Structural and Functional Analysis of the Pyridoxal Phosphate Homeostasis Protein YggS from Fusobacterium nucleatum
Abstract
:1. Introduction
2. Results and Discussion
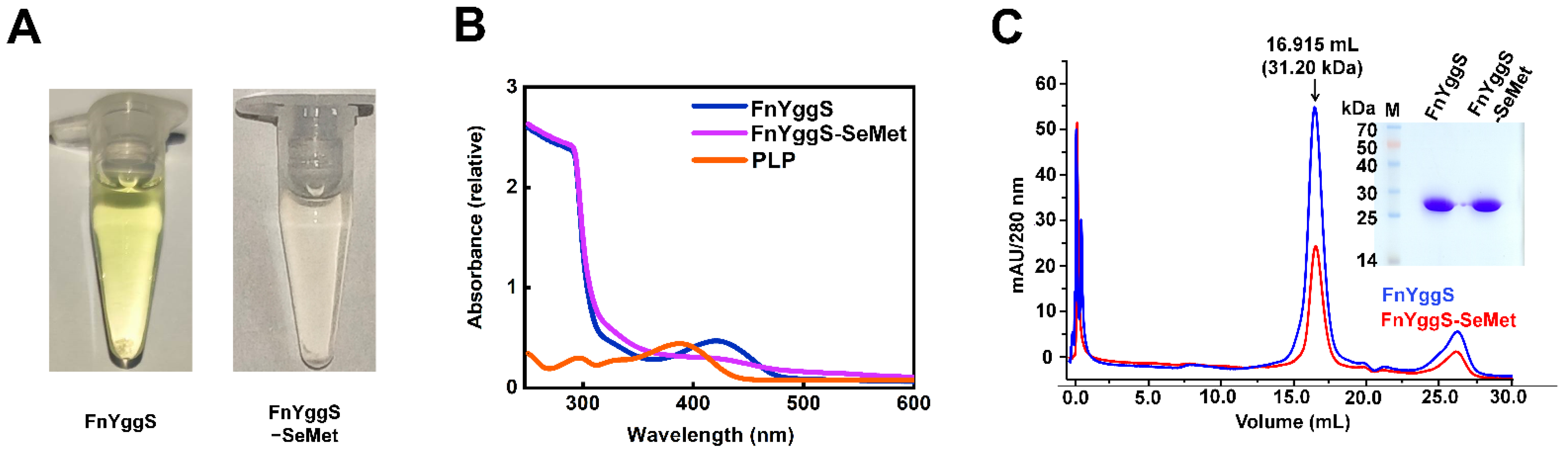
2.1. Characterization of FnYggS
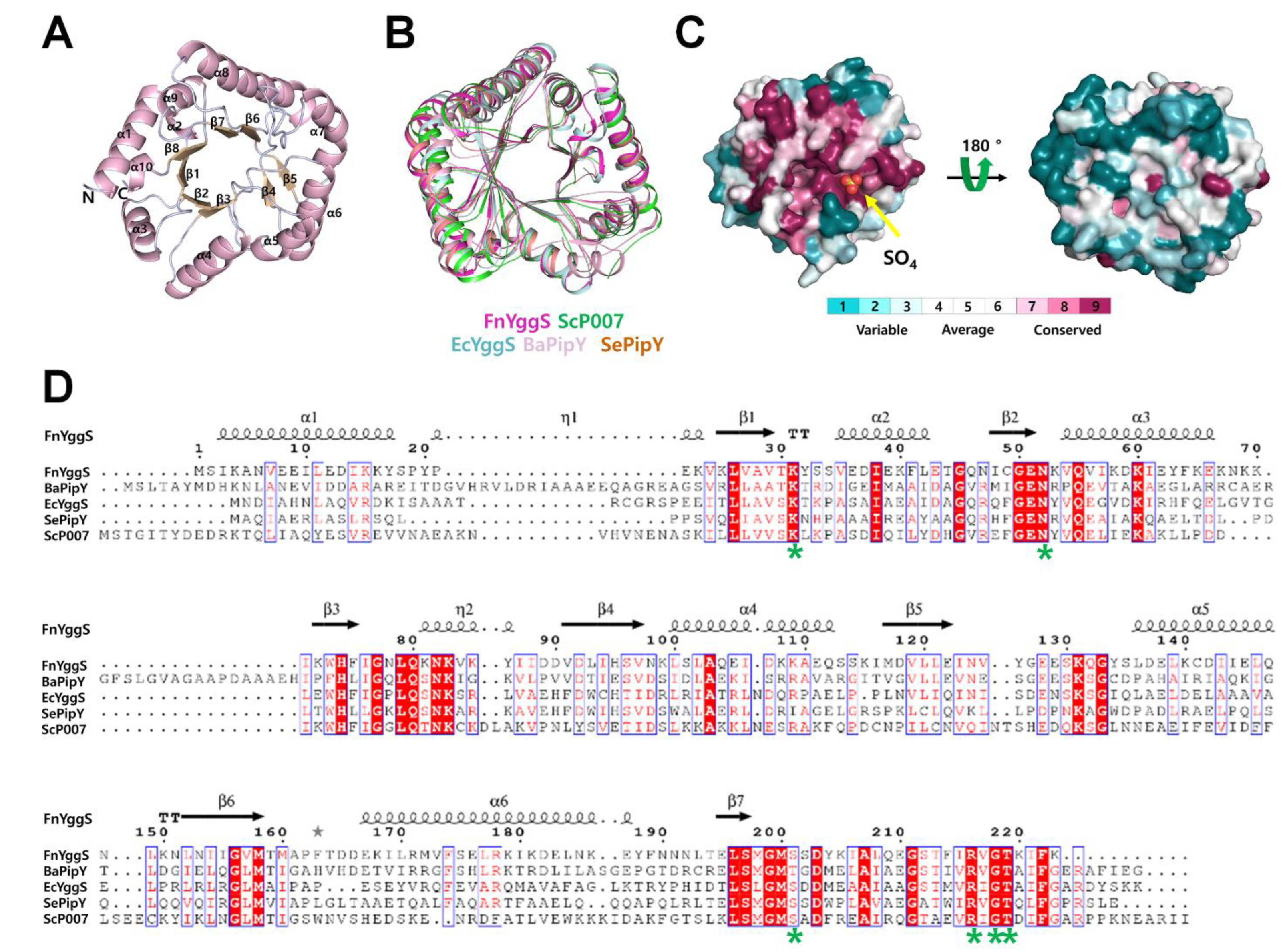
2.2. Overall Structure of FnYggS
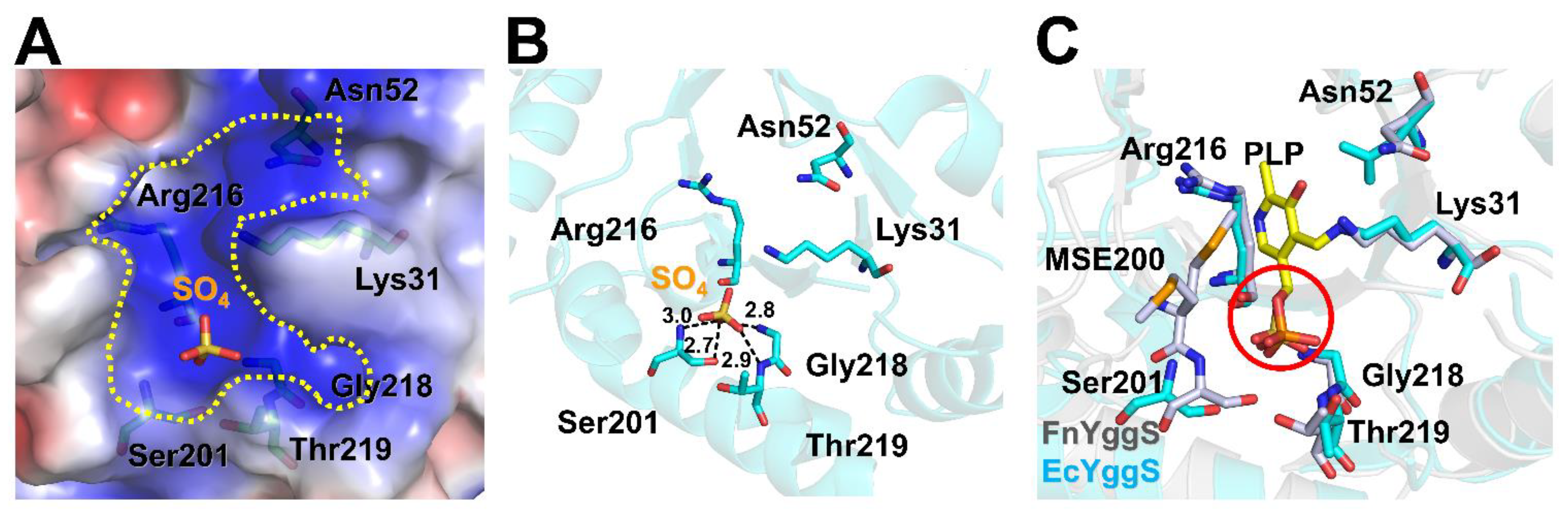
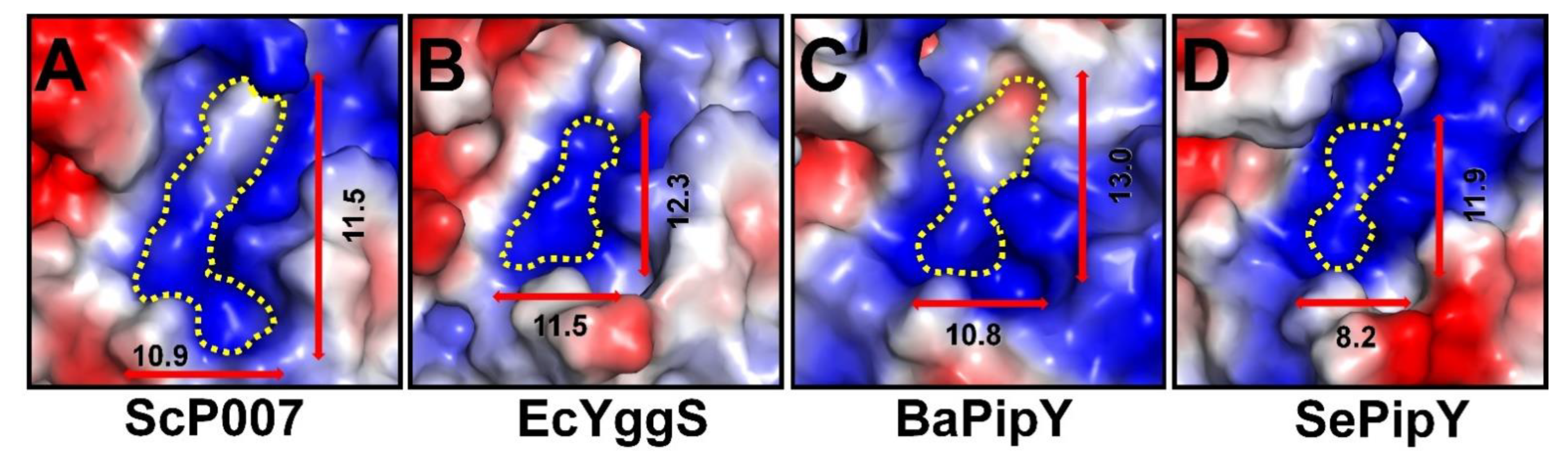
2.3. PLP-Binding Site of FnYggS
3. Materials and Methods
3.1. Protein Preparation
3.2. Analysis of PLP Binding
3.3. Crystallization, Data Collection, and Structural Determination
3.4. Size Exclusion Chromatography
3.5. Mutagenesis
3.6. Microscale Thermophoresis
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Han, Y.W. Fusobacterium nucleatum: A commensal-turned pathogen. Curr. Opin. Microbiol. 2015, 23, 141–147. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stokowa-Soltys, K.; Wojtkowiak, K.; Jagiello, K. Fusobacterium nucleatum-Friend or foe? J. Inorg. Biochem. 2021, 224, 111586. [Google Scholar] [CrossRef] [PubMed]
- Signat, B.; Roques, C.; Poulet, P.; Duffaut, D. Fusobacterium nucleatum in periodontal health and disease. Curr. Issues Mol. Biol. 2011, 13, 25–36. [Google Scholar] [PubMed]
- Han, Y.W.; Ikegami, A.; Rajanna, C.; Kawsar, H.I.; Zhou, Y.; Li, M.; Sojar, H.T.; Genco, R.J.; Kuramitsu, H.K.; Deng, C.X. Identification and characterization of a novel adhesin unique to oral fusobacteria. J. Bacteriol. 2005, 187, 5330–5340. [Google Scholar] [CrossRef] [Green Version]
