(−)-Lariciresinol Isolated from the Roots of Isatis indigotica Fortune ex Lindl. Inhibits Hepatitis B Virus by Regulating Viral Transcription
Abstract
1. Introduction
2. Results
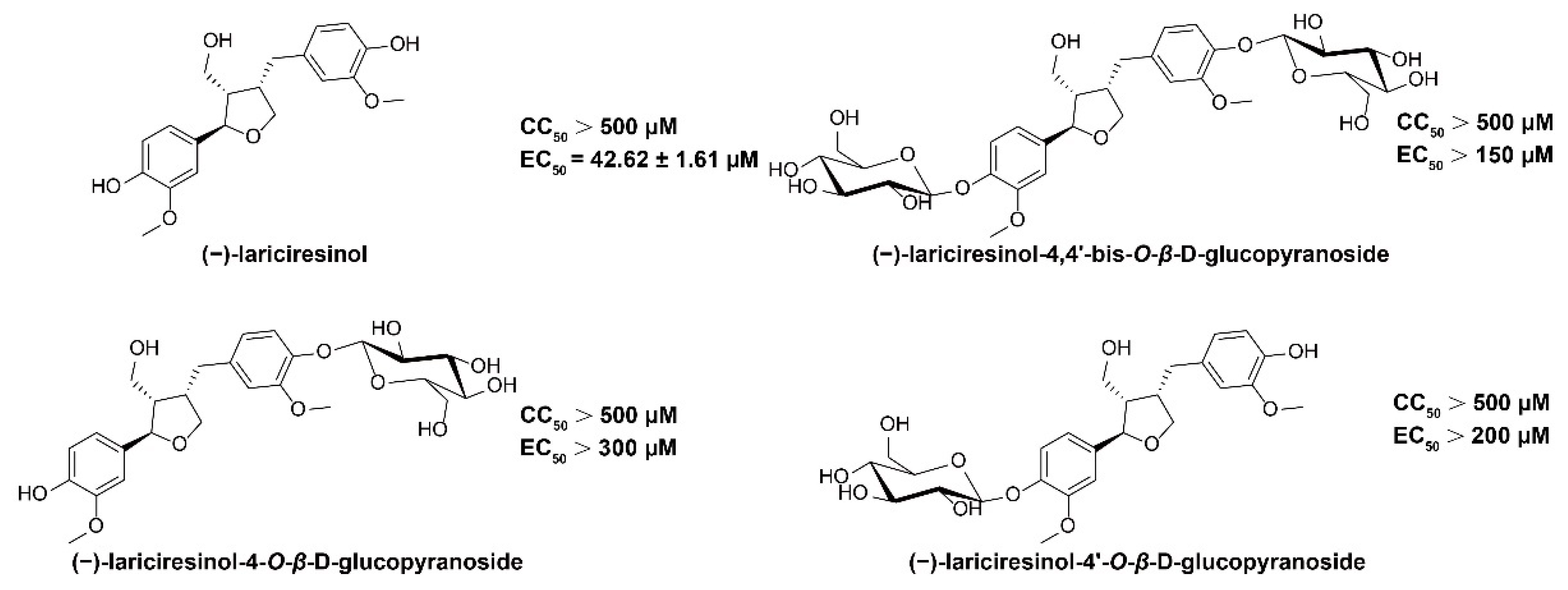
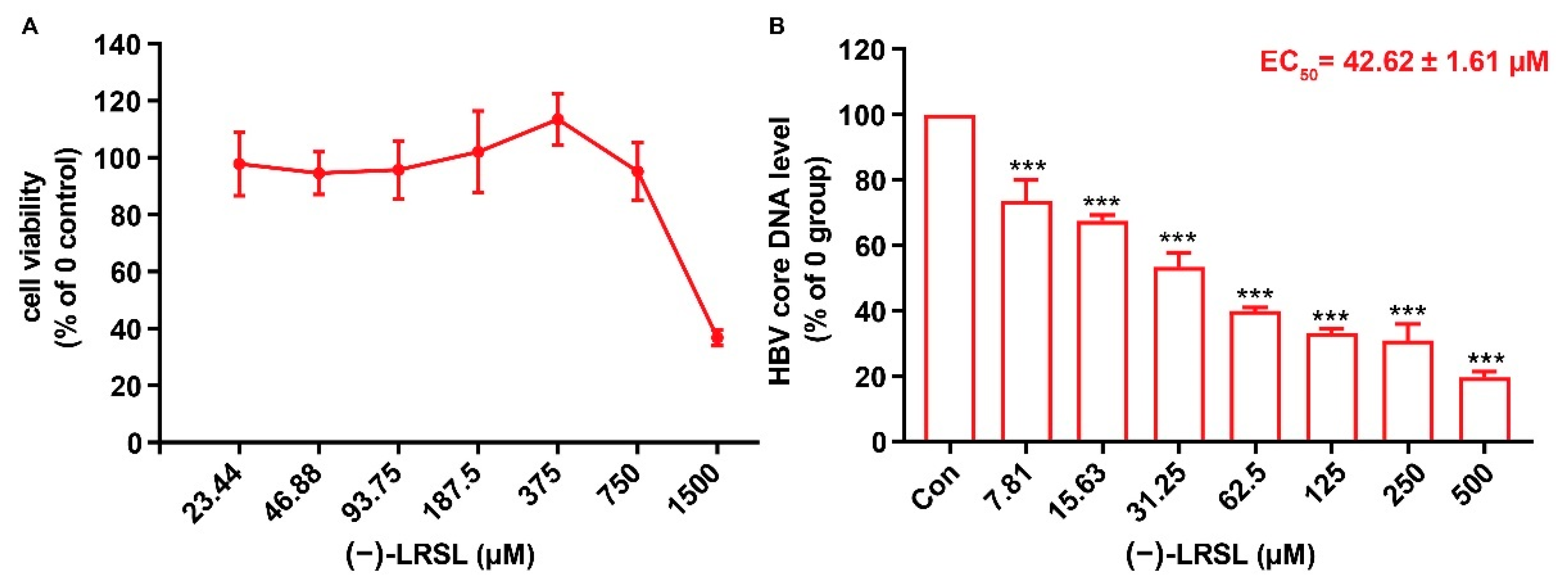
2.1. (−)-LRSL Has Anti-HBV Activity
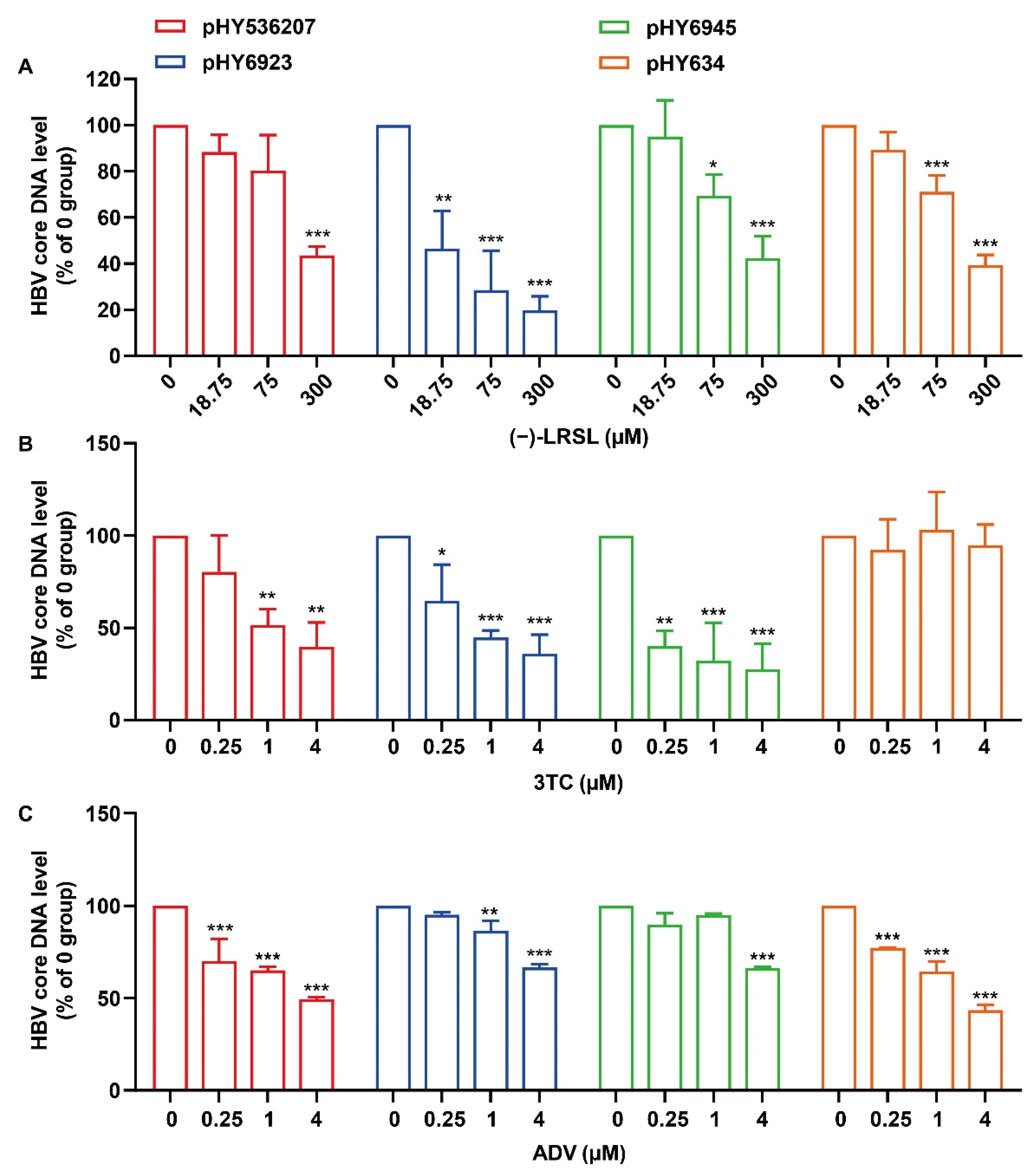
2.2. (−)-LRSL Inhibits HBV Replication Including NUC-Resistant Strains
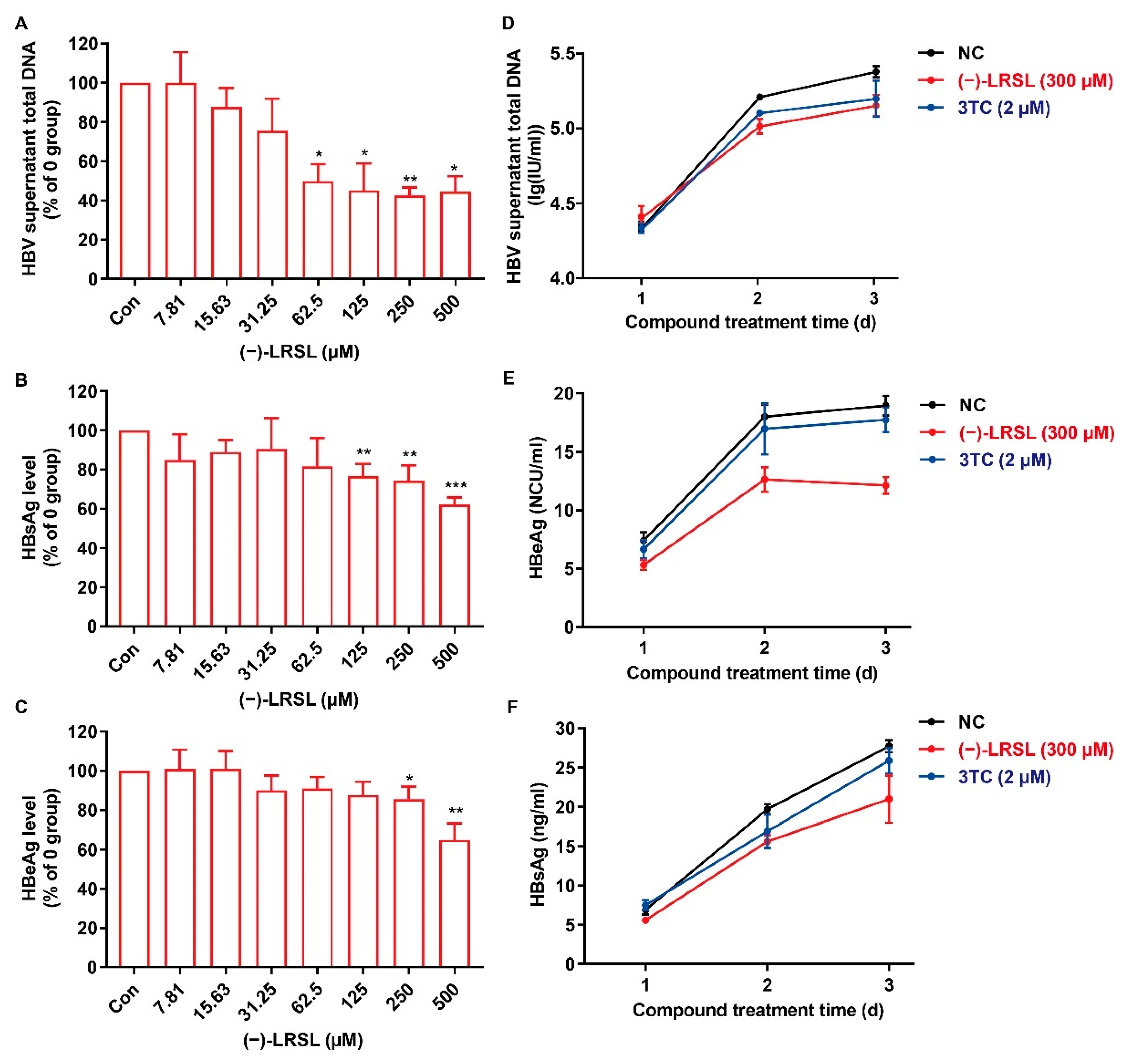
2.3. (−)-LRSL Suppressed the Secretion of HBV Antigen and Viral Particles in HepG2.2.15 Cells
2.4. (−)-LRSL Treatment Decreases the Production of HBV RNAs
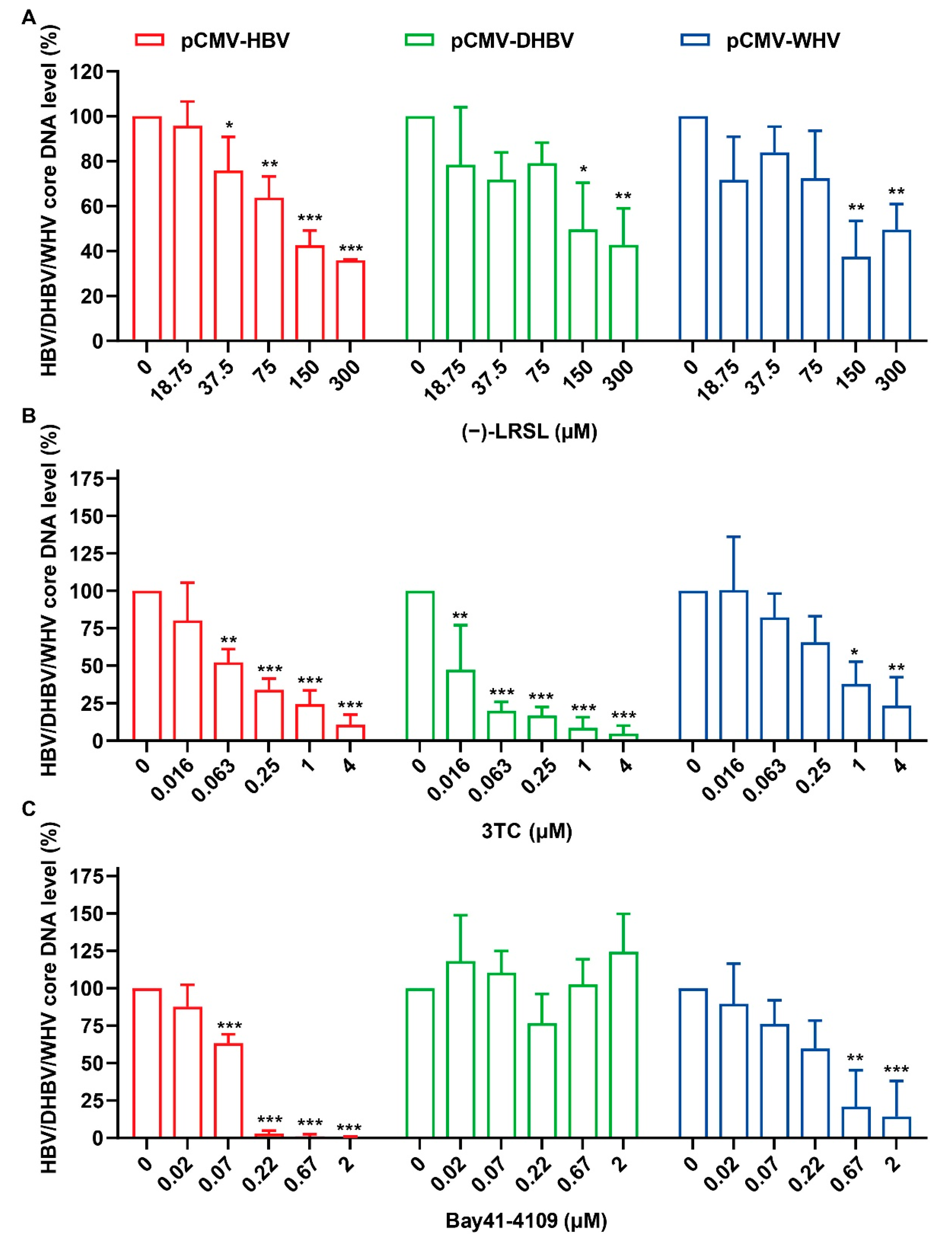
2.5. (−)-LRSL Inhibits the Replication of Different Species of Hepadnaviruses
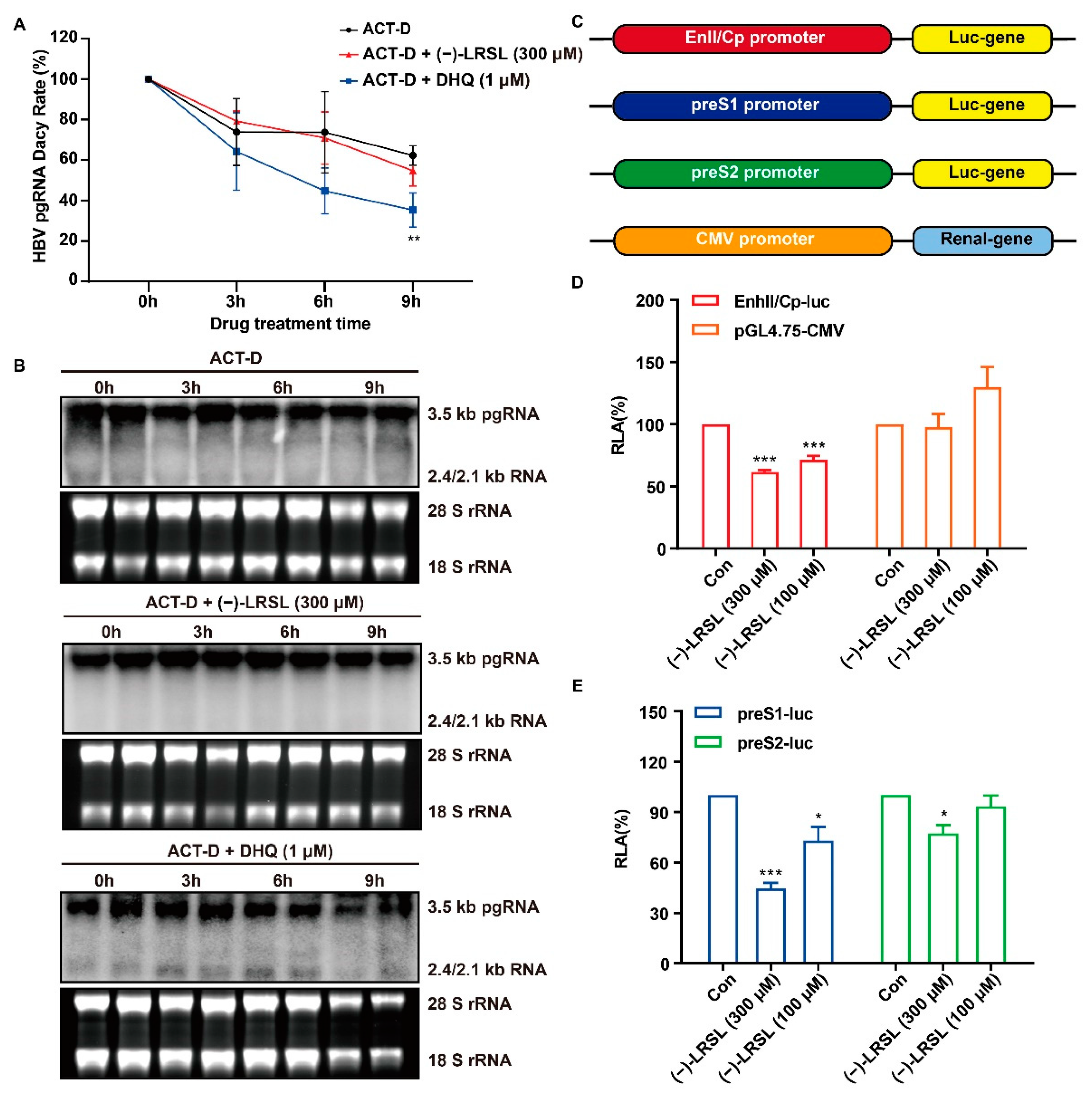
2.6. (−)-LRSL Exerts Antiviral Effects by Inhibiting HBV RNA Transcription Rather than Degradation
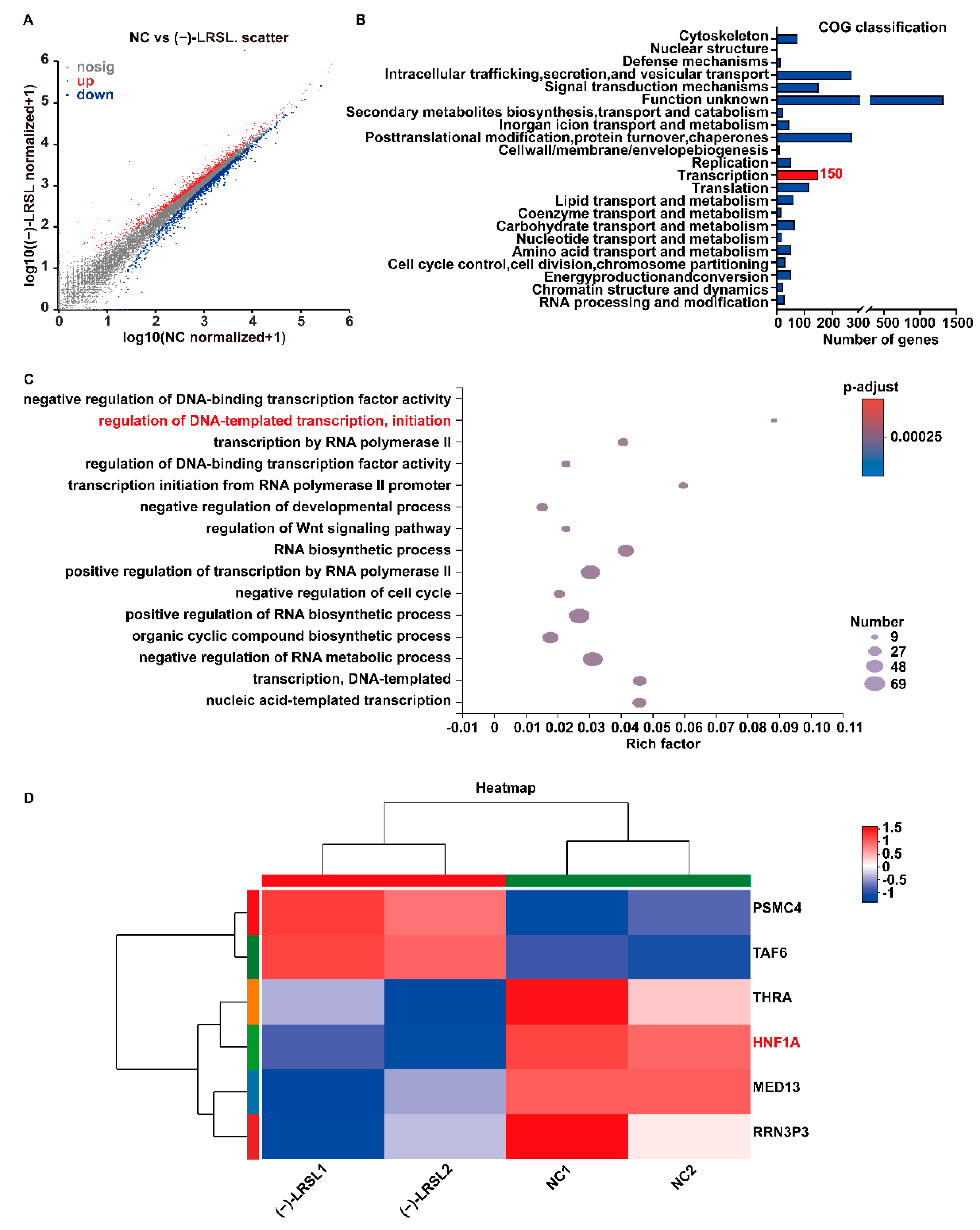
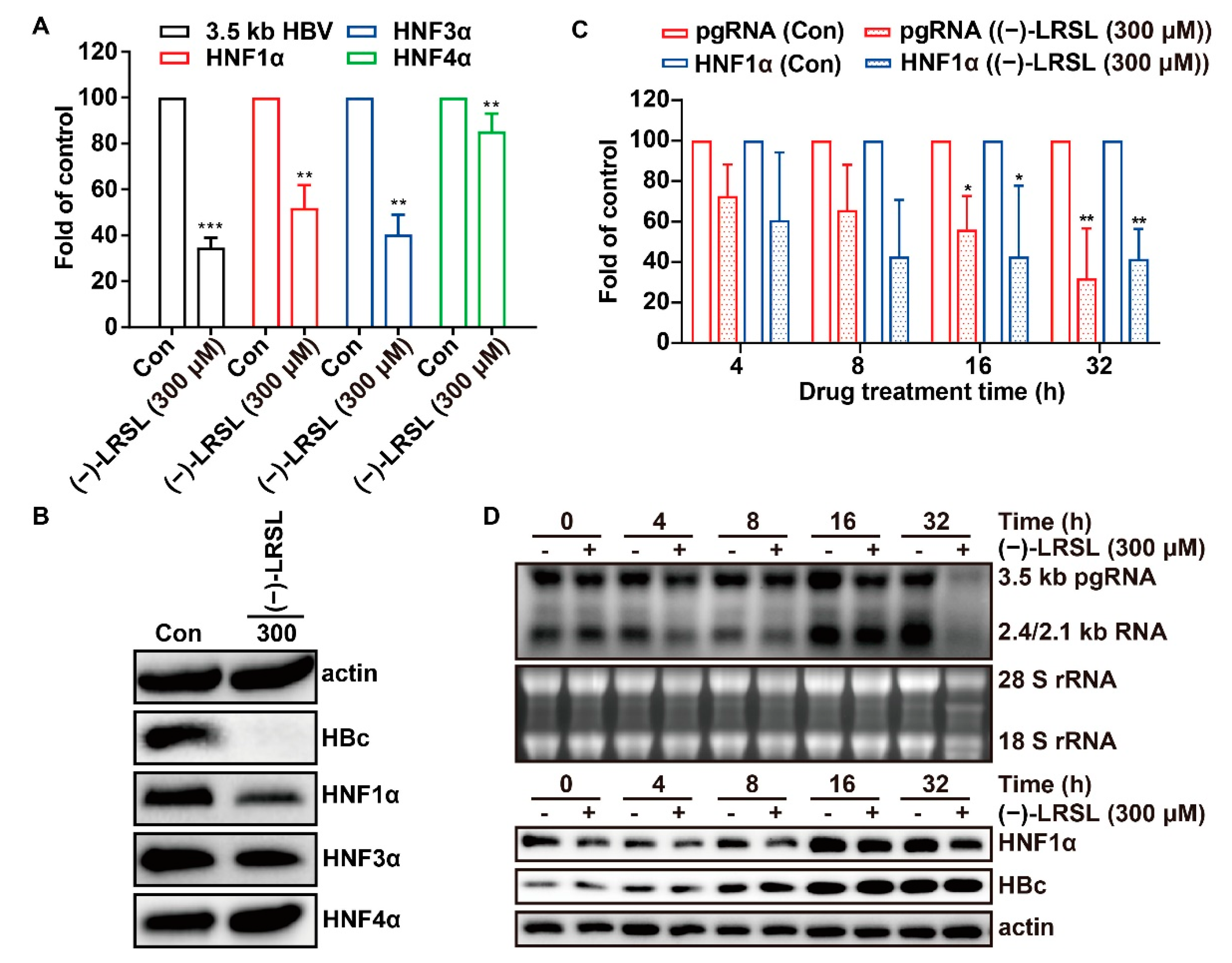
2.7. (−)-LRSL May Inhibit HBV RNA Transcription by Inhibiting HNF1α
2.8. The Inhibitory Effect of (−)-LRSL on HNF1α Is Synergistic with its Inhibitory Effect on HBV RNA
3. Discussion
4. Materials and Methods
4.1. Compounds
4.2. Cells
4.3. Plasmids
4.4. Cytotoxicity Assay
4.5. Transient-Transfection Assay
4.6. Southern Blot
4.7. Quantitative PCR (qPCR)
4.8. Northern Blot
4.9. Quantitative Reverse Transcription PCR (RT-qPCR)
4.10. HBsAg and HBeAg Quantification
4.11. Western Blot Analysis
4.12. Construction of HBV Promoter Reporter Plasmid
4.13. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Sample Availability
References
- Lucifora, J.; Arzberger, S.; Durantel, D.; Belloni, L.; Strubin, M.; Levrero, M.; Zoulim, F.; Hantz, O.; Protzer, U. Hepatitis B virus X protein is essential to initiate and maintain virus replication after infection. J. Hepatol. 2011, 55, 996–1003. [Google Scholar] [CrossRef] [PubMed]
