Fatty Acid Composition and Cytotoxic Activity of Lipid Extracts from Nannochloropsis gaditana Produced by Green Technologies
Abstract
:1. Introduction
2. Results and Discussion
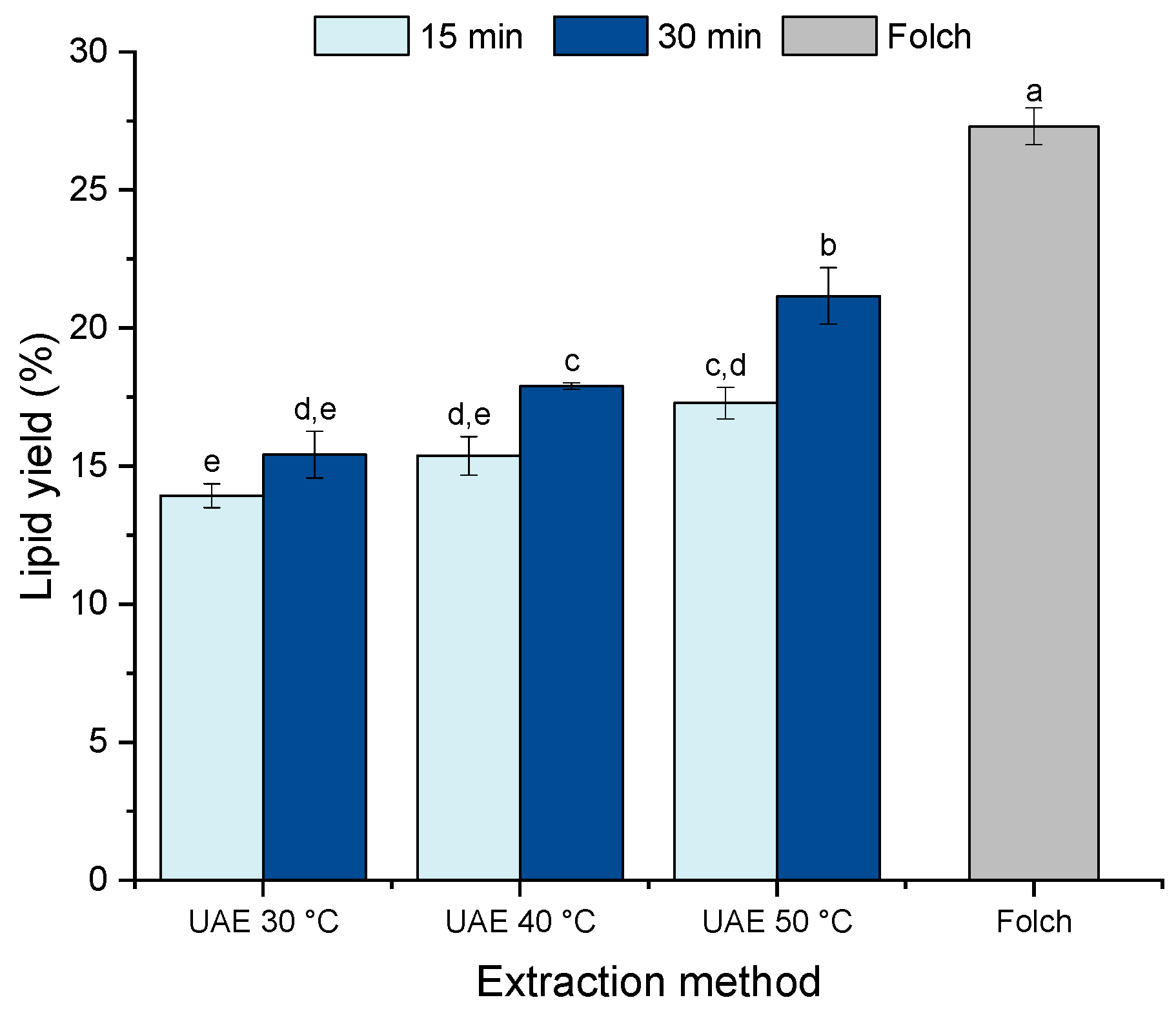
2.1. Effect of Ultrasound-Assisted Extraction on the Lipid Recovery from Nannochloropsis gaditana
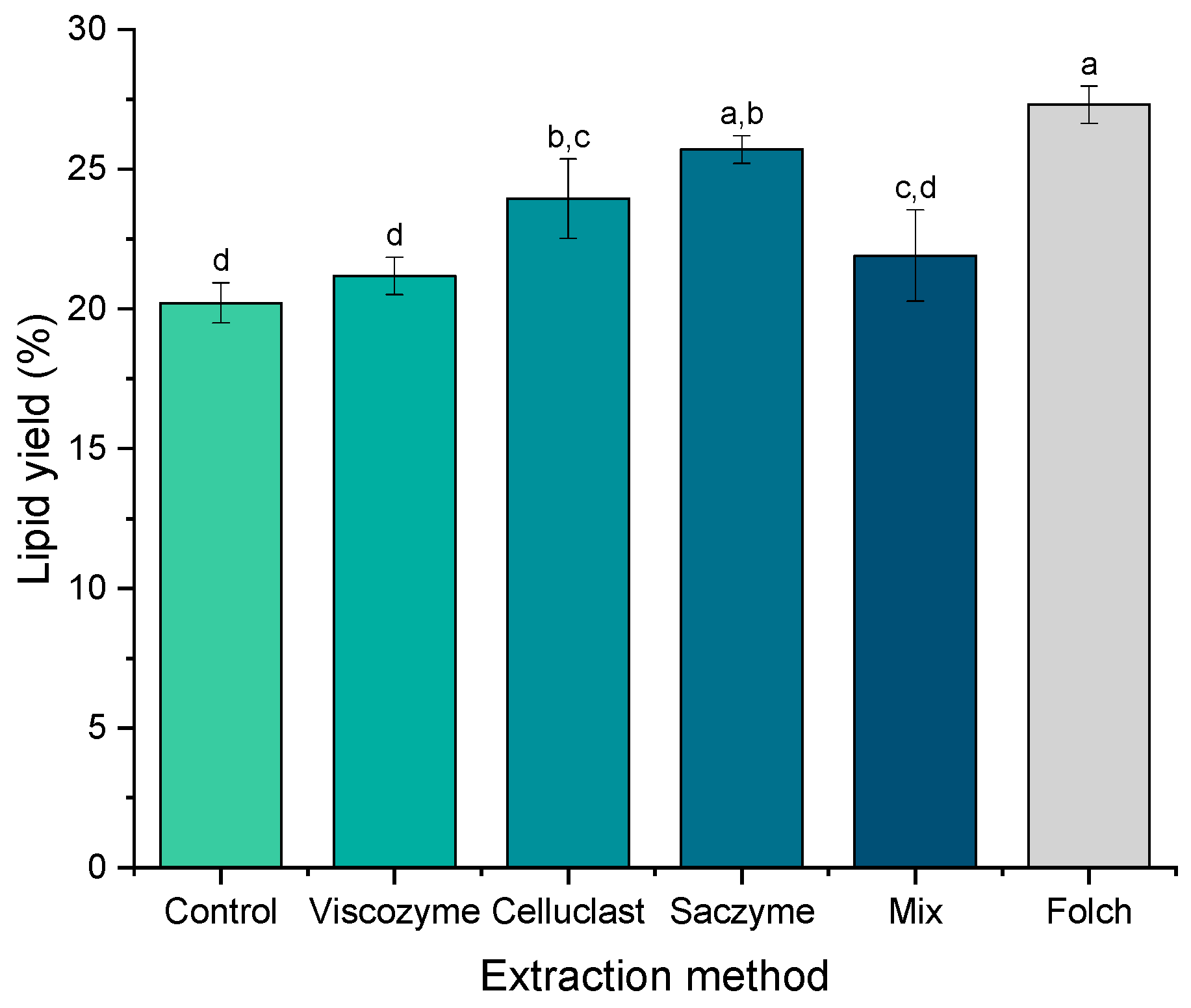
2.2. Enzyme-Based Approaches as Green Alternatives to the Conventional Extraction of Lipids from Nannochloropsis gaditana
2.3. Fatty Acid Profile of Lipid Extracts from Nannochloropsis gaditana Produced by Green Extraction Alternatives
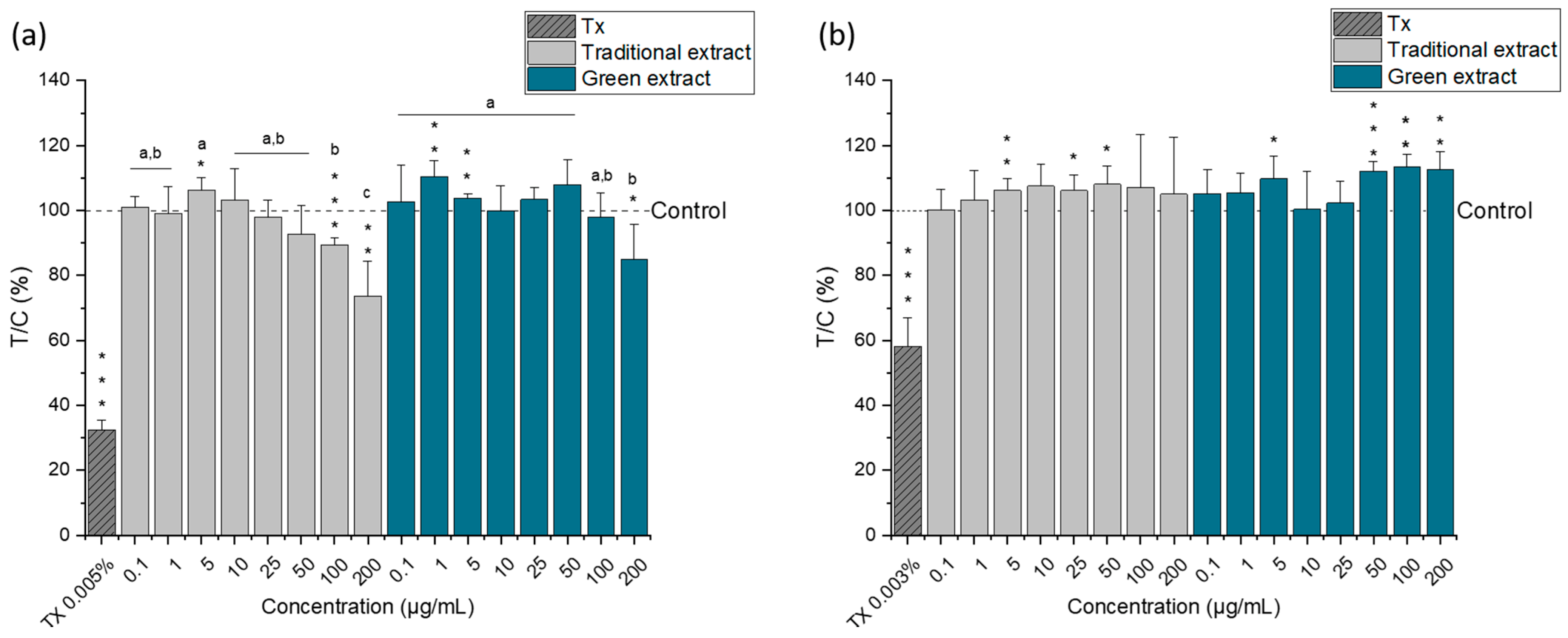
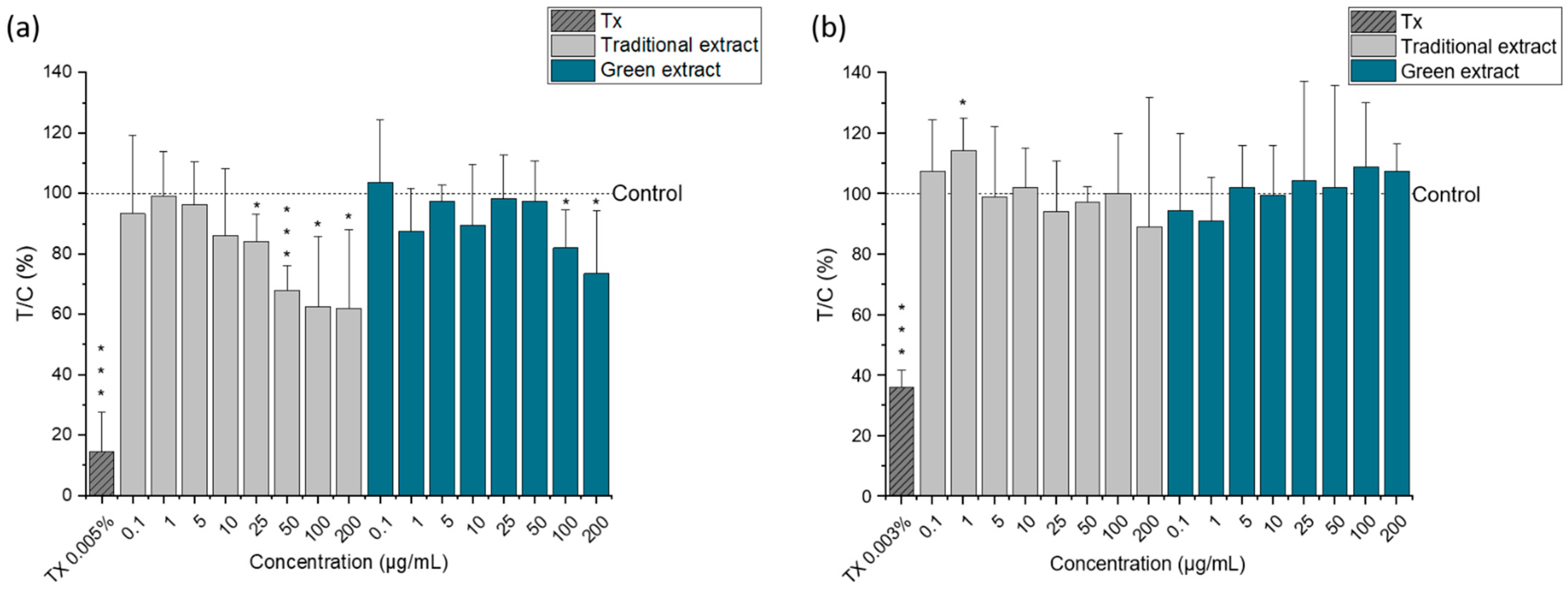
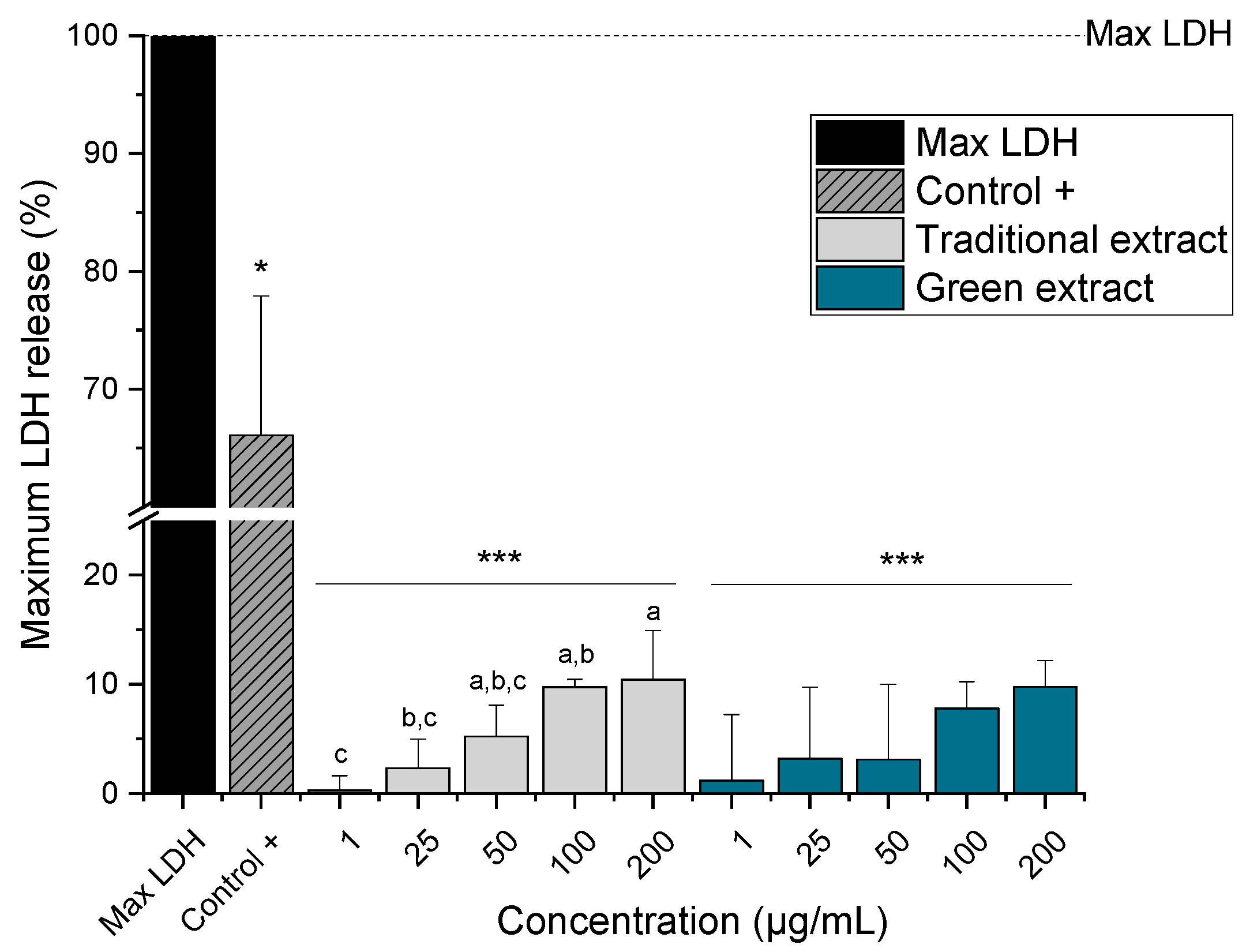
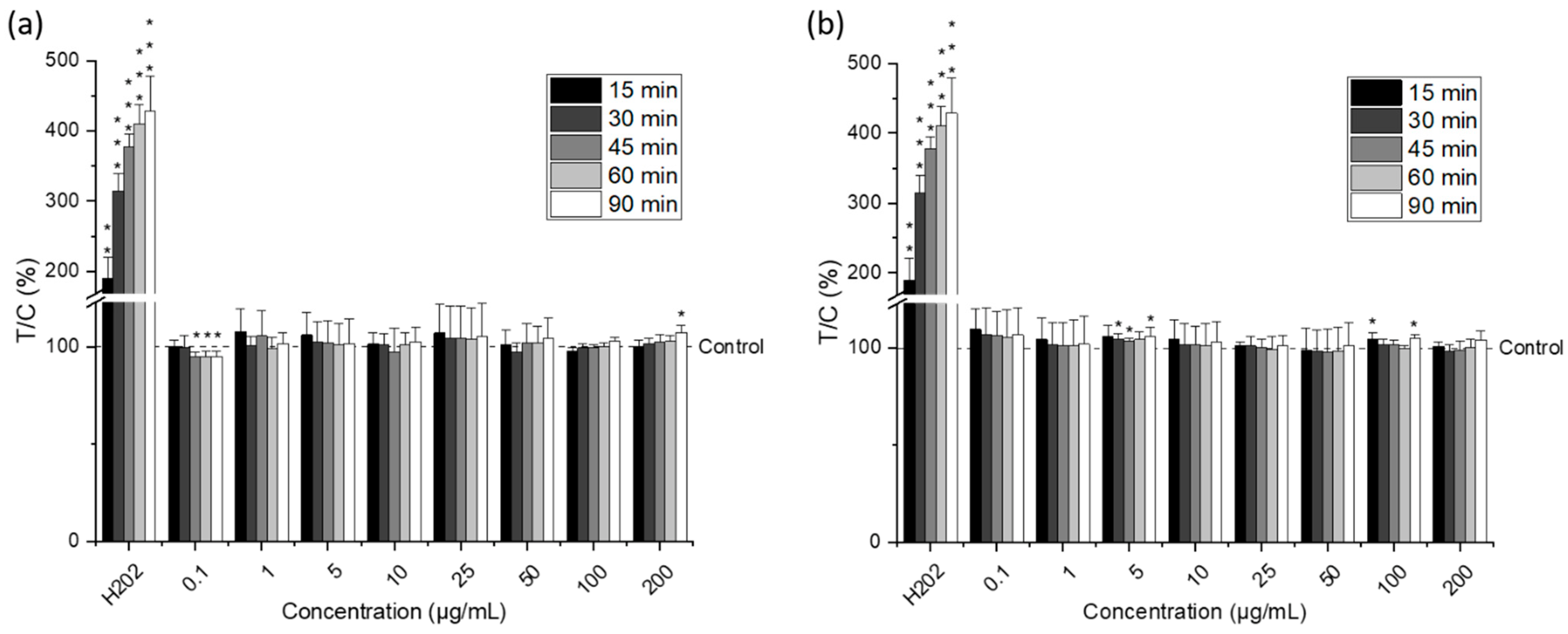
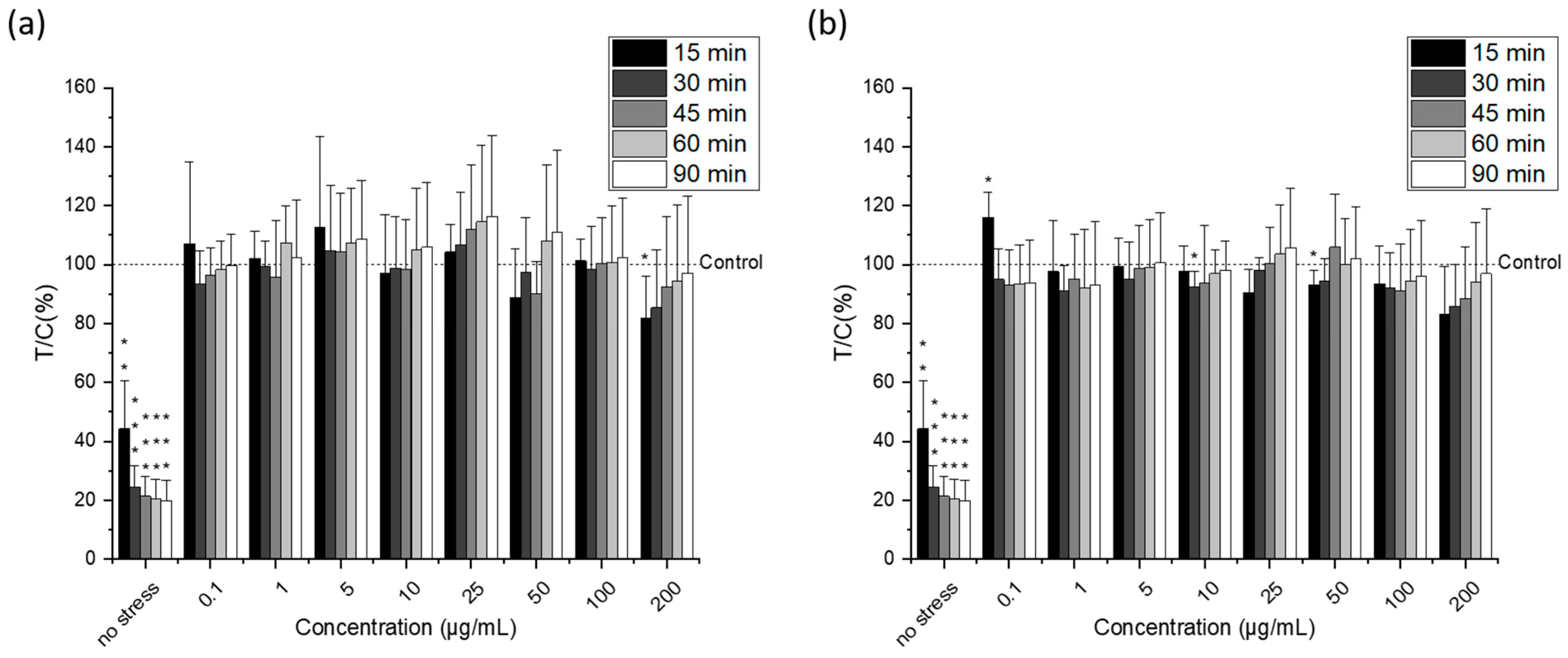
2.4. Cytotoxic Activity of Lipid Extracts from Nannochloropsis gaditana
3. Materials and Methods
3.1. Materials
3.2. Lipid Extraction of Microalgal Biomass
3.2.1. Traditional Folch Method
3.2.2. Ultrasound-Assisted Extraction
3.2.3. Enzyme-Based Extraction
3.3. Fatty Acid Composition by GC-MS
3.4. Cell Culture and Treatment
3.5. Cell Viability Assays
3.5.1. CellTiter-Blue (CTB) Assay
3.5.2. Sulforhodamine B (SRB) Assay
3.5.3. Lactate Dehydrogenase (LDH) Assay
3.6. Dichlorofluorescein (DCF) Assay
3.7. Protective Dichlorofluorescein (pDCF) Assay
3.8. Statistical Analysis
4. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Katiyar, R.; Arora, A. Health promoting functional lipids from microalgae pool: A review. Algal Res. 2020, 46, 101800. [Google Scholar] [CrossRef]
- Caporgno, M.P.; Mathys, A. Trends in Microalgae Incorporation Into Innovative Food Products With Potential Health Benefits. Front. Nutr. 2018, 5, 58. [Google Scholar] [CrossRef] [PubMed]
- Barkia, I.; Saari, N.; Manning, S.R. Microalgae for High-Value Products Towards Human Health and Nutrition. Mar. Drugs 2019, 17, 304. [Google Scholar] [CrossRef] [Green Version]
- Mehariya, S.; Goswami, R.K.; Karthikeysan, O.P.; Verma, P. Microalgae for high-value products: A way towards green nutraceutical and pharmaceutical compounds. Chemosphere 2021, 280, 130553. [Google Scholar] [CrossRef]
- Tocher, D.R.; Betancor, M.B.; Sprague, M.; Olsen, R.E.; Napier, J.A. Omega-3 long-chain polyunsaturated fatty acids, EPA and DHA: Bridging the gap between supply and demand. Nutrients 2019, 11, 89. [Google Scholar] [CrossRef] [Green Version]