- Kumar, A.; Thotakura, P.L.; Tiwary, B.K.; Krishna, R. Target identification in Fusobacterium nucleatum by subtractive genomics approach and enrichment analysis of host-pathogen protein-protein interactions. BMC Microbiol. 2016, 16, 84. [Google Scholar] [CrossRef] [Green Version]
- Strauss, J.; Kaplan, G.G.; Beck, P.L.; Rioux, K.; Panaccione, R.; Devinney, R.; Lynch, T.; Allen-Vercoe, E. Invasive potential of gut mucosa-derived Fusobacterium nucleatum positively correlates with IBD status of the host. Inflamm. Bowel. Dis. 2011, 17, 1971–1978. [Google Scholar] [CrossRef]
- Heckmann, J.G.; Lang, C.J.; Hartl, H.; Tomandl, B. Multiple brain abscesses caused by Fusobacterium nucleatum treated conservatively. Can. J. Neurol. Sci. 2003, 30, 266–268. [Google Scholar] [CrossRef]
- Han, Y.W.; Fardini, Y.; Chen, C.; Iacampo, K.G.; Peraino, V.A.; Shamonki, J.M.; Redline, R.W. Term stillbirth caused by oral Fusobacterium nucleatum. Obstet. Gynecol. 2010, 115, 442–445. [Google Scholar] [CrossRef] [Green Version]
- Wells, C.D.; Balan, V.; Smilack, J.D. Pyogenic liver abscess after colonoscopy in a patient with ulcerative colitis. Clin. Gastroenterol. Hepatol. 2005, 3, 24. [Google Scholar] [CrossRef]
- Storm, J.C.; Ford, B.A.; Streit, J.A. Myocardial infection due to Fusobacterium nucleatum. Diagn. Microbiol. Infect. Dis. 2013, 77, 373–375. [Google Scholar] [CrossRef]
- Hill, G.B. Preterm birth: Associations with genital and possibly oral microflora. Ann. Periodontol. 1998, 3, 222–232. [Google Scholar] [CrossRef]
- Liu, H.; Hong, X.L.; Sun, T.T.; Huang, X.W.; Wang, J.L.; Xiong, H. Fusobacterium nucleatum exacerbates colitis by damaging epithelial barriers and inducing aberrant inflammation. J. Dig. Dis. 2020, 21, 385–398. [Google Scholar] [CrossRef]