- Karayiannis, P. Hepatitis B virus: Virology, molecular biology, life cycle and intrahepatic spread. Hepatol. Int. 2017, 11, 500–508. [Google Scholar] [CrossRef] [PubMed]
- Tsukuda, S.; Watashi, K. Hepatitis B virus biology and life cycle. Antiviral. Res. 2020, 182, 104925. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.J.; Morris-Natschke, S.L.; Cheng, Y.Y.; Lee, K.H.; Li, R.T. Development of anti-influenza agents from natural products. Med. Res. Rev. 2020, 40, 2290–2338. [Google Scholar] [CrossRef] [PubMed]
- Li, H. Advances in anti hepatic fibrotic therapy with Traditional Chinese Medicine herbal formula. J. Ethnopharmacol. 2020, 251, 112442. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.; Shen, D.; Sun, S.; Huang, Y.; Xin, Y.; Luo, H.; Chen, Y.; Zhou, Z.; Liu, F.; Chen, X. Effect of Fufang Biejia Ruangan Tablet on lowering biochemical and virological parameters of hepatic fibrosis in patients with chronic hepatitis B: Protocol for a systematic review and meta-analysis of randomized controlled trials and cohort studies. Medicine 2019, 98, e15297. [Google Scholar] [CrossRef] [PubMed]
- Yang, N.H.; Yuan, G.S.; Zhou, Y.C.; Liu, J.W.; Huang, H.P.; Hu, C.G.; Xiong, L.; Li, Y.; Zhou, F.Y.; Yang, S.L.; et al. Entecavir combined with Fufang Biejia Ruangan tablet in treatment of chronic hepatitis B patients with liver fibrosis: 96-week efficacy analyses. Nan Fang Yi Ke Da Xue Xue Bao J. South. Med. Univ. 2016, 36, 775–779. [Google Scholar]
- Wang, H.; Yang, L.M.; Huang, L. Clinical effects of qianggan capsule on the liver tissue pathology and PDGF-BB, TGF-beta1, TIMP-1, and MMP-1 factors in patients with chronic hepatitis B. Zhongguo Zhong Xi Yi Jie He Za Zhi Zhongguo Zhongxiyi Jiehe Zazhi Chin. J. Integr. Tradit. West. Med. 2011, 31, 1337–1340. [Google Scholar]
