Bioactive Peptides and Proteins from Centipede Venoms
Abstract
1. Introduction
2. Centipede Toxins Acting on the Nervous System
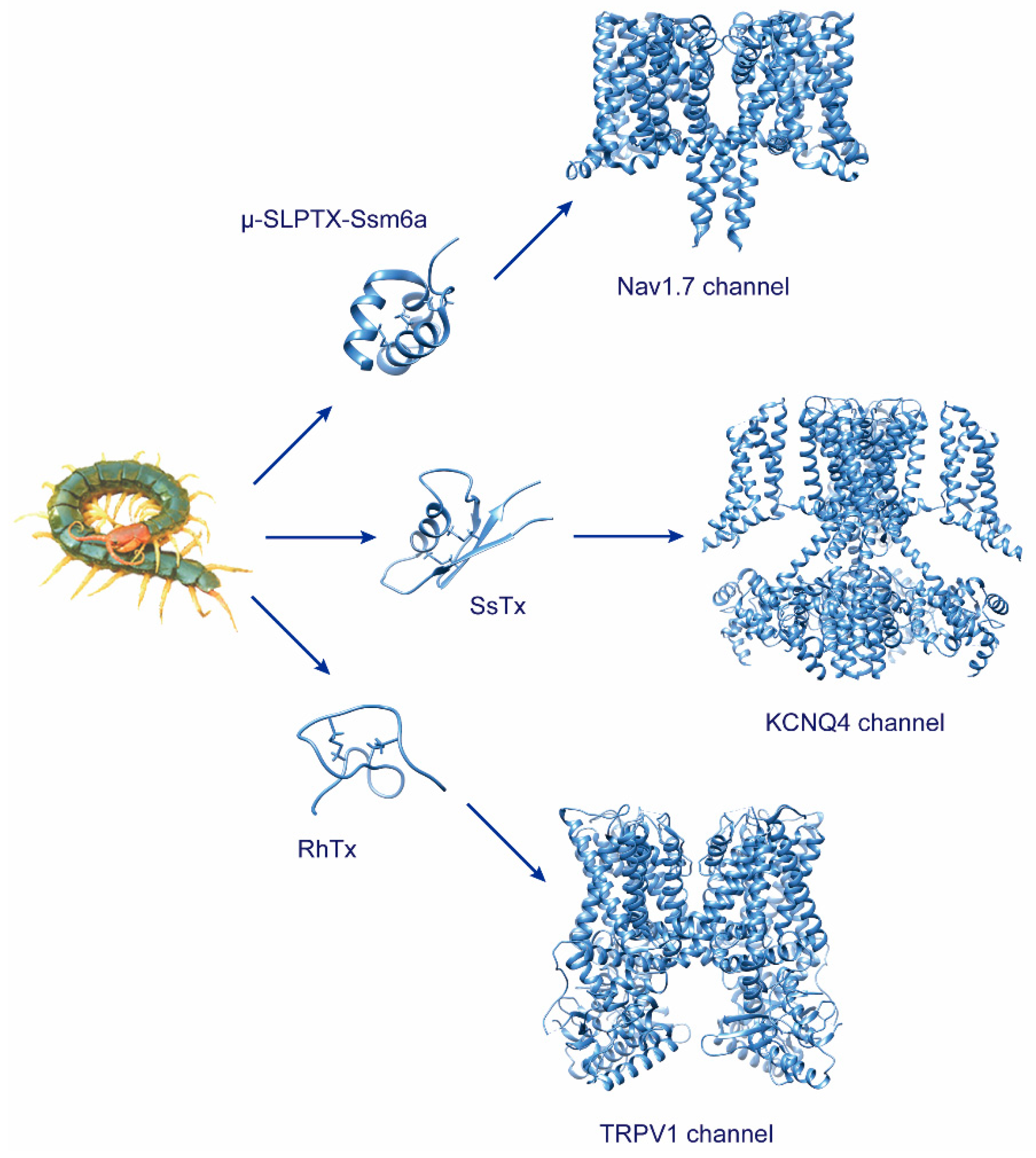
2.1. Toxins Targeting Voltage-Gated Sodium Channels
2.2. Toxins Targeting Voltage-Gated Potassium Channels
2.3. Toxins Targeting Voltage-Gated Calcium Channels
2.4. Toxins Targeting TRPV1 Channel
3. Centipede Toxins Acting on the Immune System
4. Centipede Toxins Acting on the Blood System
5. Centipede Toxins Acting on Other Systems
5.1. Metallopeptidases
5.2. Phospholipase A2
5.3. γ-Glutamyl Transpeptidase
5.4. Other Enzymes
5.5. Other Non-Enzymatic Proteins
6. Therapeutic Potential of Bioactive Peptides and Proteins from Centipede
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Darwin, C. On the Origin of Species; Routledge: London, UK, 1859. [Google Scholar]
- Zhang, Y. Why do we study animal toxins? Zool. Res. 2015, 36, 183–222. [Google Scholar] [CrossRef]
- von Reumont, B.M.; Campbell, L.I.; Jenner, R.A. Quo vadis venomics? A roadmap to neglected venomous invertebrates. Toxins 2014, 6, 3488–3551. [Google Scholar] [CrossRef] [PubMed]
- Schendel, V.; Rash, L.D.; Jenner, R.A.; Undheim, E.A.B. The Diversity of Venom: The Importance of Behavior and Venom System Morphology in Understanding Its Ecology and Evolution. Toxins 2019, 11, 666. [Google Scholar] [CrossRef] [PubMed]
- Nelsen, D.R.; Nisani, Z.; Cooper, A.M.; Fox, G.A.; Gren, E.C.; Corbit, A.G.; Hayes, W.K. Poisons, toxungens, and venoms: Redefining and classifying toxic biological secretions and the organisms that employ them. Biol. Rev. 2014, 89, 450–465. [Google Scholar] [CrossRef]
- Dumbacher, J.P.; Beehler, B.M.; Spande, T.F.; Garraffo, H.M.; Daly, J.W. Homobatrachotoxin in the genus Pitohui: Chemical defense in birds? Science 1992, 258, 799–801. [Google Scholar] [CrossRef] [PubMed]
- Kingdon, J.; Agwanda, B.; Kinnaird, M.; O’Brien, T.; Holland, C.; Gheysens, T.; Boulet-Audet, M.; Vollrath, F. A poisonous surprise under the coat of the African crested rat. Proc. Biol. Sci. 2012, 279, 675–680. [Google Scholar] [CrossRef]
- Undheim, E.A.; King, G.F. On the venom system of centipedes (Chilopoda), a neglected group of venomous animals. Toxicon 2011, 57, 512–524. [Google Scholar] [CrossRef]
- Shear, W.A.; Bonamo, P.M. Devonobiomorpha, a new order of centipeds (Chilopoda) from the Middle Devonian of Gilboa, New York State, USA, and the phylogeny of centiped orders. Am. Mus. Novit. 1988, 2927, 1–30. [Google Scholar]
- Edgecombe, G.D.; Giribet, G. Evolutionary biology of centipedes (Myriapoda: Chilopoda). Annu. Rev. Entomol. 2007, 52, 151–170. [Google Scholar] [CrossRef]
- Chen, M.; Li, J.; Zhang, F.; Liu, Z. Isolation and characterization of SsmTx-I, a Specific Kv2.1 blocker from the venom of the centipede Scolopendra Subspinipes Mutilans, L. Koch. J. Pept. Sci. 2014, 20, 159–164. [Google Scholar] [CrossRef]
- Yang, S.; Liu, Z.; Xiao, Y.; Li, Y.; Rong, M.; Liang, S.; Zhang, Z.; Yu, H.; King, G.F.; Lai, R. Chemical punch packed in venoms makes centipedes excellent predators. Mol. Cell. Proteom. 2012, 11, 640–650. [Google Scholar] [CrossRef] [PubMed]
- Eitzinger, B.; Unger, E.M.; Traugott, M.; Scheu, S. Effects of prey quality and predator body size on prey DNA detection success in a centipede predator. Mol. Ecol. 2014, 23, 3767–3776. [Google Scholar] [CrossRef] [PubMed]
- Forti, L.R.; Fischer, H.Z.; Encarnação, L.C. Treefrog Dendropsophus elegans (Wied-Neuwied, 1824) (Anura: Hylidae) as a meal to Otostigmus tibialis Brölemann, 1902 (Chilopoda: Scolopendridae) in the Tropical Rainforest in southeastern Brazil. Braz. J. Biol. 2007, 67, 583–584. [Google Scholar] [CrossRef] [PubMed]
- Luo, L.; Li, B.; Wang, S.; Wu, F.; Wang, X.; Liang, P.; Ombati, R.; Chen, J.; Lu, X.; Cui, J.; et al. Centipedes subdue giant prey by blocking KCNQ channels. Proc. Natl. Acad. Sci. USA 2018, 115, 1646–1651. [Google Scholar] [CrossRef]
- Kularatne, S.A.; Senanayake, N. Venomous snake bites, scorpions, and spiders. Handb Clin. Neurol. 2014, 120, 987–1001. [Google Scholar] [CrossRef] [PubMed]
- Undheim, E.A.; Grimm, L.L.; Low, C.F.; Morgenstern, D.; Herzig, V.; Zobel-Thropp, P.; Pineda, S.S.; Habib, R.; Dziemborowicz, S.; Fry, B.G.; et al. Weaponization of a Hormone: Convergent Recruitment of Hyperglycemic Hormone into the Venom of Arthropod Predators. Structure 2015, 23, 1283–1292. [Google Scholar] [CrossRef]
