N-Glucosylation in Corynebacterium glutamicum with YdhE from Bacillus lichenformis
Abstract
1. Introduction
2. Results and Discussion
2.1. Codon Usage Optimization of YdhE
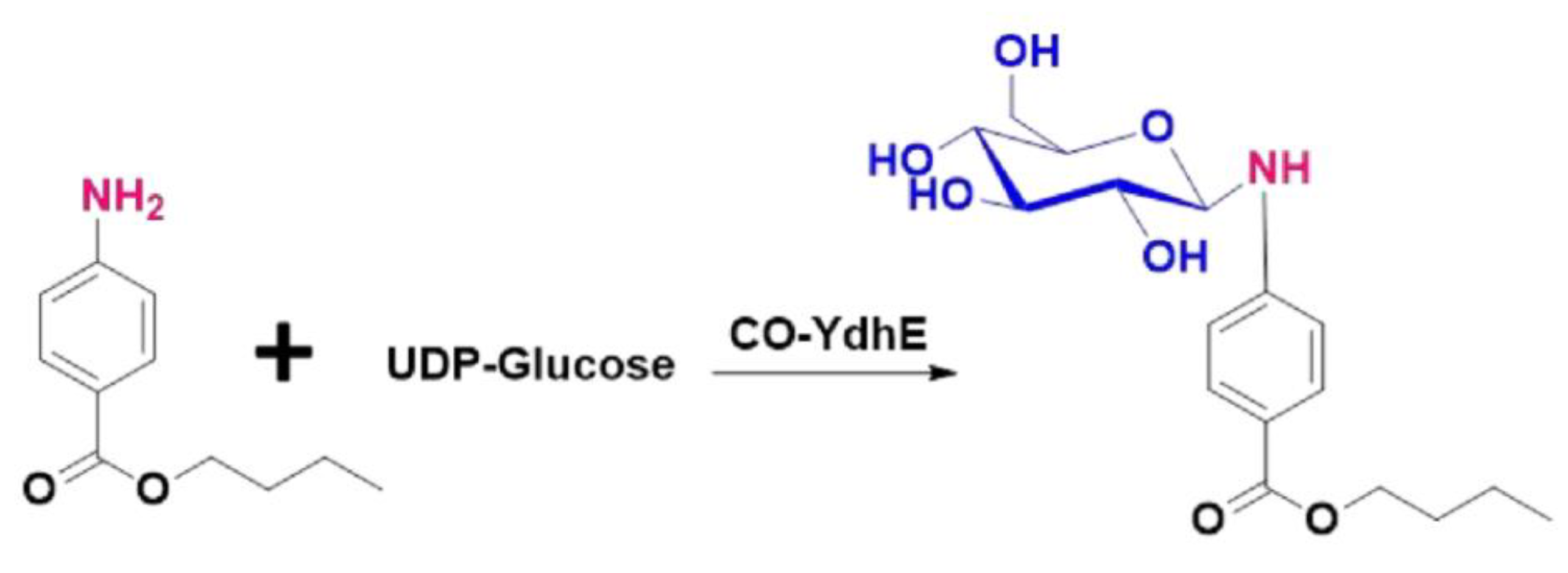
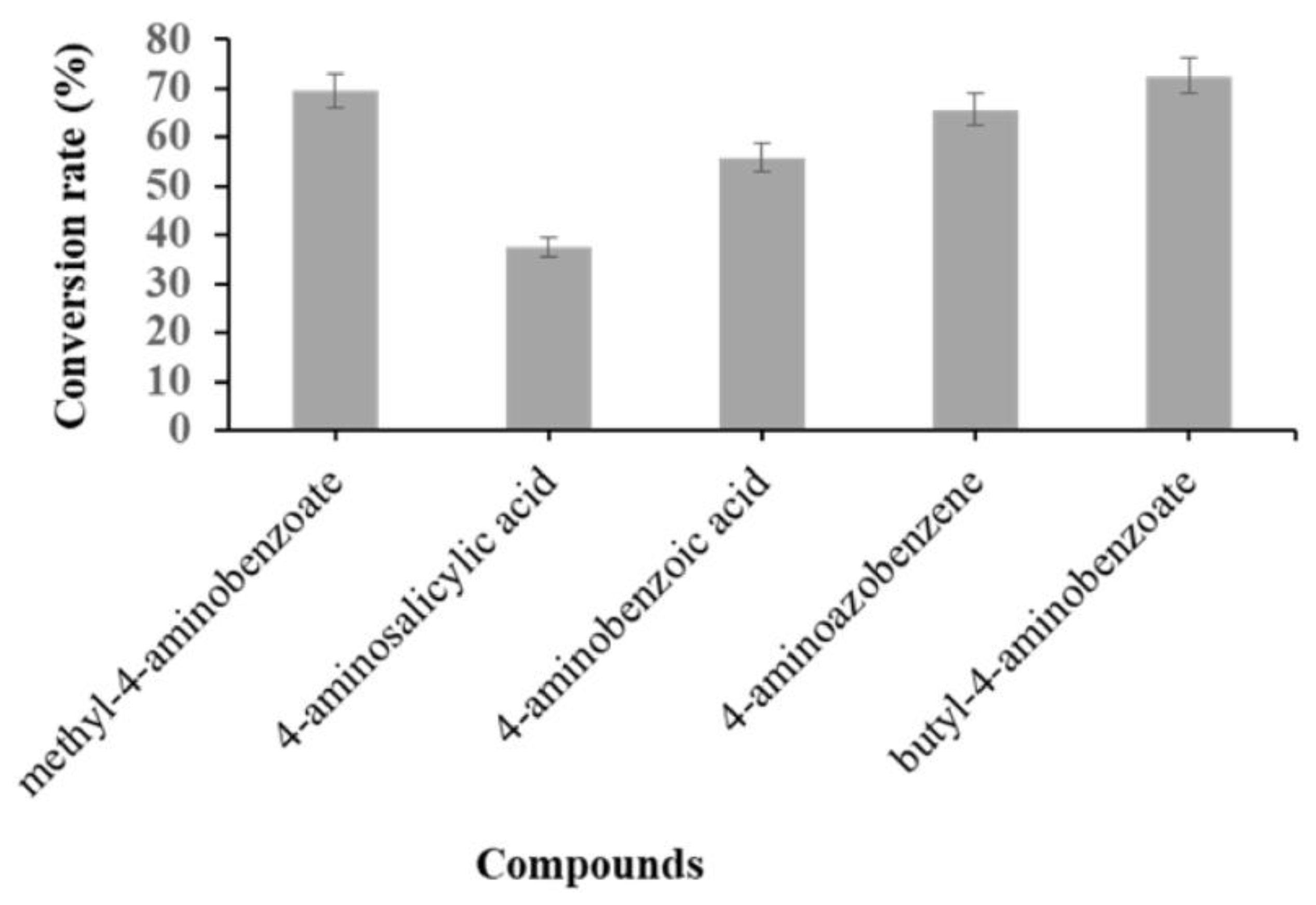
2.2. Bio-Conversion of Amine Functional Group Containing Compounds
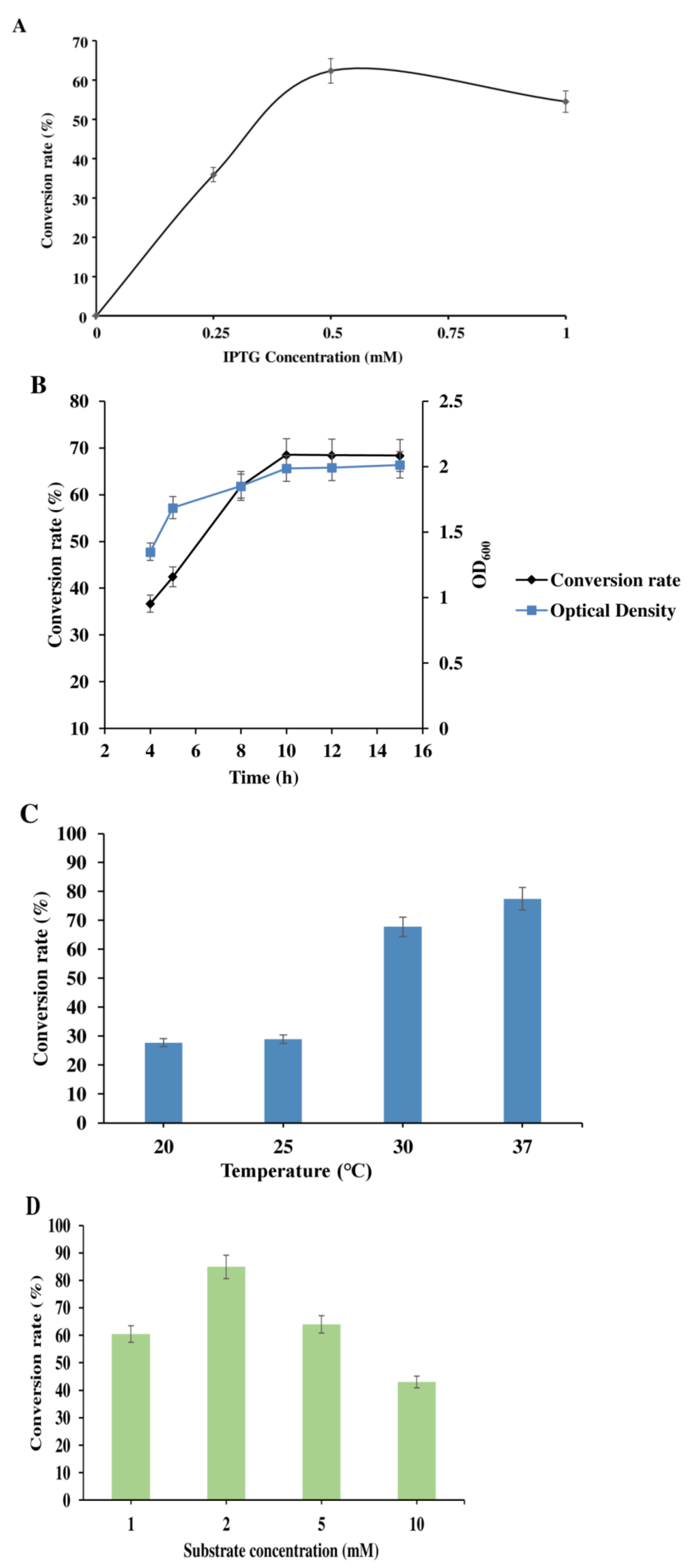
2.3. Effects of IPTG Concentrations, IPTG Induction Time, Temperature, Substrate, Glucose Concentration and Production Time on In Vivo Conversion
2.4. Elucidation of the Structure of Glycosylated Product
2.5. Determination of Water Solubility
3. Materials and Methods
3.1. Media and Bacterial Strains
3.2. Plasmid Construction, Transformation in C. glutamicum
3.3. In Vivo Screening of N-Linked Substrates
3.4. Determination of Optimal Culture Conditions
3.5. Fermentation and Analytical Procedures
3.6. Water Solubility Determination
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Hermann, T. Industrial production of amino acids by coryneform bacteria. J. Biotechnol. 2003, 104, 155–172. [Google Scholar] [CrossRef]
- Baritugo, K.A.; Kim, H.T.; David, Y.; Choi, J.I.; Hong, S.H.; Jeong, K.J.; Choi, J.H.; Joo, J.C.; Park, S.J. Metabolic engineering of Corynebacterium glutamicum for fermentative production of chemicals in biorefinery. Appl. Microbiol. Biotechnol. 2018, 102, 3915–3937. [Google Scholar] [CrossRef] [PubMed]