- Wang, H.; Zhao, Y.L.; Xu, K.C. Clinical and pathological study on effects of Qianggan Capsule combined lamivudine on hepatic fibrosis in patients with chronic hepatitis B. Zhongguo Zhong Xi Yi Jie He Za Zhi Zhongguo Zhongxiyi Jiehe Zazhi Chin. J. Integr. Tradit. West. Med 2006, 26, 978–980. [Google Scholar]
- Wang, T.; Wang, X.; Zhuo, Y.; Si, C.; Yang, L.; Meng, L.; Zhu, B. Antiviral activity of a polysaccharide from Radix Isatidis (Isatis indigotica Fortune) against hepatitis B virus (HBV) in vitro via activation of JAK/STAT signal pathway. J. Ethnopharmacol. 2020, 257, 112782. [Google Scholar] [CrossRef]
- Shan, J.; Zhao, C.; Li, Q.; Zhu, T.; Ren, J.; Li, H.; Wu, J.; Ma, H.; Qu, W.; Wang, Y. An arabinogalactan from Isatis indigotica and its adjuvant effects on H1N1 influenza and hepatitis B antigens. J. Funct. Foods 2015, 18, 631–642. [Google Scholar] [CrossRef]
- Chen, Q.; Lan, H.Y.; Peng, W.; Rahman, K.; Liu, Q.C.; Luan, X.; Zhang, H. Isatis indigotica: A review of phytochemistry, pharmacological activities and clinical applications. J. Pharm. Pharmacol. 2021, 73, 1137–1150. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.H. Naturally derived anti-hepatitis B virus agents and their mechanism of action. World J. Gastroenterol. 2016, 22, 188–204. [Google Scholar] [CrossRef]
- Xu, X.Y.; Wang, D.Y.; Li, Y.P.; Deyrup, S.T.; Zhang, H.J. Plant-derived lignans as potential antiviral agents: A systematic review. Phytochem. Rev. Proc. Phytochem. Soc. Eur. 2021, 21, 239–289. [Google Scholar] [CrossRef] [PubMed]