- De Lucca Caetano, L.H.; Nishiyama-Jr, M.Y.; de Carvalho Lins Fernandes Tavora, B.; de Oliveira, U.C.; de Loiola Meirelles Junqueira-de-Azevedo, I.; Faquim-Mauro, E.L.; Magalhaes, G.S. Recombinant Production and Characterization of a New Toxin from Cryptops iheringi Centipede Venom Revealed by Proteome and Transcriptome Analysis. Toxins 2021, 13, 858. [Google Scholar] [CrossRef]
- Undheim, E.A.B.; Jenner, R.A. Phylogenetic analyses suggest centipede venom arsenals were repeatedly stocked by horizontal gene transfer. Nat. Commun. 2021, 12, 818. [Google Scholar] [CrossRef]
- Nystrom, G.S.; Ward, M.J.; Ellsworth, S.A.; Rokyta, D.R. Sex-based venom variation in the eastern bark centipede (Hemiscolopendra marginata). Toxicon 2019, 169, 45–58. [Google Scholar] [CrossRef]
- Yang, S.; Xiao, Y.; Kang, D.; Liu, J.; Li, Y.; Undheim, E.A.; Klint, J.K.; Rong, M.; Lai, R.; King, G.F. Discovery of a selective NaV1.7 inhibitor from centipede venom with analgesic efficacy exceeding morphine in rodent pain models. Proc. Natl. Acad. Sci. USA 2013, 110, 17534–17539. [Google Scholar] [CrossRef]
- Liu, Z.C.; Zhang, R.; Zhao, F.; Chen, Z.M.; Liu, H.W.; Wang, Y.J.; Jiang, P.; Zhang, Y.; Wu, Y.; Ding, J.P.; et al. Venomic and transcriptomic analysis of centipede Scolopendra subspinipes dehaani. J. Proteome Res. 2012, 11, 6197–6212. [Google Scholar] [CrossRef] [PubMed]
- Tang, D.; Xu, J.; Li, Y.; Zhao, P.; Kong, X.; Hu, H.; Liang, S.; Tang, C.; Liu, Z. Molecular mechanisms of centipede toxin SsTx-4 inhibition of inwardly rectifying potassium channels. J. Biol. Chem. 2021, 297, 101076. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.; Yang, F.; Wei, N.; Hong, J.; Li, B.; Luo, L.; Rong, M.; Yarov-Yarovoy, V.; Zheng, J.; Wang, K.; et al. A pain-inducing centipede toxin targets the heat activation machinery of nociceptor TRPV1. Nat. Commun. 2015, 6, 8297. [Google Scholar] [CrossRef] [PubMed]
- Zhu, A.; Aierken, A.; Yao, Z.; Vu, S.; Tian, Y.; Zheng, J.; Yang, S.; Yang, F. A centipede toxin causes rapid desensitization of nociceptor TRPV1 ion channel. Toxicon 2020, 178, 41–49. [Google Scholar] [CrossRef]
- Wenhua, R.; Shuangquan, Z.; Daxiang, S.; Kaiya, Z.; Guang, Y. Induction, purification and characterization of an antibacterial peptide scolopendrin I from the venom of centipede Scolopendra subspinipes mutilans. Indian J. BioChem. Biophys. 2006, 43, 88–93. [Google Scholar]
- Peng, K.; Kong, Y.; Zhai, L.; Wu, X.; Jia, P.; Liu, J.; Yu, H. Two novel antimicrobial peptides from centipede venoms. Toxicon 2010, 55, 274–279. [Google Scholar] [CrossRef]
- Choi, H.; Hwang, J.S.; Lee, D.G. Antifungal effect and pore-forming action of lactoferricin B like peptide derived from centipede Scolopendra subspinipes mutilans. Biochim. Biophys. Acta 2013, 1828, 2745–2750. [Google Scholar] [CrossRef]
- Choi, H.; Hwang, J.S.; Lee, D.G. Identification of a novel antimicrobial peptide, scolopendin 1, derived from centipede Scolopendra subspinipes mutilans and its antifungal mechanism. Insect Mol. Biol. 2014, 23, 788–799. [Google Scholar] [CrossRef]
- Lee, H.; Hwang, J.S.; Lee, J.; Kim, J.I.; Lee, D.G. Scolopendin 2, a cationic antimicrobial peptide from centipede, and its membrane-active mechanism. Biochim. Biophys. Acta 2015, 1848, 634–642. [Google Scholar] [CrossRef]
- Lee, J.H.; Kim, I.-W.; Kim, M.-A.; Yun, E.-Y.; Nam, S.-H.; Ahn, M.-Y.; Lee, Y.-B.; Hwang, J.-S. Scolopendrasin I: A novel antimicrobial peptide isolated from the centipede Scolopendra subspinipes mutilans. Int. J. Ind. Entomol. 2015, 31, 14–19. [Google Scholar] [CrossRef][Green Version]
- Kwon, Y.N.; Lee, J.H.; Kim, I.W.; Kim, S.H.; Yun, E.Y.; Nam, S.H.; Ahn, M.Y.; Jeong, M.; Kang, D.C.; Lee, I.H.; et al. Antimicrobial activity of the synthetic peptide scolopendrasin ii from the centipede Scolopendra subspinipes mutilans. J. MicroBiol. Biotechnol. 2013, 23, 1381–1385. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.H.; Kim, I.W.; Kim, M.A.; Ahn, M.Y.; Yun, E.Y.; Hwang, J.S. Antimicrobial Activity of the Scolopendrasin V Peptide Identified from the Centipede Scolopendra subspinipes mutilans. J. MicroBiol. Biotechnol. 2017, 27, 43–48. [Google Scholar] [CrossRef]
- Lee, J.H.; Kim, I.W.; Kim, S.H.; Kim, M.A.; Yun, E.Y.; Nam, S.H.; Ahn, M.Y.; Kang, D.; Hwang, J.S. Anticancer Activity of the Antimicrobial Peptide Scolopendrasin VII Derived from the Centipede, Scolopendra subspinipes mutilans. J. MicroBiol. Biotechnol. 2015, 25, 1275–1280. [Google Scholar] [CrossRef] [PubMed]