- Becker, J.; Wittmann, C. Bio-based production of chemicals, materials and fuels -Corynebacterium glutamicum as versatile cell factory. Curr. Opin. Biotechnol. 2012, 23, 631–640. [Google Scholar] [CrossRef] [PubMed]
- Jojima, T.; Noburyu, R.; Sasaki, M.; Tajima, T.; Suda, M.; Yukawa, H.; Inui, M. Metabolic engineering for improved production of ethanol by Corynebacterium glutamicum. Appl Microbiol. Biotechnol. 2015, 99, 1165–1172. [Google Scholar] [CrossRef]
- Kind, S.; Jeong, W.K.; Schröder, H.; Wittmann, C. Systems-wide metabolic pathway engineering in Corynebacterium glutamicum for bio-based production of diaminopentane. Metab. Eng. 2010, 12, 341–351. [Google Scholar] [CrossRef]
- Kallscheuer, N.; Vogt, M.; Stenzel, A.; Gätgens, J.; Bott, M.; Marienhagen, J. Construction of a Corynebacterium glutamicum platform strain for the production of stilbenes and (2S)-flavanones. Metab. Eng. 2016, 38, 47–55. [Google Scholar] [CrossRef]
- Kogure, T.; Inui, M. Recent advances in metabolic engineering of Corynebacterium glutamicum for bioproduction of value-added aromatic chemicals and natural products. Appl. Microbiol. Biotechnol. 2018, 102, 8685–8705. [Google Scholar] [CrossRef]