- Tseng, Y.P.; Kuo, Y.H.; Hu, C.P.; Jeng, K.S.; Janmanchi, D.; Lin, C.H.; Chou, C.K.; Yeh, S.F. The role of helioxanthin in inhibiting human hepatitis B viral replication and gene expression by interfering with the host transcriptional machinery of viral promoters. Antiviral. Res. 2008, 77, 206–214. [Google Scholar] [CrossRef]
- Chen, H.; Li, J.; Wu, Q.; Niu, X.T.; Tang, M.T.; Guan, X.L.; Li, J.; Yang, R.Y.; Deng, S.P.; Su, X.J. Anti-HBV activities of Streblus asper and constituents of its roots. Fitoterapia 2012, 83, 643–649. [Google Scholar] [CrossRef]
- Turton, K.L.; Meier-Stephenson, V.; Badmalia, M.D.; Coffin, C.S.; Patel, T.R. Host Transcription Factors in Hepatitis B Virus RNA Synthesis. Viruses 2020, 12, 160. [Google Scholar] [CrossRef]
- Tang, H.; Raney, A.K.; McLachlan, A. Replication of the wild type and a natural hepatitis B virus nucleocapsid promoter variant is differentially regulated by nuclear hormone receptors in cell culture. J. Virol. 2001, 75, 8937–8948. [Google Scholar] [CrossRef][Green Version]
- Foo, N.C.; Yen, T.S. Activation of promoters for cellular lipogenic genes by hepatitis B virus large surface protein. Virology 2000, 269, 420–425. [Google Scholar] [CrossRef][Green Version]
- Xie, M.; Guo, H.; Lou, G.; Yao, J.; Liu, Y.; Sun, Y.; Yang, Z.; Zheng, M. Neddylation inhibitor MLN4924 has anti-HBV activity via modulating the ERK-HNF1α-C/EBPα-HNF4α axis. J. Cell. Mol. Med. 2021, 25, 840–854. [Google Scholar] [CrossRef]
- Xia, C.; Tang, W.; Geng, P.; Zhu, H.; Zhou, W.; Huang, H.; Zhou, P.; Shi, X. Baicalin down-regulating hepatitis B virus transcription depends on the liver-specific HNF4alpha-HNF1alpha axis. Toxicol. Appl. Pharmacol. 2020, 403, 115131. [Google Scholar] [CrossRef] [PubMed]