- Park, Y.J.; Park, B.; Lee, M.; Jeong, Y.S.; Lee, H.Y.; Sohn, D.H.; Song, J.J.; Lee, J.H.; Hwang, J.S.; Bae, Y.S. A novel antimicrobial peptide acting via formyl peptide receptor 2 shows therapeutic effects against rheumatoid arthritis. Sci. Rep. 2018, 8, 14664. [Google Scholar] [CrossRef] [PubMed]
- Kong, Y.; Shao, Y.; Chen, H.; Ming, X.; Wang, J.B.; Li, Z.Y.; Wei, J.F. A Novel Factor Xa-Inhibiting Peptide from Centipedes Venom. Int. J. Pept. Res. Ther. 2013, 19, 303–311. [Google Scholar] [CrossRef] [PubMed]
- Undheim, E.A.; Jones, A.; Clauser, K.R.; Holland, J.W.; Pineda, S.S.; King, G.F.; Fry, B.G. Clawing through evolution: Toxin diversification and convergence in the ancient lineage Chilopoda (centipedes). Mol. Biol. Evol. 2014, 31, 2124–2148. [Google Scholar] [CrossRef] [PubMed]
- Undheim, E.A.; Fry, B.G.; King, G.F. Centipede venom: Recent discoveries and current state of knowledge. Toxins 2015, 7, 679–704. [Google Scholar] [CrossRef]
- Chahl, L.A.; Kirk, E.J. Toxins which produce pain. Pain 1975, 1, 3–49. [Google Scholar] [CrossRef]
- Welsh, J.H.; Batty, C.S. 5-Hydroxytryptamine content of some arthropod venoms and venom-containing parts. Toxicon 1963, 1, 165–170. [Google Scholar] [CrossRef]
- Mohamed, A.H.; Abu-Sinna, G.; El-Shabaka, H.A.; El-Aal, A.A. Proteins, lipids, lipoproteins and some enzyme characterizations of the venom extract from the centipede Scolopendra morsitans. Toxicon 1983, 21, 371–377. [Google Scholar] [CrossRef]
- Gomes, A.; Datta, A.; Sarangi, B.; Kar, P.K.; Lahiri, S.C. Occurrence of histamine and histamine release by centipede venom. Indian J. Med. Res. 1982, 76, 888–891. [Google Scholar] [PubMed]
- Zhao, H.; Li, Y.; Wang, Y.; Zhang, J.; Ouyang, X.; Peng, R.; Yang, J. Antitumor and immunostimulatory activity of a polysaccharide-protein complex from Scolopendra subspinipes mutilans L. Koch in tumor-bearing mice. Food Chem. Toxicol. 2012, 50, 2648–2655. [Google Scholar] [CrossRef] [PubMed]
- Malta, M.B.; Lira, M.S.; Soares, S.L.; Rocha, G.C.; Knysak, I.; Martins, R.; Guizze, S.P.; Santoro, M.L.; Barbaro, K.C. Toxic activities of Brazilian centipede venoms. Toxicon 2008, 52, 255–263. [Google Scholar] [CrossRef] [PubMed]
- Kong, Y.; Huang, S.L.; Shao, Y.; Li, S.; Wei, J.F. Purification and characterization of a novel antithrombotic peptide from Scolopendra subspinipes mutilans. J. Ethnopharmacol. 2013, 145, 182–186. [Google Scholar] [CrossRef]
- You, W.K.; Sohn, Y.D.; Kim, K.Y.; Park, D.H.; Jang, Y.; Chung, K.H. Purification and molecular cloning of a novel serine protease from the centipede, Scolopendra subspinipes mutilans. Insect BioChem. Mol. Biol. 2004, 34, 239–250. [Google Scholar] [CrossRef]
- Zhao, F.; Lan, X.; Li, T.; Xiang, Y.; Zhao, F.; Zhang, Y.; Lee, W.H. Proteotranscriptomic Analysis and Discovery of the Profile and Diversity of Toxin-like Proteins in Centipede. Mol. Cell. Proteom. 2018, 17, 709–720. [Google Scholar] [CrossRef]
- Chu, Y.; Qiu, P.; Yu, R. Centipede Venom Peptides Acting on Ion Channels. Toxins 2020, 12, 230. [Google Scholar] [CrossRef]
- Dash, T.S.; Shafee, T.; Harvey, P.J.; Zhang, C.; Peigneur, S.; Deuis, J.R.; Vetter, I.; Tytgat, J.; Anderson, M.A.; Craik, D.J.; et al. A Centipede Toxin Family Defines an Ancient Class of CSalphabeta Defensins. Structure 2019, 27, 315–326.e7. [Google Scholar] [CrossRef]
- Hakim, M.A.; Yang, S.; Lai, R. Centipede venoms and their components: Resources for potential therapeutic applications. Toxins 2015, 7, 4832–4851. [Google Scholar] [CrossRef]
- Nguyen, P.T.; Yarov-Yarovoy, V. Towards Structure-Guided Development of Pain Therapeutics Targeting Voltage-Gated Sodium Channels. Front. Pharmacol. 2022, 13, 842032. [Google Scholar] [CrossRef]
- Payandeh, J.; Scheuer, T.; Zheng, N.; Catterall, W.A. The crystal structure of a voltage-gated sodium channel. Nature 2011, 475, 353–358. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Y.; Blumenthal, K.; Jackson, J.O., 2nd; Liang, S.; Cummins, T.R. The tarantula toxins ProTx-II and huwentoxin-IV differentially interact with human Nav1.7 voltage sensors to inhibit channel activation and inactivation. Mol. Pharmacol. 2010, 78, 1124–1134. [Google Scholar] [CrossRef] [PubMed]
- Wright, Z.V.F.; McCarthy, S.; Dickman, R.; Reyes, F.E.; Sanchez-Martinez, S.; Cryar, A.; Kilford, I.; Hall, A.; Takle, A.K.; Topf, M.; et al. The Role of Disulfide Bond Replacements in Analogues of the Tarantula Toxin ProTx-II and Their Effects on Inhibition of the Voltage-Gated Sodium Ion Channel Nav1.7. J. Am. Chem. Soc. 2017, 139, 13063–13075. [Google Scholar] [CrossRef] [PubMed]