- Chin, Y.W.; Park, J.B.; Park, Y.C.; Kim, K.H.; Seo, J.H. Metabolic engineering of Corynebacterium glutamicum to produce GDP-L-fucose from glucose and mannose. Bioprocess. Biosyst. Eng. 2013, 36, 749–756. [Google Scholar] [CrossRef]
- Gauttam, R.; Desiderato, C.K.; Radoš, D.; Link, H.; Seibold, G.M.; Eikmanns, B.J. Metabolic Engineering of Corynebacterium glutamicum for Production of UDP-N-Acetylglucosamine. Front. Bioeng. Biotechnol. 2021, 9, 510–748. [Google Scholar] [CrossRef]
- Heider, S.A.; Peters-Wendisch, P.; Netzer, R.; Stafnes, M.; Brautaset, T.; Wendisch, V.F. Production and glucosylation of C50 and C40 carotenoids by metabolically engineered Corynebacterium glutamicum. Appl. Microbiol. Biotechnol. 2014, 98, 1223–1235. [Google Scholar] [CrossRef]
- Xiao, J.; Muzashvili, T.S.; Georgiev, M.I. Advances in the biotechnological glycosylation of valuable flavonoids. Biotechnol. Adv. 2014, 32, 1145–1156. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.; Zhang, L.; Yue, X.J.; Tang, Y.J.; Wu, C.; Li, Y.Z. Effects of glycosylation on the bioactivity of rapamycin. Appl. Microbiol. Biotechnol. 2020, 104, 9125–9134. [Google Scholar] [CrossRef] [PubMed]