- Dai, X.Q.; Cai, W.T.; Wu, X.; Chen, Y.; Han, F.M. Protocatechuic acid inhibits hepatitis B virus replication by activating ERK1/2 pathway and down-regulating HNF4alpha and HNF1alpha in vitro. Life Sci. 2017, 180, 68–74. [Google Scholar] [CrossRef] [PubMed]
- Pan, Y.; Ke, Z.; Ye, H.; Sun, L.; Ding, X.; Shen, Y.; Zhang, R.; Yuan, J. Saikosaponin C exerts anti-HBV effects by attenuating HNF1α and HNF4α expression to suppress HBV pgRNA synthesis. Inflamm. Res. Off. J. Eur. Histamine Res. Soc. 2019, 68, 1025–1034. [Google Scholar] [CrossRef] [PubMed]
- Billioud, G.; Pichoud, C.; Puerstinger, G.; Neyts, J.; Zoulim, F. The main hepatitis B virus (HBV) mutants resistant to nucleoside analogs are susceptible in vitro to non-nucleoside inhibitors of HBV replication. Antiviral. Res. 2011, 92, 271–276. [Google Scholar] [CrossRef]
- Stray, S.J.; Zlotnick, A. BAY 41-4109 has multiple effects on Hepatitis B virus capsid assembly. J. Mol. Recognit. 2006, 19, 542–548. [Google Scholar] [CrossRef]
- Campagna, M.R.; Liu, F.; Mao, R.; Mills, C.; Cai, D.; Guo, F.; Zhao, X.; Ye, H.; Cuconati, A.; Guo, H.; et al. Sulfamoylbenzamide derivatives inhibit the assembly of hepatitis B virus nucleocapsids. J. Virol. 2013, 87, 6931–6942. [Google Scholar] [CrossRef]
- Zhou, T.; Block, T.; Liu, F.; Kondratowicz, A.S.; Sun, L.; Rawat, S.; Branson, J.; Guo, F.; Steuer, H.M.; Liang, H.; et al. HBsAg mRNA degradation induced by a dihydroquinolizinone compound depends on the HBV posttranscriptional regulatory element. Antiviral. Res. 2018, 149, 191–201. [Google Scholar] [CrossRef]
- Mao, R.; Nie, H.; Cai, D.; Zhang, J.; Liu, H.; Yan, R.; Cuconati, A.; Block, T.M.; Guo, J.T.; Guo, H. Inhibition of hepatitis B virus replication by the host zinc finger antiviral protein. PLoS Pathog. 2013, 9, e1003494. [Google Scholar] [CrossRef]
