Computational Design of Novel Allosteric Inhibitors for Plasmodium falciparum DegP
Abstract
1. Introduction
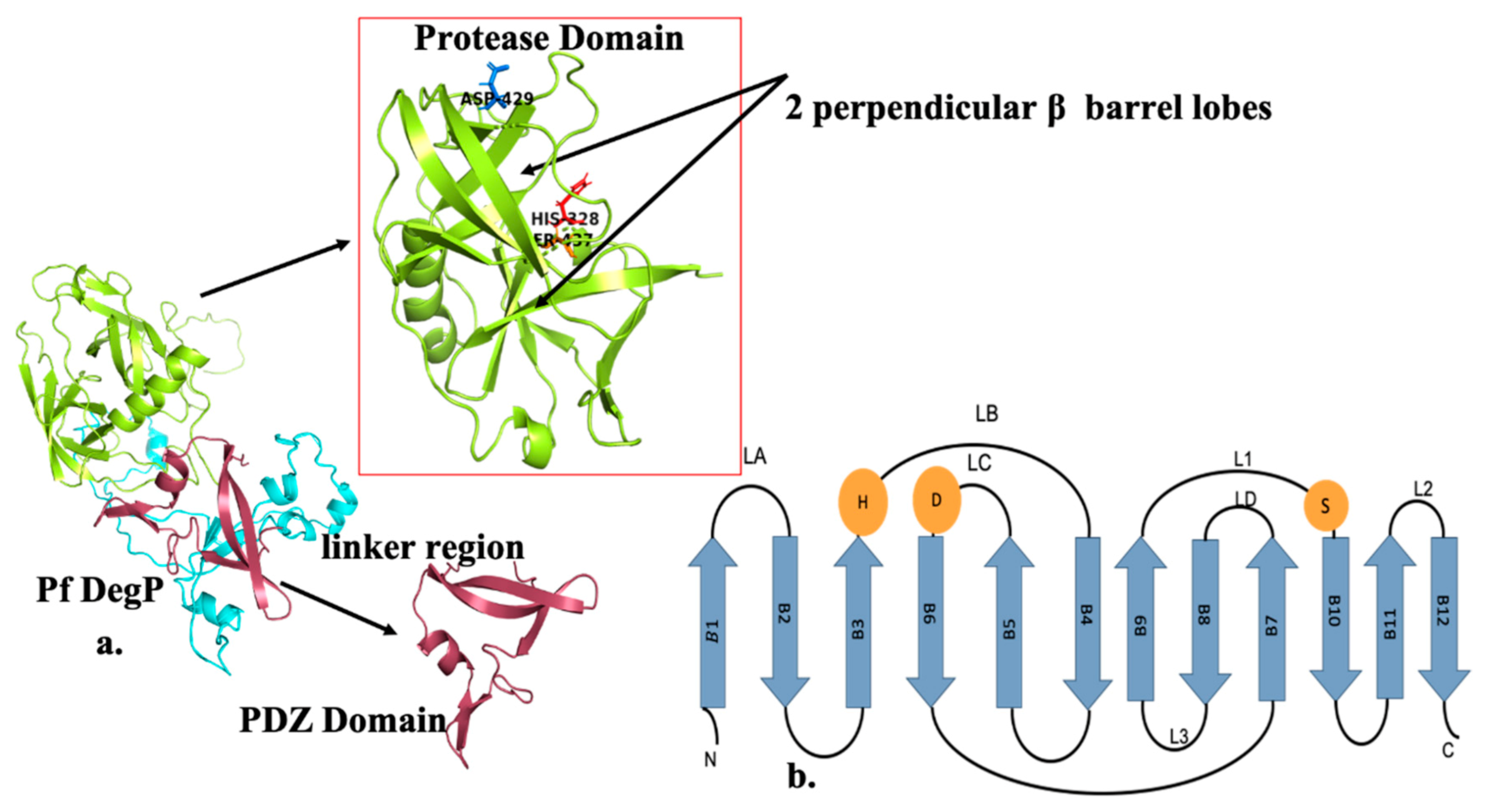
DegP Structure and Mechanism
2. Result
2.1. Conserved Domain, Evolutionary