- Han, Y.W.; Redline, R.W.; Li, M.; Yin, L.; Hill, G.B.; McCormick, T.S. Fusobacterium nucleatum induces premature and term stillbirths in pregnant mice: Implication of oral bacteria in preterm birth. Infect. Immun. 2004, 72, 2272–2279. [Google Scholar] [CrossRef] [Green Version]
- Zhang, S.; Cai, S.; Ma, Y. Association between Fusobacterium nucleatum and colorectal cancer: Progress and future directions. J. Cancer. 2018, 9, 1652–1659. [Google Scholar] [CrossRef] [Green Version]
- Yu, M.R.; Kim, H.J.; Park, H.R. Fusobacterium nucleatum Accelerates the Progression of Colitis-Associated Colorectal Cancer by Promoting EMT. Cancers 2020, 12, 2728. [Google Scholar] [CrossRef]
- Wang, H.F.; Li, L.F.; Guo, S.H.; Zeng, Q.Y.; Ning, F.; Liu, W.L.; Zhang, G. Evaluation of antibody level against Fusobacterium nucleatum in the serological diagnosis of colorectal cancer. Sci. Rep. 2016, 6, 33440. [Google Scholar] [CrossRef] [Green Version]
- Mehta, R.S.; Nishihara, R.; Cao, Y.; Song, M.; Mima, K.; Qian, Z.R.; Nowak, J.A.; Kosumi, K.; Hamada, T.; Masugi, Y.; et al. Association of Dietary Patterns with Risk of Colorectal Cancer Subtypes Classified by Fusobacterium nucleatum in Tumor Tissue. JAMA Oncol. 2017, 3, 921–927. [Google Scholar] [CrossRef] [Green Version]
- Batty, A.; Wren, M.W.; Gal, M. Fusobacterium necrophorum as the cause of recurrent sore throat: Comparison of isolates from persistent sore throat syndrome and Lemierre’s disease. J. Infect. 2005, 51, 299–306. [Google Scholar] [CrossRef]