- Li, J.; Zhou, B.; Li, C.; Chen, Q.; Wang, Y.; Li, Z.; Chen, T.; Yang, C.; Jiang, Z.; Zhong, N.; et al. Lariciresinol-4-O-beta-D-glucopyranoside from the root of Isatis indigotica inhibits influenza A virus-induced pro-inflammatory response. J. Ethnopharmacol. 2015, 174, 379–386. [Google Scholar] [CrossRef]
- Yang, Z.; Wang, Y.; Zheng, Z.; Zhao, S.; Zhao, J.; Lin, Q.; Li, C.; Zhu, Q.; Zhong, N. Antiviral activity of Isatis indigotica root-derived clemastanin B against human and avian influenza A and B viruses in vitro. Int. J. Mol. Med. 2013, 31, 867–873. [Google Scholar] [CrossRef]
- Wang, X.; Hassan, W.; Zhao, J.; Bakht, S.; Nie, Y.; Wang, Y.; Pang, Q.; Huang, Z. The impact of hepatocyte nuclear factor-1alpha on liver malignancies and cell stemness with metabolic consequences. Stem Cell Res. Ther. 2019, 10, 315. [Google Scholar] [CrossRef] [PubMed]
- Lau, H.H.; Ng, N.H.J.; Loo, L.S.W.; Jasmen, J.B.; Teo, A.K.K. The molecular functions of hepatocyte nuclear factors—In and beyond the liver. J. Hepatol. 2018, 68, 1033–1048. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.H.; Kang, H.S.; Kim, K.H. Roles of hepatocyte nuclear factors in hepatitis B virus infection. World J. Gastroenterol. 2016, 22, 7017–7029. [Google Scholar] [CrossRef] [PubMed]
- Kitanaka, S.; Miki, Y.; Hayashi, Y.; Igarashi, T. Promoter-specific repression of hepatocyte nuclear factor (HNF)-1 beta and HNF-1 alpha transcriptional activity by an HNF-1 beta missense mutant associated with Type 5 maturity-onset diabetes of the young with hepatic and biliary manifestations. J. Clin. Endocrinol. Metab. 2004, 89, 1369–1378. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Li, J.; Xu, Z.; Zheng, Y.; Johnson, D.L.; Ou, J.H. Regulation of hepatocyte nuclear factor 1 activity by wild-type and mutant hepatitis B virus X proteins. J. Virol. 2002, 76, 5875–5881. [Google Scholar] [CrossRef]