and Interlog Analysis of PfDegP
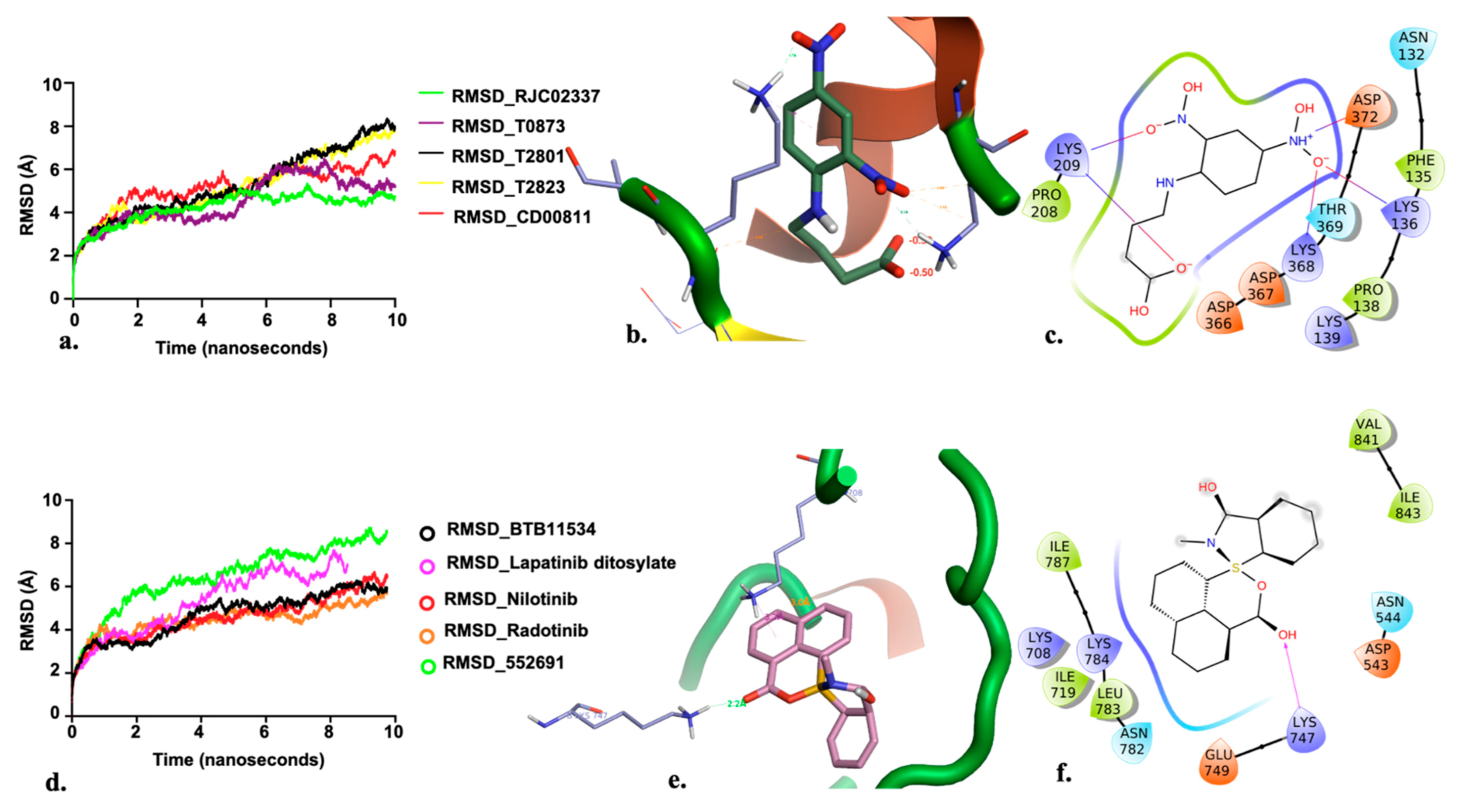
2.2. Determination of