- Li, Y.Y.; Ge, Q.X.; Cao, J.; Zhou, Y.J.; Du, Y.L.; Shen, B.; Wan, Y.J.; Nie, Y.Q. Association of Fusobacterium nucleatum infection with colorectal cancer in Chinese patients. World J. Gastroenterol. 2016, 22, 3227–3233. [Google Scholar] [CrossRef]
- Bullman, S.; Pedamallu, C.S.; Sicinska, E.; Clancy, T.E.; Zhang, X.; Cai, D.; Neuberg, D.; Huang, K.; Guevara, F.; Nelson, T.; et al. Analysis of Fusobacterium persistence and antibiotic response in colorectal cancer. Science 2017, 358, 1443–1448. [Google Scholar] [CrossRef] [Green Version]
- Merrill, A.H.; Henderson, J.M. Vitamin B6 Metabolism by Human Liver. Ann. N. Y. Acad. Sci. 1990, 585, 110–117. [Google Scholar] [CrossRef]
- Prunetti, L.; El Yacoubi, B.; Schiavon, C.R.; Kirkpatrick, E.; Huang, L.; Bailly, M.; El Badawi-Sidhu, M.; Harrison, K.; Gregory, J.F.; Fiehn, O.; et al. Evidence that COG0325 proteins are involved in PLP homeostasis. Microbiology 2016, 162, 694–706. [Google Scholar] [CrossRef]
- Kiruba, G.S.M.; Wong, M.W. Tautomeric Equilibria of Pyridoxal-5‘-phosphate (Vitamin B6) and 3-Hydroxypyridine Derivatives: A Theoretical Study of Solvation Effects. J. Org. Chem. Res. 2003, 68, 2874–2881. [Google Scholar] [CrossRef]
- Ubbink, J.B.; Serfontein, W.J.; De Villiers, L.S. Stability of pyridoxal-5-phosphate semicarbazone: Applications in plasma vitamin B6 analysis and population surveys of vitamin B6 nutritional status. J. Chromatogr. B Biomed. Appl. 1985, 342, 277–284. [Google Scholar] [CrossRef]
- Labella, J.I.; Cantos, R.; Espinosa, J.; Forcada-Nadal, A.; Rubio, V.; Contreras, A. PipY, a Member of the Conserved COG0325 Family of PLP-Binding Proteins, Expands the Cyanobacterial Nitrogen Regulatory Network. Front. Microbiol. 2017, 8, 1244. [Google Scholar] [CrossRef] [Green Version]