- Bosch, D.; Castilho, A.; Loos, A.; Schots, A.; Steinkellner, H. N-glycosylation of plant-produced recombinant proteins. Curr. Pharm. Des. 2013, 19, 5503–5512. [Google Scholar] [CrossRef] [PubMed]
- Langenhan, J.M.; Peters, N.R.; Guzei, I.A.; Hoffmann, F.M.; Thorson, J.S. Enhancing the anticancer properties of cardiac glycosides by neoglycorandomization. Proc. Natl. Acad. Sci. USA 2005, 102, 12305–12310. [Google Scholar] [CrossRef] [PubMed]
- Manach, C.; Donovan, J.L. Pharmacokinetics and metabolism of dietary flavonoids in humans. Free. Radic. Res. 2004, 38, 771–785. [Google Scholar] [CrossRef] [PubMed]
- Rayyan, S.; Fossen, T.; Andersen, Ø. Flavone C-glycosides from leaves of Oxalis triangularis. J. Agric. Food Chem. 2005, 53, 10057–10060. [Google Scholar] [CrossRef]
- Pandey, R.P.; Bashyal, P.; Parajuli, P.; Yamaguchi, T.; Sohng, J.K. Two Trifunctional Leloir Glycosyltransferases as Biocatalysts for Natural Products Glycodiversification. Org. Lett. 2019, 21, 8058–8064. [Google Scholar] [CrossRef]
- FDA Code of Federal Regulations, Title 21, vol. 4, revised April 1 2020, Sec. 216.24. Available online: https://www.accessdata.fda.gov/scripts/cdrh/cfdocs/cfcfr/CFRSearch.cfm?fr=216.24. (accessed on 25 April 2022).