- González-Morales, L.; Pedraza-Escalona, M.; Diego-Garcia, E.; Restano-Cassulini, R.; Batista, C.V.; Gutiérrez Mdel, C.; Possani, L.D. Proteomic characterization of the venom and transcriptomic analysis of the venomous gland from the Mexican centipede Scolopendra viridis. J. Proteom. 2014, 111, 224–237. [Google Scholar] [CrossRef]
- Rong, M.; Yang, S.; Wen, B.; Mo, G.; Kang, D.; Liu, J.; Lin, Z.; Jiang, W.; Li, B.; Du, C.; et al. Peptidomics combined with cDNA library unravel the diversity of centipede venom. J. Proteom. 2015, 114, 28–37. [Google Scholar] [CrossRef]
- Du, C.; Li, J.; Shao, Z.; Mwangi, J.; Xu, R.; Tian, H.; Mo, G.; Lai, R.; Yang, S. Centipede KCNQ Inhibitor SsTx Also Targets K(V)1.3. Toxins 2019, 11, 76. [Google Scholar] [CrossRef] [PubMed]
- Ramu, Y.; Lu, Z. A family of orthologous proteins from centipede venoms inhibit the hKir6.2 channel. Sci. Rep. 2019, 9, 14088. [Google Scholar] [CrossRef] [PubMed]
- Aguilar, M.B.; Perez-Reyes, L.I.; Lopez, Z.; de la Cotera, E.P.; Falcon, A.; Ayala, C.; Galvan, M.; Salvador, C.; Escobar, L.I. Peptide sr11a from Conus spurius is a novel peptide blocker for Kv1 potassium channels. Peptides 2010, 31, 1287–1291. [Google Scholar] [CrossRef] [PubMed]
- Julius, D. TRP channels and pain. Annu. Rev. Cell Dev. Biol. 2013, 29, 355–384. [Google Scholar] [CrossRef]
- Luo, L.; Wang, Y.; Li, B.; Xu, L.; Kamau, P.M.; Zheng, J.; Yang, F.; Yang, S.; Lai, R. Molecular basis for heat desensitization of TRPV1 ion channels. Nat. Commun. 2019, 10, 2134. [Google Scholar] [CrossRef]
- Farsky, S.H.; Antunes, E.; Mello, S.B. Pro and antiinflammatory properties of toxins from animal venoms. Curr. Drug Targets Inflamm. Allergy 2005, 4, 401–411. [Google Scholar] [CrossRef] [PubMed]
- Harzsch, S. Phylogenetic comparison of serotonin-immunoreactive neurons in representatives of the Chilopoda, Diplopoda, and Chelicerata: Implications for arthropod relationships. J. Morphol. 2004, 259, 198–213. [Google Scholar] [CrossRef] [PubMed]
- Rates, B.; Bemquerer, M.P.; Richardson, M.; Borges, M.H.; Morales, R.A.; De Lima, M.E.; Pimenta, A.M. Venomic analyses of Scolopendra viridicornis nigra and Scolopendra angulata (Centipede, Scolopendromorpha): Shedding light on venoms from a neglected group. Toxicon 2007, 49, 810–826. [Google Scholar] [CrossRef] [PubMed]
- Dos Santos, A.T.; Cruz, G.S.; Baptista, G.R. Anti-inflammatory activities of arthropod peptides: A systematic review. J. Venom. Anim. Toxins Trop. Dis. 2021, 27, e20200152. [Google Scholar] [CrossRef]
- Koh, C.Y.; Kini, R.M. Anticoagulants from hematophagous animals. Expert. Rev. Hematol. 2008, 1, 135–139. [Google Scholar] [CrossRef]
- Champagne, D.E. Antihemostatic strategies of blood-feeding arthropods. Curr. Drug Targets Cardiovasc. Haematol. Disord. 2004, 4, 375–396. [Google Scholar] [CrossRef]
- Gan, W.; Deng, L.; Yang, C.; He, Q.; Hu, J.; Yin, H.; Jin, X.; Lu, C.; Wu, Y.; Peng, L. An anticoagulant peptide from the human hookworm, Ancylostoma duodenale that inhibits coagulation factors Xa and XIa. FEBS Lett. 2009, 583, 1976–1980. [Google Scholar] [CrossRef]
- Wu, Y.; Li, J.; Wang, J.; Si, Q.; Zhang, J.; Jiang, Y.; Chu, L. Anti-atherogenic effects of centipede acidic protein in rats fed an atherogenic diet. J. Ethnopharmacol. 2009, 122, 509–516. [Google Scholar] [CrossRef]
- Ward, M.J.; Rokyta, D.R. Venom-gland transcriptomics and venom proteomics of the giant Florida blue centipede, Scolopendra viridis. Toxicon 2018, 152, 121–136. [Google Scholar] [CrossRef]
- Ellsworth, S.A.; Nystrom, G.S.; Ward, M.J.; Freitas de Sousa, L.A.; Hogan, M.P.; Rokyta, D.R. Convergent recruitment of adamalysin-like metalloproteases in the venom of the red bark centipede (Scolopocryptops sexspinosus). Toxicon 2019, 168, 1–15. [Google Scholar] [CrossRef]
- Francischetti, I.M.; Mather, T.N.; Ribeiro, J.M. Cloning of a salivary gland metalloprotease and characterization of gelatinase and fibrin(ogen)lytic activities in the saliva of the Lyme disease tick vector Ixodes scapularis. Biochem. Biophys. Res. Commun. 2003, 305, 869–875. [Google Scholar] [CrossRef]
- de Graaf, D.C.; Aerts, M.; Brunain, M.; Desjardins, C.A.; Jacobs, F.J.; Werren, J.H.; Devreese, B. Insights into the venom composition of the ectoparasitoid wasp Nasonia vitripennis from bioinformatic and proteomic studies. Insect Mol. Biol. 2010, 19 (Suppl. 1), 11–26. [Google Scholar] [CrossRef] [PubMed]