- Darin, N.; Reid, E.; Prunetti, L.; Samuelsson, L.; Husain, R.A.; Wilson, M.; El Yacoubi, B.; Footitt, E.; Chong, W.K.; Wilson, L.C.; et al. Mutations in PROSC Disrupt Cellular Pyridoxal Phosphate Homeostasis and Cause Vitamin-B6-Dependent Epilepsy. Am. J. Hum. Genet. 2016, 99, 1325–1337. [Google Scholar] [CrossRef] [Green Version]
- Tremiño, L.; Forcada-Nadal, A.; Contreras, A.; Rubio, V. Studies on cyanobacterial protein PipY shed light on structure, potential functions, and vitamin B6-dependent epilepsy. FEBS Lett. 2017, 591, 3431–3442. [Google Scholar] [CrossRef]
- Eswaramoorthy, S.; Gerchman, S.; Graziano, V.; Kycia, H.; Studier, F.W.; Swaminathan, S. Structure of a yeast hypothetical protein selected by a structural genomics approach. Acta Crystallogr. D Biol. Crystallogr. 2002, 59, 127–135. [Google Scholar] [CrossRef]
- Knight, A.M.; Nobili, A.; van den Bergh, T.; Genz, M.; Joosten, H.-J.; Albrecht, D.; Riedel, K.; Pavlidis, I.V.; Bornscheuer, U.T. Bioinformatic analysis of fold-type III PLP-dependent enzymes discovers multimeric racemases. Appl. Microbiol. Biotechnol. 2016, 101, 1499–1507. [Google Scholar] [CrossRef] [Green Version]
- Denesyuk, A.I.; Denessiouk, K.A.; Korpela, T.; Johnson, M.S. Phosphate group binding “cup” of PLP-dependent and non-PLP-dependent enzymes: Leitmotif and variations. Biochim. Biophys. Acta-Proteins Proteom. 2003, 1647, 234–238. [Google Scholar] [CrossRef]
- Holm, L.; Park, J. DaliLite workbench for protein structure comparison. Bioinformatics 2000, 16, 566–567. [Google Scholar] [CrossRef] [PubMed]