- Mura, P.; Maestrelli, F.; Cirri, M.; Nerli, G.; di Cesare Mannelli, L.; Ghelardini, C.; Mennini, N. Improvement of butamben anesthetic efficacy by the development of deformable liposomes bearing the drug as cyclodextrin complex. Pharmaceutics. 2021, 13, 872. [Google Scholar] [CrossRef]
- Chen, T.; Zhu, N.; Xia, H. Aerobic production of succinate from arabinose by metabolically engineered Corynebacterium glutamicum. Bioresour. Technol. 2014, 151, 411–414. [Google Scholar] [CrossRef]
- Liu, G.; Wu, J.; Yang, H.; Bao, Q. Codon Usage Patterns in Corynebacterium glutamicum: Mutational Bias, Natural Selection and Amino Acid Conservation. Comp. Funct. Genom. 2010, 2010, 343–569. [Google Scholar] [CrossRef]
- Wang, X.; Peng, F.; Dong, G.; Sun, Y.; Dai, X.; Yang, Y.; Liu, X.; Bai, Z. Identification and Validation of Appropriate Reference Genes for qRT-PCR Analysis in Corynebacterium glutamicum. FEMS Microbiol. Lett. 2018, 8, 365. [Google Scholar] [CrossRef] [PubMed]
- Xie, K.; Chen, R.; Chen, D.; Li, J.; Wang, R.; Yang, L.; Dai, J. Enzymatic N-Glycosylation of Diverse Arylamine Aglycones by a Promiscuous Glycosyltransferase from Carthamus tinctorius. Adv. Synth. Catal. 2017, 359, 603. [Google Scholar] [CrossRef]
- Wu, C.Z.; Jang, J.H.; Woo, M.; Ahn, J.S.; Kim, J.S.; Hong, Y.S. Enzymatic glycosylation of nonbenzoquinone geldanamycin analogs via Bacillus UDP-glycosyltransferase. Appl. Environ. Microbiol. 2012, 78, 7680–7686. [Google Scholar] [CrossRef] [PubMed]
- Yim, S.S.; An, S.J.; Choi, J.W.; Ryu, A.J.; Jeong, K.J. High-level secretory production of recombinant single-chain variable fragment (ScFv) in Corynebacterium glutamicum. Appl. Microbiol. Biotechnol. 2014, 98, 273–284. [Google Scholar] [CrossRef]
- Jiang, Y.; Qian, F.; Yang, J.; Liu, Y.; Dong, F.; Xu, C.; Sun, B.; Chen, B.; Xu, X.; Li, Y.; et al. CRISPR-Cpf1 assisted genome editing of Corynebacterium glutamicum. Nat. Commun. 2017, 8, 151–179. [Google Scholar] [CrossRef] [PubMed]
- Park, C.M.; Kim, G.M.; Cha, G.S. Biotransformation of Flavonoids by Newly Isolated and Characterized Lactobacillus pentosus NGI01 Strain from Kimchi. Microorganisms. 2021, 9, 1075. [Google Scholar] [CrossRef] [PubMed]
- Thapa, S.B.; Pandey, R.P.; Bashyal, P.; Yamaguchi, T.; Sohng, J.K. Cascade biocatalysis systems for bioactive naringenin glucosides and quercetin rhamnoside production from sucrose. Appl. Microbiol. Biotechnol. 2019, 103, 7953–7969. [Google Scholar] [CrossRef]
- Teles, Y.; Souza, M.; Souza, M. Sulphated flavonoids: Biosynthesis, structures, and biological activities. Molecules. 2018, 23, 2480. [Google Scholar] [CrossRef]
- Xie, L.; Zhang, L.; Wang, C.; Wang, X.; Xu, Y.; Yu, H.; Wu, P.; Li, S.; Han, L.; Gunatilaka, A.; et al. Methylglucosylation of aromatic amino and phenolic moieties of drug-like biosynthons by combinatorial biosynthesis. Proc. Natl. Acad. Sci. USA 2018, 115, 980–989. [Google Scholar] [CrossRef]
- Deng, X.; Wang, Z.; Liu, J.; Xiong, S.; Xiong, R.; Cao, X.; Chen, Y.; Zheng, X.; Tang, G. Design, synthesis and biological evaluation of flavonoid salicylate derivatives as potential anti-tumor agents. RSC Adv. 2017, 7, 38171–38178. [Google Scholar] [CrossRef]