- Martins, D.A.; Custódio, L.; Barreira, L.; Pereira, H.; Ben-Hamadou, R.; Varela, J.; Abu-Salah, K.M. Alternative sources of n-3 long-chain polyunsaturated fatty acids in marine microalgae. Mar. Drugs 2013, 11, 2259–2281. [Google Scholar] [CrossRef] [Green Version]
- Ranjith Kumar, R.; Hanumantha Rao, P.; Arumugam, M. Lipid Extraction Methods from Microalgae: A Comprehensive Review. Front. Energy Res. 2015, 2, 61. [Google Scholar] [CrossRef] [Green Version]
- Zhou, J.; Wang, M.; Saraiva, J.A.; Martins, A.P.; Pinto, C.A.; Prieto, M.A.; Simal-Gandara, J.; Cao, H.; Xiao, J.; Barba, F.J. Extraction of lipids from microalgae using classical and innovative approaches. Food Chem. 2022, 384, 132236. [Google Scholar] [CrossRef]
- Figueiredo, A.R.P.; da Costa, E.; Silva, J.; Domingues, M.R.; Domingues, P. The effects of different extraction methods of lipids from Nannochloropsis oceanica on the contents of omega-3 fatty acids. Algal Res. 2019, 41, 101556. [Google Scholar] [CrossRef]
- Castejón, N.; Señoráns, F.J. Simultaneous extraction and fractionation of omega-3 acylglycerols and glycolipids from wet microalgal biomass of Nannochloropsis gaditana using pressurized liquids. Algal Res. 2019, 37, 74–82. [Google Scholar] [CrossRef]
- Zhang, R.; Parniakov, O.; Grimi, N.; Lebovka, N.; Marchal, L.; Vorobiev, E. Emerging techniques for cell disruption and extraction of valuable bio-molecules of microalgae Nannochloropsis sp. Bioprocess Biosyst. Eng. 2019, 42, 173–186. [Google Scholar] [CrossRef] [PubMed]
- Zuorro, A.; Miglietta, S.; Familiari, G.; Lavecchia, R. Enhanced lipid recovery from Nannochloropsis microalgae by treatment with optimized cell wall degrading enzyme mixtures. Bioresour. Technol. 2016, 212, 35–41. [Google Scholar] [CrossRef] [PubMed]
- Bharte, S.; Desai, K. Techniques for harvesting, cell disruption and lipid extraction of microalgae for biofuel production. Biofuels 2021, 12, 285–305. [Google Scholar] [CrossRef]
- Demuez, M.; Mahdy, A.; Tomás-Pejó, E.; González-Fernández, C.; Ballesteros, M. Enzymatic cell disruption of microalgae biomass in biorefinery processes. Biotechnol. Bioeng. 2015, 112, 1955–1966. [Google Scholar] [CrossRef]