- Zheng, Y.; Li, J.; Ou, J.H. Regulation of hepatitis B virus core promoter by transcription factors HNF1 and HNF4 and the viral X protein. J. Virol. 2004, 78, 6908–6914. [Google Scholar] [CrossRef]
- Bar-Yishay, I.; Shaul, Y.; Shlomai, A. Hepatocyte metabolic signalling pathways and regulation of hepatitis B virus expression. Liv. Int. 2011, 31, 282–290. [Google Scholar] [CrossRef]
- Lin, J.; Gu, C.; Shen, Z.; Liu, Y.; Wang, W.; Tao, S.; Cui, X.; Liu, J.; Xie, Y. Hepatocyte nuclear factor 1alpha downregulates HBV gene expression and replication by activating the NF-kappaB signaling pathway. PLoS ONE 2017, 12, e0174017. [Google Scholar]
- Yamauchi, S.; Kumamoto, M.; Ochi, Y.; Nishiwaki, H.; Shuto, Y. Structure-plant growth inhibitory activity relationship of lariciresinol. J. Agric. Food Chem. 2013, 61, 12297–12306. [Google Scholar] [CrossRef]
- Zuo, L.; Li, J.B.; Xu, J.; Yang, J.Z.; Zhang, D.M.; Tong, Y.L. Studies on chemical constituents in root of Isatis indigotica. China J. Chin. Mater. Med. 2007, 32, 688–691. [Google Scholar]
- He, L.W.; Li, X.; Chen, J.W.; Sun, D.D.; Jü, W.Z.; Wang, K.C. Chemical constituents from water extract of Radix isatidis. Acta Pharm. Sin. 2006, 41, 1193–1196. [Google Scholar]
- Zhang, J.M.; Yao, X.; Wang, Y.X.; Liu, F.; Ma, Z.M.; Weng, X.H.; Wen, Y.M. High replicative full-length lamivudine-resistant hepatitis B virus isolated during acute exacerbations. J. Med. Virol. 2005, 77, 203–208. [Google Scholar] [CrossRef] [PubMed]
- Guo, H.; Jiang, D.; Zhou, T.; Cuconati, A.; Block, T.M.; Guo, J.T. Characterization of the intracellular deproteinized relaxed circular DNA of hepatitis B virus: An intermediate of covalently closed circular DNA formation. J. Virol. 2007, 81, 12472–12484. [Google Scholar] [CrossRef]