- Yuan, S.; Yin, S.; Liu, M.; Kong, J.Q. Isolation and characterization of a multifunctional flavonoid glycosyltransferase from Ornithogalum caudatum with glycosidase activity. Sci. Rep. 2018, 8, 5886. [Google Scholar] [CrossRef] [PubMed]
- Darsandhari, S.; Pandey, R.P.; Shrestha, B.; Parajuli, P.; Liou, K.; Sohng, J.K. One-Pot Multienzyme Cofactors Recycling (OPME-CR) System for Lactose and Non-natural Saccharide Conjugated Polyphenol Production. J. Agric. Food Chem. 2018, 66, 7965–7974. [Google Scholar] [CrossRef] [PubMed]
- Schmölzer, K.; Gutmann, A.; Diricks, M.; Desmet, T.; Nidetzky, B. Sucrose synthase: A unique glycosyltransferase for biocatalytic glycosylation process development. Biotechnol. Adv. 2016, 34, 88–111. [Google Scholar] [CrossRef] [PubMed]
- Lee, B.H.; Pyo, M.K.; Lee, J.H.; Choi, S.H.; Shin, T.J.; Lee, S.M.; Lim, Y.; Han, Y.S.; Paik, H.D.; Cho, S.G.; et al. Differential Regulations of Quercetin and Its Glycosides on Ligand-Gated Ion Channels. Biol. Pharm. Bull. 2008, 31, 611–617. [Google Scholar] [CrossRef][Green Version]
- Kim, B.; Park, H.; Na, D.; Lee, S.Y. Metabolic engineering of Escherichia coli for the production of phenol from glucose. Biotechnol. J. 2014, 9, 621–629. [Google Scholar] [CrossRef]
- Shomar, H.; Gontier, S.; van den Broek, N.J.F.; Tejeda Mora, H.; Noga, M.J.; Hagedoorn, P.L.; Bokinsky, G. Metabolic engineering of a carbapenem antibiotic synthesis pathway in Escherichia coli. Nat. Chem. Biol. 2018, 14, 794–800. [Google Scholar] [CrossRef]



| Strains and Plasmids | Relevant Characteristics | Source or References |
|---|---|---|
| Strains | ||
| E. coli XL1Blue | Δ(mcrA)183 Δ(mcrCB-hsdSMR-mrr)173 endA1 supE44 thi-1 recA1 gyrA1 gyrA96 relA1 lac [F’ proAB lacIqZΔM15 Tn10 9Tetr)] | Stratagene |
| C. glutamicum | Wild-type strain, ATCC 13032 | ATCC |
| Plasmids | ||
| pGEM®-T easy vector | E. coli general cloning vector, Ampr | Promega (USA) |
| pSK003 | KmR; C. glutamicum/E.coli shuttle vector. (pSod, pBL1, oriVC.g., oriVE.c). | Our lab |
| pSKSM | pSK003+ pTac +LacI | This study |
| pSKSM−YdhE | pSK003+ pTac +LacI+YdhE | This study |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Amoah, O.J.; Nguyen, H.T.; Sohng, J.K. N-Glucosylation in Corynebacterium glutamicum with YdhE from Bacillus lichenformis. Molecules 2022, 27, 3405. https://doi.org/10.3390/molecules27113405
Amoah OJ, Nguyen HT, Sohng JK. N-Glucosylation in Corynebacterium glutamicum with YdhE from Bacillus lichenformis. Molecules. 2022; 27(11):3405. https://doi.org/10.3390/molecules27113405
Chicago/Turabian StyleAmoah, Obed Jackson, Hue Thi Nguyen, and Jae Kyung Sohng. 2022. "N-Glucosylation in Corynebacterium glutamicum with YdhE from Bacillus lichenformis" Molecules 27, no. 11: 3405. https://doi.org/10.3390/molecules27113405
APA StyleAmoah, O. J., Nguyen, H. T., & Sohng, J. K. (2022). N-Glucosylation in Corynebacterium glutamicum with YdhE from Bacillus lichenformis. Molecules, 27(11), 3405. https://doi.org/10.3390/molecules27113405