Binding Sites within the 3D Structure of PfDegP
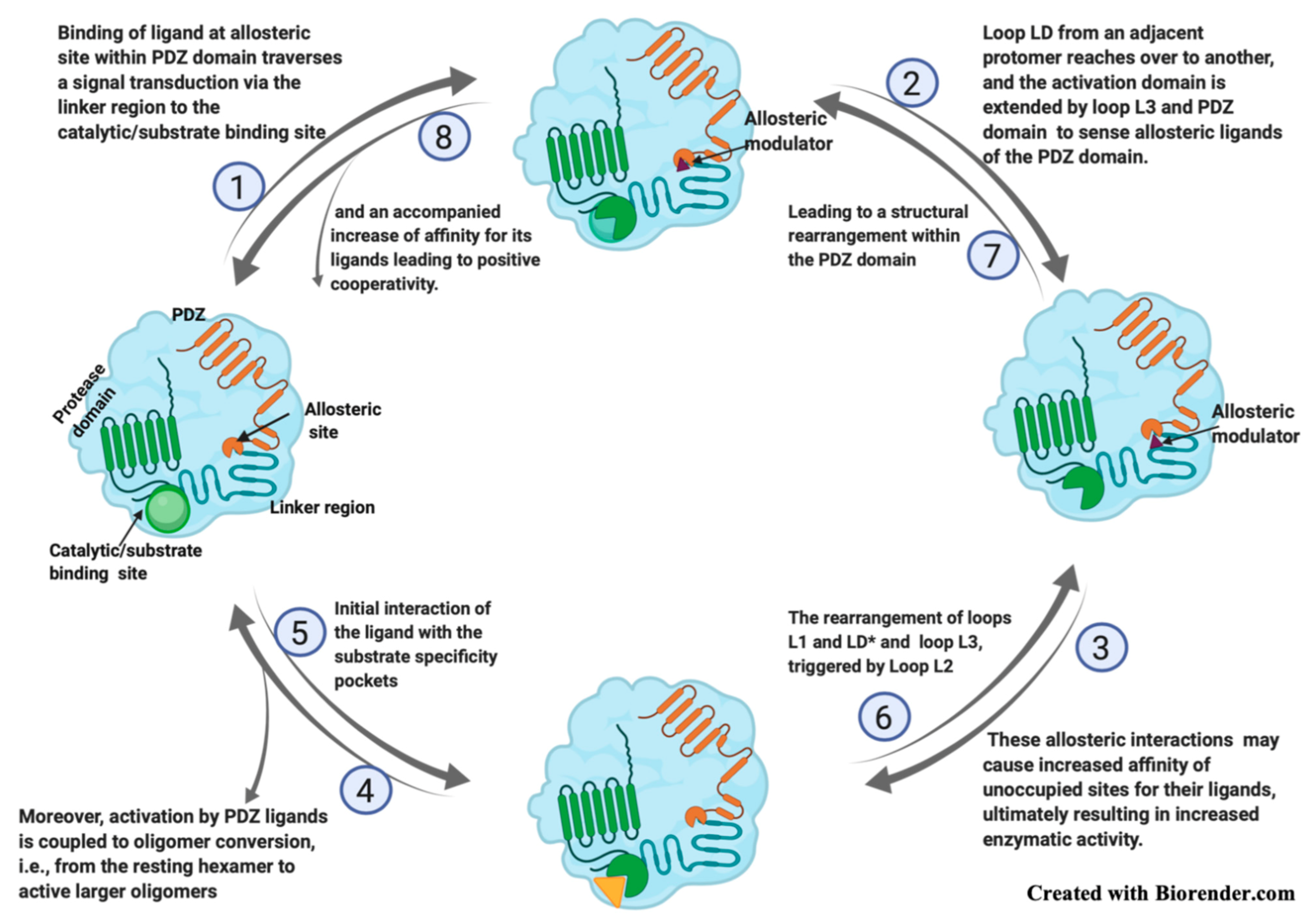