- Letsiou, S.; Kalliampakou, K.; Gardikis, K.; Mantecon, L.; Infante, C.; Chatzikonstantinou, M.; Labrou, N.E.; Flemetakis, E. Skin protective effects of Nannochloropsis gaditana extract on H2O2-stressed human dermal fibroblasts. Front. Mar. Sci. 2017, 4, 221. [Google Scholar] [CrossRef] [Green Version]
- Carrasco-Reinado, R.; Escobar-Niño, A.; Fajardo, C.; Morano, I.M.; Amil-Ruiz, F.; Martinez-Rodríguez, G.; Fuentes-Almagro, C.; Capilla, V.; Tomás-Cobos, L.; Soriano-Romaní, L.; et al. Development of New Antiproliferative Compound against Human Tumor Cells from the Marine Microalgae Nannochloropsis gaditana by Applied Proteomics. Int. J. Mol. Sci. 2021, 22, 96. [Google Scholar] [CrossRef]
- Ávila-Román, J.; Talero, E.; de Los Reyes, C.; Zubía, E.; Motilva, V.; García-Mauriño, S. Cytotoxic Activity of Microalgal-derived Oxylipins against Human Cancer Cell lines and their Impact on ATP Levels. Nat. Prod. Commun. 2016, 11, 1871–1875. [Google Scholar] [CrossRef] [Green Version]
- De los Reyes, C.; Ávila-Román, J.; Ortega, M.J.; de la Jara, A.; García-Mauriño, S.; Motilva, V.; Zubía, E. Oxylipins from the microalgae Chlamydomonas debaryana and Nannochloropsis gaditana and their activity as TNF-α inhibitors. Phytochemistry 2014, 102, 152–161. [Google Scholar] [CrossRef]
- Ávila-Román, J.; Talero, E.; de los Reyes, C.; García-Mauriño, S.; Motilva, V. Microalgae-derived oxylipins decrease inflammatory mediators by regulating the subcellular location of NFκB and PPAR-γ. Pharmacol. Res. 2018, 128, 220–230. [Google Scholar] [CrossRef]
- Dos Santos, R.R.; Moreira, D.M.; Kunigami, C.N.; Aranda, D.A.G.; Teixeira, C.M.L.L. Comparison between several methods of total lipid extraction from Chlorella vulgaris biomass. Ultrason. Sonochem. 2015, 22, 95–99. [Google Scholar] [CrossRef]
- Gerde, J.A.; Montalbo-Lomboy, M.; Yao, L.; Grewell, D.; Wang, T. Evaluation of microalgae cell disruption by ultrasonic treatment. Bioresour. Technol. 2012, 125, 175–181. [Google Scholar] [CrossRef] [PubMed]
- Greenly, J.M.; Tester, J.W. Ultrasonic cavitation for disruption of microalgae. Bioresour. Technol. 2015, 184, 276–279. [Google Scholar] [CrossRef] [PubMed]
- Señoráns, M.; Castejón, N.; Señoráns, F.J. Advanced Extraction of Lipids with DHA from Isochrysis galbana with enzymatic pre-treatment combined with pressurized liquids and ultrasound assisted extractions. Molecules 2020, 25, 3310. [Google Scholar] [CrossRef] [PubMed]