- He, S.; Yuan, C.; Bai, X.; Bu, T.; Zhang, J.; Wang, L.; Quan, C.; Xu, Y. Pyridoxal phosphate homeostasis protein FnYggS from Fusobacterium nucleatum: Purification, crystallization, and X-ray crystallographic analysis. Biodesign 2022, 10, 34–38. [Google Scholar] [CrossRef]
- Otwinowski, Z.; Minor, W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997, 276, 307–326. [Google Scholar] [CrossRef] [PubMed]
- Emsley, P.; Cowtan, K. Coot: Model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004, 60, 2126–2132. [Google Scholar] [CrossRef] [Green Version]
- Liebschner, D.; Afonine, P.V.; Baker, M.L.; Bunkoczi, G.; Chen, V.B.; Croll, T.I.; Hintze, B.; Hung, L.W.; Jain, S.; McCoy, A.J.; et al. Macromolecular structure determination using X-rays, neutrons and electrons: Recent developments in Phenix. Acta Crystallogr. D Struct. Biol. 2019, 75, 861–877. [Google Scholar] [CrossRef] [Green Version]
- Williams, C.J.; Headd, J.J.; Moriarty, N.W.; Prisant, M.G.; Videau, L.L.; Deis, L.N.; Verma, V.; Keedy, D.A.; Hintze, B.J.; Chen, V.B.; et al. MolProbity: More and better reference data for improved all-atom structure validation. Protein Sci. 2018, 27, 293–315. [Google Scholar] [CrossRef]
- Ashkenazy, H.; Abadi, S.; Martz, E.; Chay, O.; Mayrose, I.; Pupko, T.; Ben-Tal, N. ConSurf 2016: An improved methodology to estimate and visualize evolutionary conservation in macromolecules. Nucleic Acids Res. 2016, 44, W344–W350. [Google Scholar] [CrossRef] [Green Version]
- The PyMOL Molecular Graphics System, Version 1.8; Schrödinger: New York, NY, USA, 2002; Available online: https://pymol.org/2/ (accessed on 1 June 2022).