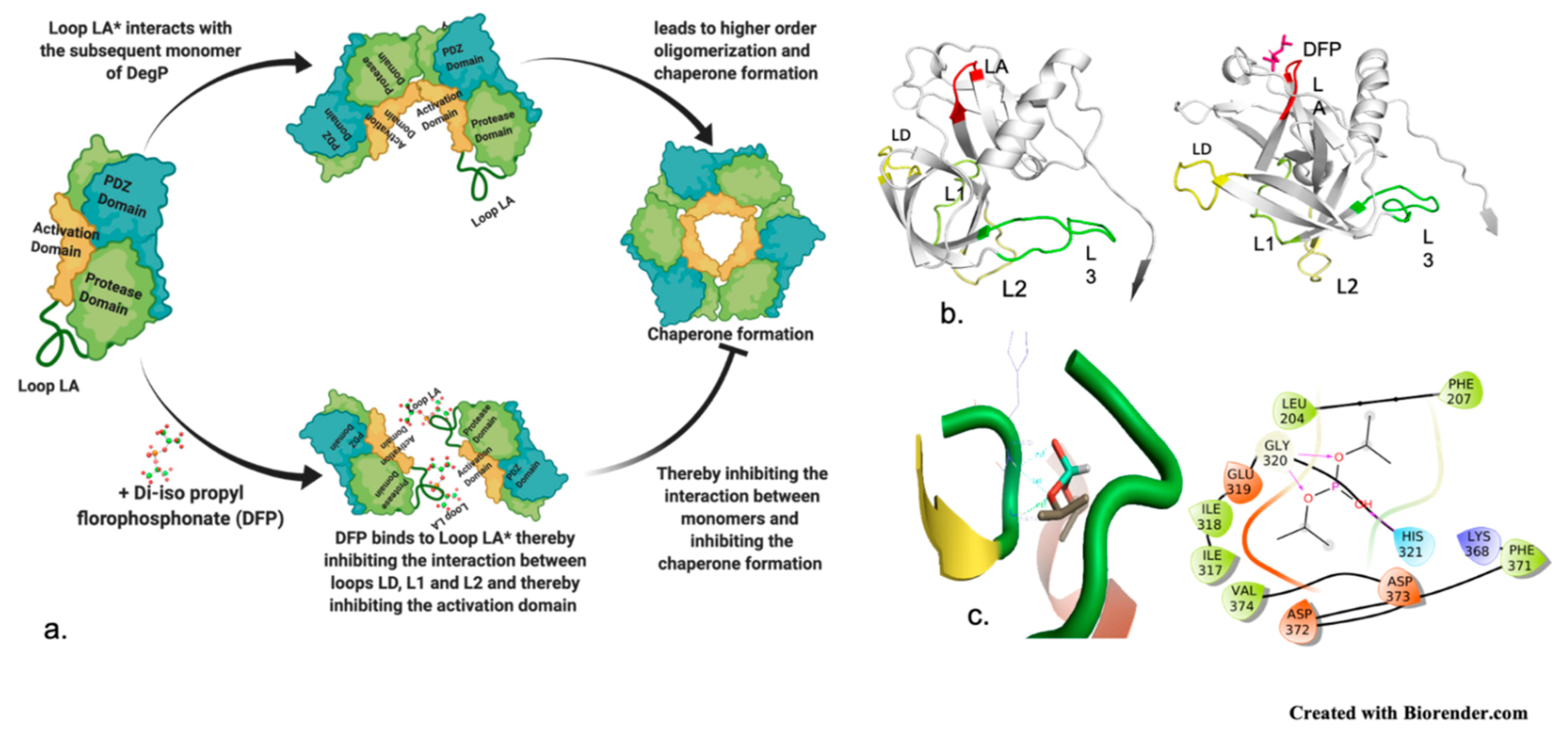
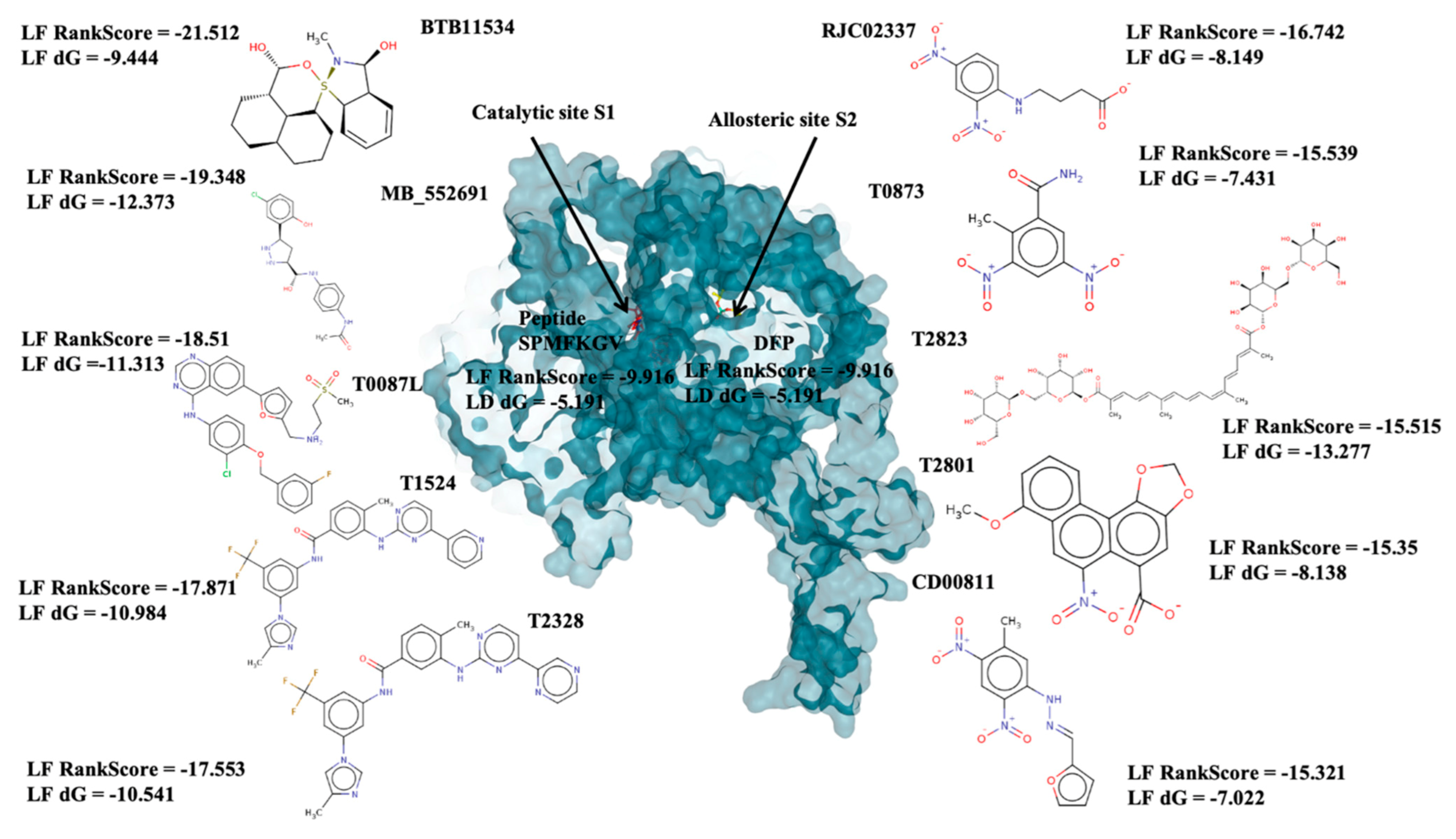
2.3. Allosteric Site
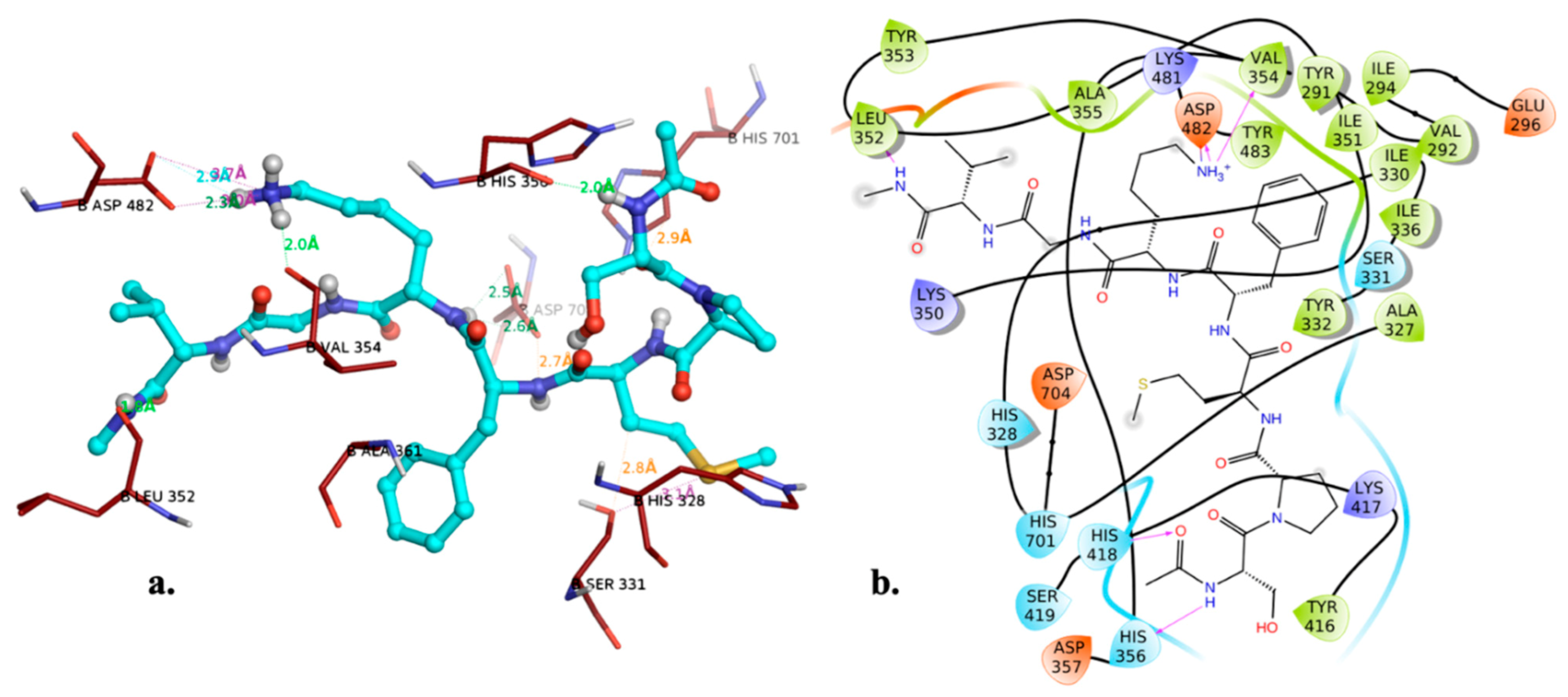