- González-Balderas, R.M.; Velásquez-Orta, S.B.; Valdez-Vazquez, I.; Orta Ledesma, M.T. Intensified recovery of lipids, proteins, and carbohydrates from wastewater-grown microalgae Desmodesmus sp. by using ultrasound or ozone. Ultrason. Sonochem. 2020, 62, 104852. [Google Scholar] [CrossRef]
- Scholz, M.J.; Weiss, T.L.; Jinkerson, R.E.; Jing, J.; Roth, R.; Goodenough, U.; Posewitz, M.C.; Gerken, H.G. Ultrastructure and composition of the Nannochloropsis gaditana cell wall. Eukaryot. Cell 2014, 13, 1450–1464. [Google Scholar] [CrossRef] [Green Version]
- Blanco-Llamero, C.; García-García, P.; Señoráns, F.J. Combination of Synergic Enzymes and Ultrasounds as an Effective Pretreatment Process to Break Microalgal Cell Wall and Enhance Algal Oil Extraction. Foods 2021, 10, 1928. [Google Scholar] [CrossRef]
- Miranda, I.; Simões, R.; Medeiros, B.; Nampoothiri, K.M.; Sukumaran, R.K.; Rajan, D.; Pereira, H.; Ferreira-Dias, S. Valorization of lignocellulosic residues from the olive oil industry by production of lignin, glucose and functional sugars. Bioresour. Technol. 2019, 292, 121936. [Google Scholar] [CrossRef]
- Neiva, D.M.; Costa, R.A.; Gominho, J.; Ferreira-Dias, S.; Pereira, H. Fractionation and valorization of industrial bark residues by autohydrolysis and enzymatic saccharification. Bioresour. Technol. Rep. 2020, 11, 100441. [Google Scholar] [CrossRef]
- Molino, A.; Martino, M.; Larocca, V.; Di Sanzo, G.; Spagnoletta, A.; Marino, T.; Karatza, D.; Iovine, A.; Mehariya, S.; Musmarra, D. Eicosapentaenoic Acid Extraction from Nannochloropsis gaditana Using Carbon Dioxide at Supercritical Conditions. Mar. Drugs 2019, 17, 132. [Google Scholar] [CrossRef] [Green Version]
- Taher, H.; Giwa, A.; Abusabiekeh, H.; Al-Zuhair, S. Biodiesel production from Nannochloropsis gaditana using supercritical CO2 for lipid extraction and immobilized lipase transesterification: Economic and environmental impact assessments. Fuel Process. Technol. 2020, 198, 106249. [Google Scholar] [CrossRef]