- Wong, E.S.; Papenfuss, A.T.; Whittington, C.M.; Warren, W.C.; Belov, K. A limited role for gene duplications in the evolution of platypus venom. Mol. Biol. Evol. 2012, 29, 167–177. [Google Scholar] [CrossRef]
- Brust, A.; Sunagar, K.; Undheim, E.A.; Vetter, I.; Yang, D.C.; Casewell, N.R.; Jackson, T.N.; Koludarov, I.; Alewood, P.F.; Hodgson, W.C.; et al. Differential evolution and neofunctionalization of snake venom metalloprotease domains. Mol. Cell. Proteom. 2013, 12, 651–663. [Google Scholar] [CrossRef] [PubMed]
- Ruder, T.; Sunagar, K.; Undheim, E.A.; Ali, S.A.; Wai, T.C.; Low, D.H.; Jackson, T.N.; King, G.F.; Antunes, A.; Fry, B.G. Molecular phylogeny and evolution of the proteins encoded by coleoid (cuttlefish, octopus, and squid) posterior venom glands. J. Mol. Evol. 2013, 76, 192–204. [Google Scholar] [CrossRef] [PubMed]
- Weston, A.J.; Chung, R.; Dunlap, W.C.; Morandini, A.C.; Marques, A.C.; Moura-da-Silva, A.M.; Ward, M.; Padilla, G.; da Silva, L.F.; Andreakis, N.; et al. Proteomic characterisation of toxins isolated from nematocysts of the South Atlantic jellyfish Olindias sambaquiensis. Toxicon 2013, 71, 11–17. [Google Scholar] [CrossRef]
- Jenner, R.A.; von Reumont, B.M.; Campbell, L.I.; Undheim, E.A.B. Parallel Evolution of Complex Centipede Venoms Revealed by Comparative Proteotranscriptomic Analyses. Mol. Biol. Evol. 2019, 36, 2748–2763. [Google Scholar] [CrossRef]
- Fry, B.G.; Roelants, K.; Champagne, D.E.; Scheib, H.; Tyndall, J.D.; King, G.F.; Nevalainen, T.J.; Norman, J.A.; Lewis, R.J.; Norton, R.S.; et al. The toxicogenomic multiverse: Convergent recruitment of proteins into animal venoms. Annu. Rev. Genom. Hum. Genet. 2009, 10, 483–511. [Google Scholar] [CrossRef]
- Gonzalez-Morales, L.; Diego-Garcia, E.; Segovia, L.; Gutierrez Mdel, C.; Possani, L.D. Venom from the centipede Scolopendra viridis Say: Purification, gene cloning and phylogenetic analysis of a phospholipase A2. Toxicon 2009, 54, 8–15. [Google Scholar] [CrossRef]
- Condrea, E.; Devries, A.; Mager, J. Hemolysis and splitting of human erythrocyte phospholipids by snake venoms. Biochim. Biophys. Acta 1964, 84, 60–73. [Google Scholar] [CrossRef]
- Xie, C.; Bittenbinder, M.A.; Slagboom, J.; Arrahman, A.; Bruijns, S.; Somsen, G.W.; Vonk, F.J.; Casewell, N.R.; García-Vallejo, J.J.; Kool, J. Erythrocyte haemotoxicity profiling of snake venom toxins after nanofractionation. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 2021, 1176, 122586. [Google Scholar] [CrossRef] [PubMed]
- Condrea, E.; Mammon, Z.; Aloof, S.; Devries, A. Susceptibility of Erythrocytes of Various Animal Species to the Hemolytic and Phospholipid Splitting Action of Snake Venom. Biochim. Biophys. Acta 1964, 84, 365–375. [Google Scholar] [CrossRef]
- Courtay, C.; Oster, T.; Michelet, F.; Visvikis, A.; Diederich, M.; Wellman, M.; Siest, G. Gamma-glutamyltransferase: Nucleotide sequence of the human pancreatic cDNA. Evidence for a ubiquitous gamma-glutamyltransferase polypeptide in human tissues. Biochem. Pharmacol. 1992, 43, 2527–2533. [Google Scholar] [CrossRef]
- Balasubramanian, P.G.; Beckmann, A.; Warnken, U.; Schnölzer, M.; Schüler, A.; Bornberg-Bauer, E.; Holstein, T.W.; Özbek, S. Proteome of Hydra nematocyst. J. Biol. Chem. 2012, 287, 9672–9681. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Zhao, L.; Jiang, L.; Meng, E.; Zhang, Y.; Xiong, X.; Liang, S. Transcriptome analysis revealed novel possible venom components and cellular processes of the tarantula Chilobrachys jingzhao venom gland. Toxicon 2008, 52, 794–806. [Google Scholar] [CrossRef] [PubMed]
- Fernandes-Pedrosa Mde, F.; Junqueira-de-Azevedo Ide, L.; Gonçalves-de-Andrade, R.M.; Kobashi, L.S.; Almeida, D.D.; Ho, P.L.; Tambourgi, D.V. Transcriptome analysis of Loxosceles laeta (Araneae, Sicariidae) spider venomous gland using expressed sequence tags. BMC Genom. 2008, 9, 279. [Google Scholar] [CrossRef] [PubMed]
- Kramer, K.J.; Muthukrishnan, S. Insect chitinases: Molecular biology and potential use as biopesticides. Insect Biochem. Mol. Biol. 1997, 27, 887–900. [Google Scholar] [CrossRef]
- Vincent, B.; Kaeslin, M.; Roth, T.; Heller, M.; Poulain, J.; Cousserans, F.; Schaller, J.; Poirié, M.; Lanzrein, B.; Drezen, J.M.; et al. The venom composition of the parasitic wasp Chelonus inanitus resolved by combined expressed sequence tags analysis and proteomic approach. BMC Genom. 2010, 11, 693. [Google Scholar] [CrossRef]