- Sievers, F.; Wilm, A.; Dineen, D.; Gibson, T.J.; Karplus, K.; Li, W.; Lopez, R.; McWilliam, H.; Remmert, M.; Soding, J.; et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 2011, 7, 539. [Google Scholar] [CrossRef]

| Data Collection | FnYggS |
|---|---|
| Diffraction source | Beamline 5A, PLS-II |
| Wavelength | 0.9793 |
| Detector | ADSC Q315r CCD |
| Rotation range per image (°) | 1 |
| Total rotation range (°) | 360 |
| Exposure time per image (s) | 0.5 |
| Space group | P21 |
| Cell dimensions | |
| a, b, c (Å) | 37.929, 146.375, 74.128 |
| Resolution (Å) | 36.59–2.08 |
| Completeness | 96.03 (86.73) |
| Redundancy | 4.2 (2.7) |
| I/σ(I) | 27.1304 (4.33) |
| Rsym (%) | 27.1 (60.7) |
| Refinement statistics | |
| Rwork (%) | 18.39 (19.59) |
| Rfree (%) | 21.77 (22.86) |
| B-factor (Averaged) | |
| Protein | 28.83 |
| R.m.s. deviations | |
| Bond lengths (Å) | 0.009 |
| Bond angles (°) | 1.23 |
| Ramachandran plot (%) | |
| favored | 97.59 |
| allowed | 2.26 |
| disallowed regions | 0.15 |
| PDB code | 7YGF |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
He, S.; Chen, Y.; Wang, L.; Bai, X.; Bu, T.; Zhang, J.; Lu, M.; Ha, N.-C.; Quan, C.; Nam, K.H.; et al. Structural and Functional Analysis of the Pyridoxal Phosphate Homeostasis Protein YggS from Fusobacterium nucleatum. Molecules 2022, 27, 4781. https://doi.org/10.3390/molecules27154781
He S, Chen Y, Wang L, Bai X, Bu T, Zhang J, Lu M, Ha N-C, Quan C, Nam KH, et al. Structural and Functional Analysis of the Pyridoxal Phosphate Homeostasis Protein YggS from Fusobacterium nucleatum. Molecules. 2022; 27(15):4781. https://doi.org/10.3390/molecules27154781
Chicago/Turabian StyleHe, Shanru, Yuanyuan Chen, Lulu Wang, Xue Bai, Tingting Bu, Jie Zhang, Ming Lu, Nam-Chul Ha, Chunshan Quan, Ki Hyun Nam, and et al. 2022. "Structural and Functional Analysis of the Pyridoxal Phosphate Homeostasis Protein YggS from Fusobacterium nucleatum" Molecules 27, no. 15: 4781. https://doi.org/10.3390/molecules27154781
APA StyleHe, S., Chen, Y., Wang, L., Bai, X., Bu, T., Zhang, J., Lu, M., Ha, N.-C., Quan, C., Nam, K. H., & Xu, Y. (2022). Structural and Functional Analysis of the Pyridoxal Phosphate Homeostasis Protein YggS from Fusobacterium nucleatum. Molecules, 27(15), 4781. https://doi.org/10.3390/molecules27154781