- Ho, B.C.H.; Kamal, S.M.M.; Danquah, M.K.; Harun, R. Optimization of Subcritical Water Extraction (SWE) of Lipid and Eicosapentaenoic Acid (EPA) from Nannochloropsis gaditana. Biomed. Res. Int. 2018, 2018, 8273581. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Blanco-Llamero, C.; Señoráns, F.J. Biobased Solvents for Pressurized Liquid Extraction of Nannochloropsis gaditana Omega-3 Lipids. Mar. Drugs 2021, 19, 107. [Google Scholar] [CrossRef] [PubMed]
- Tang, Y.; Zhang, Y.; Rosenberg, J.N.; Sharif, N.; Betenbaugh, M.J.; Wang, F. Efficient lipid extraction and quantification of fatty acids from algal biomass using accelerated solvent extraction (ASE). RSC Adv. 2016, 6, 29127–29134. [Google Scholar] [CrossRef]
- Jiménez Callejón, M.J.; Robles Medina, A.; Macías Sánchez, M.D.; González Moreno, P.A.; Navarro López, E.; Esteban Cerdán, L.; Molina Grima, E. Supercritical fluid extraction and pressurized liquid extraction processes applied to eicosapentaenoic acid-rich polar lipid recovery from the microalga Nannochloropsis sp. Algal Res. 2022, 61, 102586. [Google Scholar] [CrossRef]
- Heredia, V.; Pruvost, J.; Gonçalves, O.; Drouin, D.; Marchal, L. Lipid recovery from Nannochloropsis gaditana using the wet pathway: Investigation of the operating parameters of bead milling and centrifugal extraction. Algal Res. 2021, 56, 102318. [Google Scholar] [CrossRef]
- Jiménez Callejón, M.J.; Robles Medina, A.; Macías Sánchez, M.D.; Esteban Cerdán, L.; González Moreno, P.A.; Navarro López, E.; Hita Peña, E.; Grima, E.M. Obtaining highly pure EPA-rich lipids from dry and wet Nannochloropsis gaditana microalgal biomass using ethanol, hexane and acetone. Algal Res. 2020, 45, 101729. [Google Scholar] [CrossRef]
- Hita Peña, E.; Robles Medina, A.; Jiménez Callejón, M.J.; Macías Sánchez, M.D.; Esteban Cerdán, L.; González Moreno, P.A.; Molina Grima, E. Extraction of free fatty acids from wet Nannochloropsis gaditana biomass for biodiesel production. Renew. Energy 2015, 75, 366–373. [Google Scholar] [CrossRef]
- Angles, E.; Jaouen, P.; Pruvost, J.; Marchal, L. Wet lipid extraction from the microalga Nannochloropsis sp.: Disruption, physiological effects and solvent screening. Algal Res. 2017, 21, 27–34. [Google Scholar] [CrossRef]
- Castejón, N.; Luna, P.; Señoráns, F.J. Alternative oil extraction methods from Echium plantagineum L. seeds using advanced techniques and green solvents. Food Chem. 2018, 244, 75–82. [Google Scholar] [CrossRef]