- Chung, L.P.; Keshav, S.; Gordon, S. Cloning the human lysozyme cDNA: Inverted Alu repeat in the mRNA and in situ hybridization for macrophages and Paneth cells. Proc. Natl. Acad. Sci. USA 1988, 85, 6227–6231. [Google Scholar] [CrossRef]
- Low, D.H.; Sunagar, K.; Undheim, E.A.; Ali, S.A.; Alagon, A.C.; Ruder, T.; Jackson, T.N.; Pineda Gonzalez, S.; King, G.F.; Jones, A.; et al. Dracula’s children: Molecular evolution of vampire bat venom. J. Proteom. 2013, 89, 95–111. [Google Scholar] [CrossRef]
- Girish, K.S.; Shashidharamurthy, R.; Nagaraju, S.; Gowda, T.V.; Kemparaju, K. Isolation and characterization of hyaluronidase a “spreading factor” from Indian cobra (Naja naja) venom. Biochimie 2004, 86, 193–202. [Google Scholar] [CrossRef] [PubMed]
- Violette, A.; Leonardi, A.; Piquemal, D.; Terrat, Y.; Biass, D.; Dutertre, S.; Noguier, F.; Ducancel, F.; Stöcklin, R.; Križaj, I.; et al. Recruitment of glycosyl hydrolase proteins in a cone snail venomous arsenal: Further insights into biomolecular features of Conus venoms. Mar. Drugs 2012, 10, 258–280. [Google Scholar] [CrossRef] [PubMed]
- Kuhn-Nentwig, L.; Schaller, J.; Nentwig, W. Biochemistry, toxicology and ecology of the venom of the spider Cupiennius salei (Ctenidae). Toxicon 2004, 43, 543–553. [Google Scholar] [CrossRef] [PubMed]
- Bak, T.G. Studies on glucose dehydrogenase of Aspergillus oryzae: II. Purification and physical and chemical properties. Biochim. Biophys. Acta 1967, 139, 277–293. [Google Scholar] [CrossRef]
- Undheim, E.A.; Georgieva, D.N.; Thoen, H.H.; Norman, J.A.; Mork, J.; Betzel, C.; Fry, B.G. Venom on ice: First insights into Antarctic octopus venoms. Toxicon 2010, 56, 897–913. [Google Scholar] [CrossRef] [PubMed]
- Heitz, J.R.; Norment, B.R. Characteristics of an alkaline phosphatase activity in brown recluse venom. Toxicon 1974, 12, 181–187. [Google Scholar] [CrossRef]
- Rodrigues, M.C.; Guimarães, L.H.; Liberato, J.L.; de Lourdes Teixeira de Moraes Polizeli, M.; dos Santos, W.F. Acid and alkaline phosphatase activities of a fraction isolated from Parawixia bistriata spider venom. Toxicon 2006, 47, 854–858. [Google Scholar] [CrossRef]
- Sulkowski, E.; Bjork, W.; Laskowski, M., Sr. A specific and nonspecific alkaline monophosphatase in the venom of Bothrops atrox and their occurrence in the purified venom phosphodiesterase. J. Biol. Chem. 1963, 238, 2477–2486. [Google Scholar] [CrossRef]
- Tu, A.T.; Chua, A. Acid and alkaline phosphomonoesterase activities in snake venoms. Comp. BioChem. Physiol 1966, 17, 297–307. [Google Scholar] [CrossRef]
- Cociancich, S.; Goyffon, M.; Bontems, F.; Bulet, P.; Bouet, F.; Menez, A.; Hoffmann, J. Purification and characterization of a scorpion defensin, a 4kDa antibacterial peptide presenting structural similarities with insect defensins and scorpion toxins. Biochem. Biophys. Res. Commun. 1993, 194, 17–22. [Google Scholar] [CrossRef]
- Dai, C.; Ma, Y.; Zhao, Z.; Zhao, R.; Wang, Q.; Wu, Y.; Cao, Z.; Li, W. Mucroporin, the first cationic host defense peptide from the venom of Lychas mucronatus. Antimicrob. Agents Chemother. 2008, 52, 3967–3972. [Google Scholar] [CrossRef] [PubMed]
- Du, Q.; Hou, X.; Wang, L.; Zhang, Y.; Xi, X.; Wang, H.; Zhou, M.; Duan, J.; Wei, M.; Chen, T.; et al. AaeAP1 and AaeAP2: Novel antimicrobial peptides from the venom of the scorpion, Androctonus aeneas: Structural characterisation, molecular cloning of biosynthetic precursor-encoding cDNAs and engineering of analogues with enhanced antimicrobial and anticancer activities. Toxins 2015, 7, 219–237. [Google Scholar] [CrossRef] [PubMed]
- Primon-Barros, M.; Jose Macedo, A. Animal Venom Peptides: Potential for New Antimicrobial Agents. Curr. Top. Med. Chem. 2017, 17, 1119–1156. [Google Scholar] [CrossRef] [PubMed]
- Shen, C.; Liu, M.; Mackeigan, D.T.; Chen, Z.Y.; Chen, P.; Karakas, D.; Li, J.; Norris, P.A.A.; Li, J.; Deng, Y.; et al. Viper venoms drive the macrophages and hepatocytes to sequester and clear platelets: Novel mechanism and therapeutic strategy for venom-induced thrombocytopenia. Arch. Toxicol. 2021, 95, 3589–3599. [Google Scholar] [CrossRef]
- Al-Sadoon, M.K.; Rabah, D.M.; Badr, G. Enhanced anticancer efficacy of snake venom combined with silica nanoparticles in a murine model of human multiple myeloma: Molecular targets for cell cycle arrest and apoptosis induction. Cell. Immunol. 2013, 284, 129–138. [Google Scholar] [CrossRef]