- Guo, J.T.; Pryce, M.; Wang, X.; Barrasa, M.I.; Hu, J.; Seeger, C. Conditional replication of duck hepatitis B virus in hepatoma cells. J. Virol. 2003, 77, 1885–1893. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.X.; Yang, L.; Wang, H.Q.; Zhao, X.Q.; Liu, T.; Li, Y.H.; Zeng, Q.X.; Li, Y.H.; Song, D.Q. Synthesis and Evolution of Berberine Derivatives as a New Class of Antiviral Agents against Enterovirus 71 through the MEK/ERK Pathway and Autophagy. Molecules 2018, 23, 2084. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, L.; Wang, H.; Yan, H.; Wang, K.; Wu, S.; Li, Y. (−)-Lariciresinol Isolated from the Roots of Isatis indigotica Fortune ex Lindl. Inhibits Hepatitis B Virus by Regulating Viral Transcription. Molecules 2022, 27, 3223. https://doi.org/10.3390/molecules27103223
Yang L, Wang H, Yan H, Wang K, Wu S, Li Y. (−)-Lariciresinol Isolated from the Roots of Isatis indigotica Fortune ex Lindl. Inhibits Hepatitis B Virus by Regulating Viral Transcription. Molecules. 2022; 27(10):3223. https://doi.org/10.3390/molecules27103223
Chicago/Turabian StyleYang, Lu, Huiqiang Wang, Haiyan Yan, Kun Wang, Shuo Wu, and Yuhuan Li. 2022. "(−)-Lariciresinol Isolated from the Roots of Isatis indigotica Fortune ex Lindl. Inhibits Hepatitis B Virus by Regulating Viral Transcription" Molecules 27, no. 10: 3223. https://doi.org/10.3390/molecules27103223
APA StyleYang, L., Wang, H., Yan, H., Wang, K., Wu, S., & Li, Y. (2022). (−)-Lariciresinol Isolated from the Roots of Isatis indigotica Fortune ex Lindl. Inhibits Hepatitis B Virus by Regulating Viral Transcription. Molecules, 27(10), 3223. https://doi.org/10.3390/molecules27103223