- Kumar, P.; Nagarajan, A.; Uchil, P.D. Analysis of cell viability by the lactate dehydrogenase assay. Cold Spring Harb. Protoc. 2018, 6, pdb.prot095497. [Google Scholar] [CrossRef]
- Çetin, Z.; Saygili, E.İ.; Benlier, N.; Ozkur, M.; Sayin, S. Omega-3 polyunsaturated fatty acids and cancer. In Nutraceuticals and Cancer Signaling: Clinical Aspects and Mode of Action; Jafari, S.M., Nabavi, S.M., Silva, A.S., Eds.; Springer International Publishing: Cham, Switzerland, 2021; pp. 591–631. [Google Scholar]
- D’Eliseo, D.; Velotti, F. Omega-3 Fatty acids and cancer cell cytotoxicity: Implications for multi-targeted cancer therapy. J. Clin. Med. 2016, 5, 15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cockbain, A.J.; Toogood, G.J.; Hull, M.A. Omega-3 polyunsaturated fatty acids for the treatment and prevention of colorectal cancer. Gut 2012, 61, 135–149. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miccadei, S.; Masella, R.; Mileo, A.M.; Gessani, S. ω3 Polyunsaturated fatty acids as immunomodulators in colorectal cancer: New potential role in adjuvant therapies. Front. Immunol. 2016, 7, 486. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aldoori, J.; Cockbain, A.J.; Toogood, G.J.; Hull, M.A. Omega-3 polyunsaturated fatty acids: Moving towards precision use for prevention and treatment of colorectal cancer. Gut 2022, 71, 822. [Google Scholar] [CrossRef]
- Kubota, H.; Matsumoto, H.; Higashida, M.; Murakami, H.; Nakashima, H.; Oka, Y.; Okumura, H.; Yamamura, M.; Nakamura, M.; Hirai, T. Eicosapentaenoic acid modifies cytokine activity and inhibits cell proliferation in an oesophageal cancer cell line. Anticancer Res. 2013, 33, 4319–4324. [Google Scholar]
- Fukui, M.; Kang, K.S.; Okada, K.; Zhu, B.T. EPA, an omega-3 fatty acid, induces apoptosis in human pancreatic cancer cells: Role of ROS accumulation, caspase-8 activation, and autophagy induction. J. Cell. Biochem. 2013, 114, 192–203. [Google Scholar] [CrossRef]
- Yin, Y.; Sui, C.; Meng, F.; Ma, P.; Jiang, Y. The omega-3 polyunsaturated fatty acid docosahexaenoic acid inhibits proliferation and progression of non-small cell lung cancer cells through the reactive oxygen species-mediated inactivation of the PI3K /Akt pathway. Lipids Health Dis. 2017, 16, 87. [Google Scholar] [CrossRef] [Green Version]