- McDermott, A. News Feature: Venom back in vogue as a wellspring for drug candidates. Proc. Natl. Acad. Sci. USA 2020, 117, 10100–10104. [Google Scholar] [CrossRef]
- Ahorukomeye, P.; Disotuar, M.M.; Gajewiak, J.; Karanth, S.; Watkins, M.; Robinson, S.D.; Florez Salcedo, P.; Smith, N.A.; Smith, B.J.; Schlegel, A.; et al. Fish-hunting cone snail venoms are a rich source of minimized ligands of the vertebrate insulin receptor. Elife 2019, 8, e41574. [Google Scholar] [CrossRef]
- Cook Sangar, M.L.; Girard, E.J.; Hopping, G.; Yin, C.; Pakiam, F.; Brusniak, M.Y.; Nguyen, E.; Ruff, R.; Gewe, M.M.; Byrnes-Blake, K.; et al. A potent peptide-steroid conjugate accumulates in cartilage and reverses arthritis without evidence of systemic corticosteroid exposure. Sci. Transl. Med. 2020, 12, eaay1041. [Google Scholar] [CrossRef]
- Jimenez, R.; Ikonomopoulou, M.P.; Lopez, J.A.; Miles, J.J. Immune drug discovery from venoms. Toxicon 2018, 141, 18–24. [Google Scholar] [CrossRef]
- Hübner, C.A.; Jentsch, T.J. Ion channel diseases. Hum. Mol. Genet. 2002, 11, 2435–2445. [Google Scholar] [CrossRef]
- Coughlin, S.R. Thrombin signalling and protease-activated receptors. Nature 2000, 407, 258–264. [Google Scholar] [CrossRef] [PubMed]
- Di Cera, E. Serine proteases. IUBMB Life 2009, 61, 510–515. [Google Scholar] [CrossRef] [PubMed]
- Macfarlane, S.R.; Seatter, M.J.; Kanke, T.; Hunter, G.D.; Plevin, R. Proteinase-activated receptors. Pharmacol. Rev. 2001, 53, 245–282. [Google Scholar]
- Molinari, F.; Meskanaite, V.; Munnich, A.; Sonderegger, P.; Colleaux, L. Extracellular proteases and their inhibitors in genetic diseases of the central nervous system. Hum. Mol. Genet. 2003, 12, R195–R200. [Google Scholar] [CrossRef] [PubMed]
- Salvador, L.A.; Taori, K.; Biggs, J.S.; Jakoncic, J.; Ostrov, D.A.; Paul, V.J.; Luesch, H. Potent elastase inhibitors from cyanobacteria: Structural basis and mechanisms mediating cytoprotective and anti-inflammatory effects in bronchial epithelial cells. J. Med. Chem. 2013, 56, 1276–1290. [Google Scholar] [CrossRef] [PubMed]
- Turk, B. Targeting proteases: Successes, failures and future prospects. Nat. Rev. Drug Discov. 2006, 5, 785–799. [Google Scholar] [CrossRef]
- Puente, X.S.; Sánchez, L.M.; Overall, C.M.; López-Otín, C. Human and mouse proteases: A comparative genomic approach. Nat. Rev. Genet. 2003, 4, 544–558. [Google Scholar] [CrossRef]
- Cushman, D.W.; Ondetti, M.A. History of the design of captopril and related inhibitors of angiotensin converting enzyme. Hypertension 1991, 17, 589–592. [Google Scholar] [CrossRef]
- Li, B.; Lyu, P.; Xi, X.; Ge, L.; Mahadevappa, R.; Shaw, C.; Kwok, H.F. Triggering of cancer cell cycle arrest by a novel scorpion venom-derived peptide-Gonearrestide. J. Cell. Mol. Med. 2018, 22, 4460–4473. [Google Scholar] [CrossRef]
- Swenson, S.; Minea, R.O.; Tuan, C.D.; Thein, T.Z.; Chen, T.C.; Markland, F.S. A Novel Venom-Derived Peptide for Brachytherapy of Glioblastoma: Preclinical Studies in Mice. Molecules 2018, 23, 2918. [Google Scholar] [CrossRef]
- Miljanich, G.P. Ziconotide: Neuronal calcium channel blocker for treating severe chronic pain. Curr. Med. Chem. 2004, 11, 3029–3040. [Google Scholar] [CrossRef] [PubMed]
- Kornalík, F. The influence of snake venom enzymes on blood coagulation. Pharmacol. Ther. 1985, 29, 353–405. [Google Scholar] [CrossRef]
- Giordano, A.; Musumeci, G.; D’Angelillo, A.; Rossini, R.; Zoccai, G.B.; Messina, S.; Coscioni, E.; Romano, S.; Romano, M.F. Effects of Glycoprotein IIb/IIIa Antagonists: Anti Platelet Aggregation And Beyond. Curr. Drug Metab. 2016, 17, 194–203. [Google Scholar] [CrossRef] [PubMed]

| Venom Components | Centipede Species | Activities | Component Source | References |
|---|---|---|---|---|
| μ-SLPTX-Ssm1a | S. subspinipes mutilans | TTX-S NaV channel inhibitor | Venom | [12] |
| μ-SLPTX-Ssm6a | S. subspinipes mutilans | NaV1.7 channel inhibitor | Venom | [21] |
| ω-SLPTX-Ssm1a | S. subspinipes | Activator of Cav channels in DRG | Venom | [12] |
| ω-SLPTX-Ssm2a | S. subspinipes | Inhibitor of CaV channels in DRG | Venom | [12] |
| κ-SLPTX-Ssm1a | S. subspinipes mutilans | Inhibitor of KV channels in DRG | Venom | [12] |
| κ-SLPTX-Ssm2a | S. subspinipes mutilans | Inhibitor of KV channels in DRG | Venom | [12] |
| κ-SLPTX-Ssm3a | S. subspinipes mutilans | Inhibitor of KV channels in DRG | Venom | [12] |
