1/2, min) | Auranamide | 7.14 | 4.23 | 4.50 |
| Patriscabratine | 5.48 | 3.93 | 3.90 |
| PDB | CMP | 2D | 3D |
|---|---|---|---|
| 4IQK | AUR | 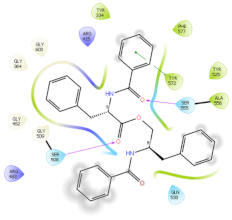 |  |
| 4IQK | PAT | 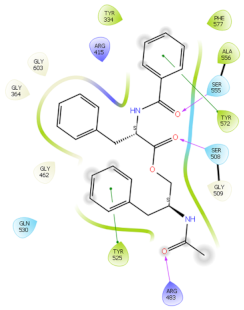 |  |
| 3ZRL | AUR | 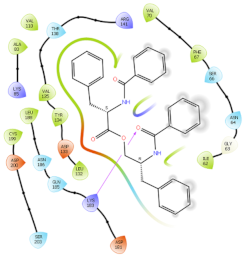 | 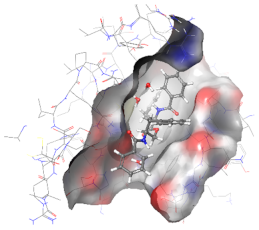 |
| 3ZRL | PAT | 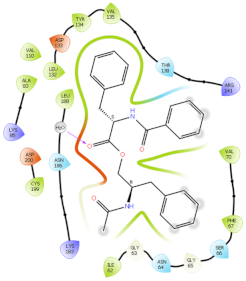 | 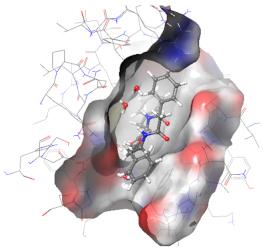 |
| PDB | Cpd | Docking Score | ΔG Bind | ΔG Coulomb | ΔG Covalent | ΔG Hbond | ΔG Lipo | ΔG VdW |
|---|---|---|---|---|---|---|---|---|
| 4IQK | AUR | −6.768 | −72.34 | −21.68 | 2.09 | −1.05 | −24.17 | −50.48 |
| PAT | −6.167 | −69.44 | −23.39 | 2.10 | −1.19 | −21.96 | −48.61 | |
| 3ZRL | AUR | −5.518 | −49.69 | −17.57 | 6.23 | −1.62 | −22.12 | −52.74 |
| PAT | −5.270 | −45.50 | −21.71 | 13.30 | −1.09 | −25.05 | −51.89 |
| Cpd | CNS | Mass | logPo/w | logS | logHERG | PCaco | logKhsa | %HOA | PSA | RO5 |
|---|---|---|---|---|---|---|---|---|---|---|
| AUR | −2 | 506.6 | 7.149 | −8.1 | −8.622 | 1335 | 1.373 | 100 | 101 | 2 |
| PAT | −2 | 444.5 | 4.836 | −5.2 | −5.807 | 425 | 0.545 | 100 | 102 | 0 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mak, K.-K.; Zhang, S.; Low, J.S.; Balijepalli, M.K.; Sakirolla, R.; Dinkova-Kostova, A.T.; Epemolu, O.; Mohd, Z.; Pichika, M.R. Anti-Inflammatory Effects of Auranamide and Patriscabratine—Mechanisms and In Silico Studies. Molecules 2022, 27, 4992. https://doi.org/10.3390/molecules27154992
Mak K-K, Zhang S, Low JS, Balijepalli MK, Sakirolla R, Dinkova-Kostova AT, Epemolu O, Mohd Z, Pichika MR. Anti-Inflammatory Effects of Auranamide and Patriscabratine—Mechanisms and In Silico Studies. Molecules. 2022; 27(15):4992. https://doi.org/10.3390/molecules27154992
Chicago/Turabian StyleMak, Kit-Kay, Shiming Zhang, Jun Sheng Low, Madhu Katyayani Balijepalli, Raghavendra Sakirolla, Albena T. Dinkova-Kostova, Ola Epemolu, Zulkefeli Mohd, and Mallikarjuna Rao Pichika. 2022. "Anti-Inflammatory Effects of Auranamide and Patriscabratine—Mechanisms and In Silico Studies" Molecules 27, no. 15: 4992. https://doi.org/10.3390/molecules27154992
APA StyleMak, K.-K., Zhang, S., Low, J. S., Balijepalli, M. K., Sakirolla, R., Dinkova-Kostova, A. T., Epemolu, O., Mohd, Z., & Pichika, M. R. (2022). Anti-Inflammatory Effects of Auranamide and Patriscabratine—Mechanisms and In Silico Studies. Molecules, 27(15), 4992. https://doi.org/10.3390/molecules27154992