- Tanaka, A.; Yamamoto, A.; Murota, K.; Tsujiuchi, T.; Iwamori, M.; Fukushima, N. Polyunsaturated fatty acids induce ovarian cancer cell death through ROS-dependent MAP kinase activation. Biochem. Biophys. Res. Commun. 2017, 493, 468–473. [Google Scholar] [CrossRef]
- Oono, K.; Ohtake, K.; Watanabe, C.; Shiba, S.; Sekiya, T.; Kasono, K. Contribution of Pyk2 pathway and reactive oxygen species (ROS) to the anti-cancer effects of eicosapentaenoic acid (EPA) in PC3 prostate cancer cells. Lipids Health Dis. 2020, 19, 15. [Google Scholar] [CrossRef] [Green Version]
- Folch, J.; Lees, M.; Stanley, G.S. A simple method for the isolation and purification of total lipides from animal tissues. J. Biol. Chem. 1957, 226, 497–509. [Google Scholar] [CrossRef]
- Skehan, P.; Storeng, R.; Scudiero, D.; Monks, A.; McMahon, J.; Vistica, D.; Warren, J.T.; Bokesch, H.; Kenney, S.; Boyd, M.R. New colorimetric cytotoxicity assay for anticancer-drug screening. J. Natl. Cancer Inst. 1990, 82, 1107–1112. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Joseph, J.A. Quantifying cellular oxidative stress by dichlorofluorescein assay using microplate reader. Free Radic. Biol. Med. 1999, 27, 612–616. [Google Scholar] [CrossRef]
| Fatty Acid | RT (min) | % Fatty Acids | |||||
|---|---|---|---|---|---|---|---|
| Folch | Control | Viscozyme | Celluclast | Saczyme | Mix | ||
| 12:0 | 6.2 | 0.4 ± 0.0 | 0.6 ± 0.1 | 0.5 ± 0.0 | 0.5 ± 0.1 | 0.4 ± 0.1 | 0.6 ± 0.0 |
| 14:0 | 10.0 | 4.3 ± 0.1 | 4.5 ± 0.1 | 4.3 ± 0.1 | 4.4 ± 0.1 | 4.3 ± 0.2 | 4.4 ± 0.1 |
| 16:1 n-7 | 14.6 | 26.1 ± 0.6 a | 22.9 ± 0.8 b | 24.6 ± 0.7 a,b | 24.0 ± 0.8 a,b | 24.9 ± 1.1 a,b | 23.2 ± 0.6 b |
| 16:0 | 15.1 | 23.0 ± 0.8 a | 18.3 ± 1.3 b | 21.7 ± 1.3 a,b | 19.7 ± 1.6 a,b | 21.5 ± 1.7 a,b | 19.1 ± 0.7 a,b |
| 17:1 | 16.3 | 0.7 ± 0.0 | 0.7 ± 0.2 | 0.5 ± 0.2 | 0.7 ± 0.1 | 0.5 ± 0.3 | 0.3 ± 0.1 |
| 18:2 isomer n.i. | 17.1 | 0.7 ± 0.1 | 0.5 ± 0.2 | 0.7 ± 0.1 | 0.6 ± 0.1 | 0.7 ± 0.0 | 0.7 ± 0.0 |
| 18:2 n-6 | 19.7 | 3.2 ± 0.1 b | 3.5 ± 0.0 a | 3.5 ± 0.0 a | 3.5 ± 0.0 a | 3.5 ± 0.0 a | 3.5 ± 0.0 a |
| 18:1 n-9 | 19.8 | 4.8 ± 0.1 | 4.7 ± 0.0 | 4.7 ± 0.0 | 4.8 ± 0.0 | 4.8 ± 0.0 | 4.7 ± 0.1 |
| 18:1 n-7 | 20.0 | 0.7 ± 0.1 | 0.6 ± 0.0 | 0.5 ± 0.1 | 0.6 ± 0.0 | 0.5 ± 0.1 | 0.5 ± 0.0 |
| 18:0 | 20.6 | 0.2 ± 0.0 a | 0.2 ± 0.0 b | 0.2 ± 0.0 b | 0.2 ± 0.0 b | 0.2 ± 0.0 b | 0.2 ± 0.0 b |