| SSD559 | S. subspinipes dehaani | Inhibitor of KV channels in DRG | Venom | [22] |
| SsTx | S. subspinipes mutilans | Inhibitor of KCNQ4 and KV1.3 channel | Venom | [15] |
| SsTx-4 | S. subspinipes mutilans | Inhibitor of Kir1.1, Kir4.1 and Kir6.2/SUR1 channels | Venom | [23] |
| SSD1052 | S. subspinipes dehaani | Inhibitor of CaV channels in DRG | Venom | [22] |
| RhTx | S. subspinipes mutilans | TRPV1 channel activator | Venom | [24] |
| RhTx2 | S. subspinipes mutilans | TRPV1 channel activator | Venom | [25] |
| Scolopendrin I | S. subspinipes mutilans | Antimicrobial activity | Venom | [26] |
| Scolopin 1 | S. subspinipes mutilans | Antimicrobial activity | Venom | [27] |
| Scolopin 2 | S. subspinipes mutilans | Antimicrobial activity | Venom | [27] |
| LBLP | S. subspinipes mutilans | Antifungal activity | Whole centipede | [28] |
| Scolopendin 1 | S. subspinipes mutilans | Antimicrobial activity | Whole centipede | [29] |
| Scolopendin 2 | S. subspinipes mutilans | Antimicrobial activity | Whole centipede | [30] |
| Scolopendrasin I | S. subspinipes mutilans | Antimicrobial activity | Whole centipede | [31] |
| Scolopendrasin II | S. subspinipes mutilans | Antimicrobial activity | Whole centipede | [32] |
| Scolopendrasin V | S. subspinipes mutilans | Antimicrobial activity | Whole centipede | [33] |
| Scolopendrasin VII | S. subspinipes mutilans | Antimicrobial activity; Anticancer activity | Whole centipede | [34] |
| Scolopendrasin IX | S. subspinipes mutilans | Anti-inflammatory activity | Whole centipede | [35] |
| TNGYT | S. subspinipes mutilans | FXa inhibitor | Venom | [36] |
| SSD14 | S. subspinipes dehaani | γ-Glutamyl Transpeptidase, platelet aggregation and hemolytic activities | Venom | [22] |
| Trypsin-like S1 family | S. subspinipes dehaani | Serine peptidases, potentially involved in activation of toxins | Venom | [37,38] |
| Subtilisin-like S8 family | E. rubripes C. westwoodi S. subspinipes dehaani | Serine peptidases, potentially involved in activation of toxins | Venom | [37,38] |
| β-pore-forming toxins | Scolopendrids | In cell membranes, it has the potential to cause cytotoxicity by forming polymeric pores structures in cell membranes | Venom | [37,38] |
| CAP (cysteine rich proteins) protein | Scolopendrids | Unknown (CAP1, CAP3); CaV channel antagonist and trypsin inhibitor (CAP2) | Venom | [37,38] |
| Serotonin | S. viridicornis | Analgesic activity | Venom | [39,40] |
| Histamine | S. subspinipes | Analgesic activity | Venom | [41,42] |
| Transferrin | E. rubripes S. morsitans | Potential antimicrobial activity | Venom | [37,38] |
| Polysaccharide–protein complex | S. subspinipes mutilans | Inhibitor of tumor cells | Whole centipede | [43] |
| Hyaluronidase | Scolopendrids | Glycosaminoglycan degradation; potential for spreading of venom components | Venom | [37,44] |
| Cystatin type-1 | E. rubripes | Potential peptidase inhibitors | Venom | [37,38] |
| Antithrombotic peptide SQL | E. rubripes | Inhibitor of platelet aggregation | Whole centipede | [45] |
| Lysozyme C | Scolopendrids | Potential antimicrobial activity | Venom | [37,38] |
| Scolonase | S. subspinipes mutilans | Fibrinolytic activity; Serine peptidase | Whole centipede | [46] |
| Phospholipase A2 | S. viridis S. subspinipes dehaan S. viridicornis O. pradoi | Hydrolysis of glycerophospholipids; involved in anti-inflammatory, hemolysis, neurotoxicity, and cardiotoxicity | Venom | [15,47,48] |
| CentiPAD | T. longicornis L. forficatus | Peptidylarginine deiminase, potentially involved in post-translational modification of toxins | Venom | [19] |
| LDLA protein | Scolopendrids | Unknown | Venom | [37,38] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Han, Y.; Kamau, P.M.; Lai, R.; Luo, L. Bioactive Peptides and Proteins from Centipede Venoms. Molecules 2022, 27, 4423. https://doi.org/10.3390/molecules27144423
Han Y, Kamau PM, Lai R, Luo L. Bioactive Peptides and Proteins from Centipede Venoms. Molecules. 2022; 27(14):4423. https://doi.org/10.3390/molecules27144423
Chicago/Turabian StyleHan, Yalan, Peter Muiruri Kamau, Ren Lai, and Lei Luo. 2022. "Bioactive Peptides and Proteins from Centipede Venoms" Molecules 27, no. 14: 4423. https://doi.org/10.3390/molecules27144423
APA StyleHan, Y., Kamau, P. M., Lai, R., & Luo, L. (2022). Bioactive Peptides and Proteins from Centipede Venoms. Molecules, 27(14), 4423. https://doi.org/10.3390/molecules27144423