| 20:4 n-6 | 24.2 | 5.9 ± 0.7 b | 7.1 ± 0.0 a | 6.8 ± 0.1 a,b | 7.0 ± 0.2 a | 6.8 ± 0.2 a,b | 7.3 ± 0.1 a |
| 20:5 n-3 | 24.5 | 29.3 ± 0.8 | 34.5 ± 1.9 | 30.6 ± 1.9 | 32.7 ± 2.1 | 30.2 ± 2.4 | 33.8 ± 1.1 |
| 20:5 isomer n.i. | 24.6 | 0.6 ± 0.2 | 0.9 ± 0.1 | 1.1 ± 0.1 | 1.1 ± 0.2 | 1.1 ± 0.5 | 1.2 ± 0.4 |
| 20:5 isomer n.i. | 25.8 | n.d. | 1.1 ± 0.1 | 0.6 ± 0.2 | 0.7 ± 0.4 | 0.6 ± 0.2 | 0.4 ± 0.2 |
| SFA | 27.9 ± 1.0 a | 23.5 ± 1.3 b | 26.5 ± 1.4 a,b | 24.7 ± 1.7 a,b | 26.4 ± 1.9 a,b | 24.3 ± 0.8 a,b | |
| MUFA | 32.4 ± 0.9 a | 28.9 ± 1.2 b | 30.4 ± 1.1 a,b | 30.0 ± 1.1 a,b | 30.8 ± 1.7 a,b | 28.8 ± 1.2 b | |
| PUFA | 39.7 ± 2.0 b | 47.6 ± 2.5 a | 43.1 ± 2.5 a,b | 45.6 ± 2.8 a,b | 42.8 ± 3.6 a,b | 47.0 ± 1.7 a | |
| n-3 | 29.3 | 34.5 | 30.6 | 32.7 | 30.2 | 33.8 | |
| n-6 | 9.1 | 10.6 | 10.3 | 10.5 | 10.2 | 10.8 | |
| n-6/n-3 ratio | 0.3 | 0.3 | 0.3 | 0.3 | 0.3 | 0.3 | |
| Product Name | Component Name | Side Activities | Activity | pH Range | T a Range |
|---|---|---|---|---|---|
| Viscozyme® L | Beta-glucanase (endo-1,3(4)-) | xylanase, cellulase and hemicellulase | 100 FBG/g | 3.3–5.5 | 40–50 °C |
| Celluclast® 1.5 L | Cellulase | no reported | 700 EGU/g | 4.0–6.0 | 50–60 °C |
| Saczyme® Yield | Glucoamylase (glucan 1,4-alpha-glucosidase) | alpha-amylase, cellulase, beta-glucosidase, cellulose and 1,4-beta-cellobiosidase | 900 AGU/g | 3.5–5.5 | 30–60 °C |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Castejón, N.; Marko, D. Fatty Acid Composition and Cytotoxic Activity of Lipid Extracts from Nannochloropsis gaditana Produced by Green Technologies. Molecules 2022, 27, 3710. https://doi.org/10.3390/molecules27123710
Castejón N, Marko D. Fatty Acid Composition and Cytotoxic Activity of Lipid Extracts from Nannochloropsis gaditana Produced by Green Technologies. Molecules. 2022; 27(12):3710. https://doi.org/10.3390/molecules27123710
Chicago/Turabian StyleCastejón, Natalia, and Doris Marko. 2022. "Fatty Acid Composition and Cytotoxic Activity of Lipid Extracts from Nannochloropsis gaditana Produced by Green Technologies" Molecules 27, no. 12: 3710. https://doi.org/10.3390/molecules27123710
APA StyleCastejón, N., & Marko, D. (2022). Fatty Acid Composition and Cytotoxic Activity of Lipid Extracts from Nannochloropsis gaditana Produced by Green Technologies. Molecules, 27(12), 3710. https://doi.org/10.3390/molecules27123710