2.4. Catalytic Site Binders
2.5. Compound Annotations and Design of New Chemotypes
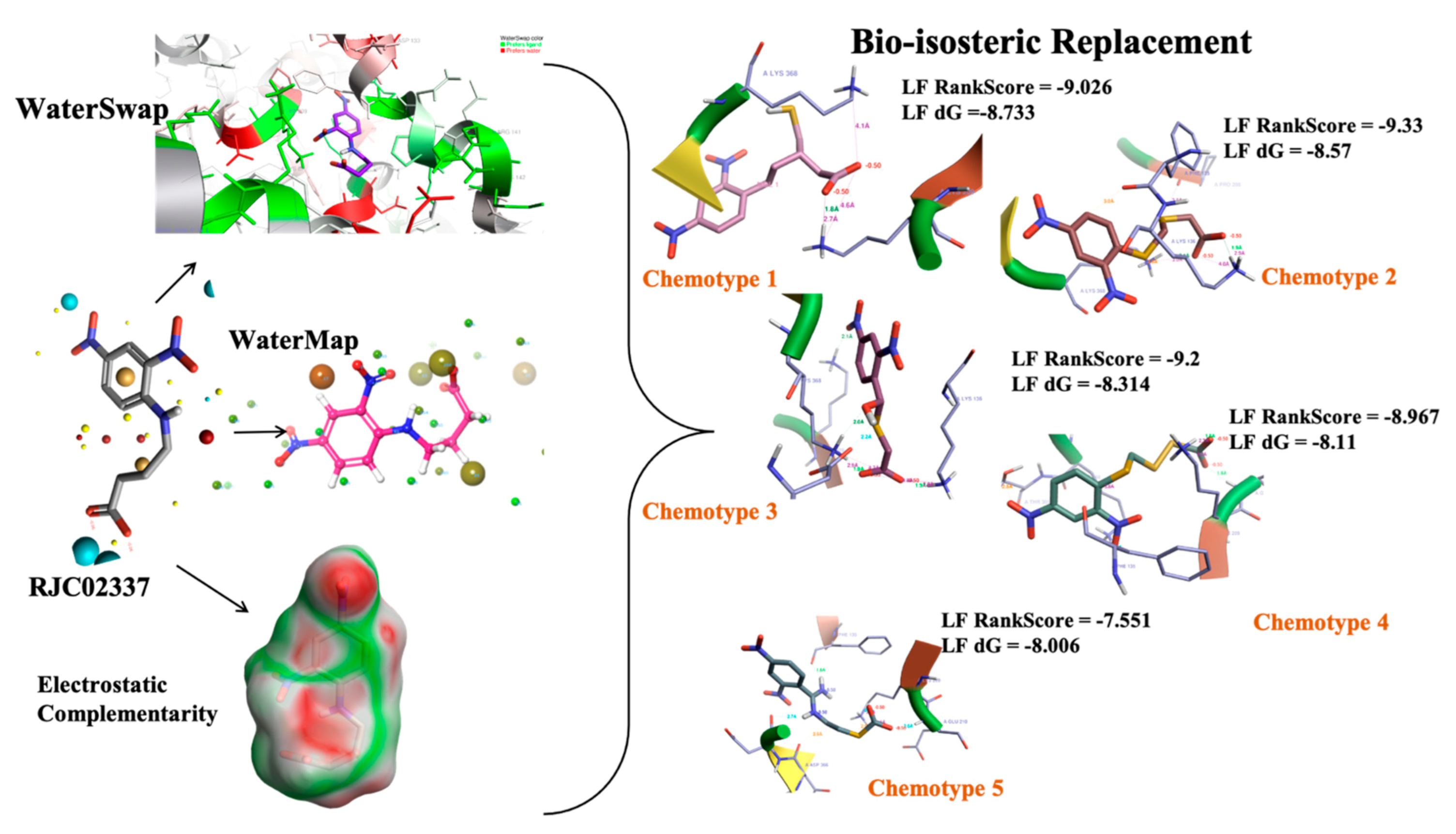
2.5.1. Absolute Binding Free Energy Calculation of Allosteric Modulator
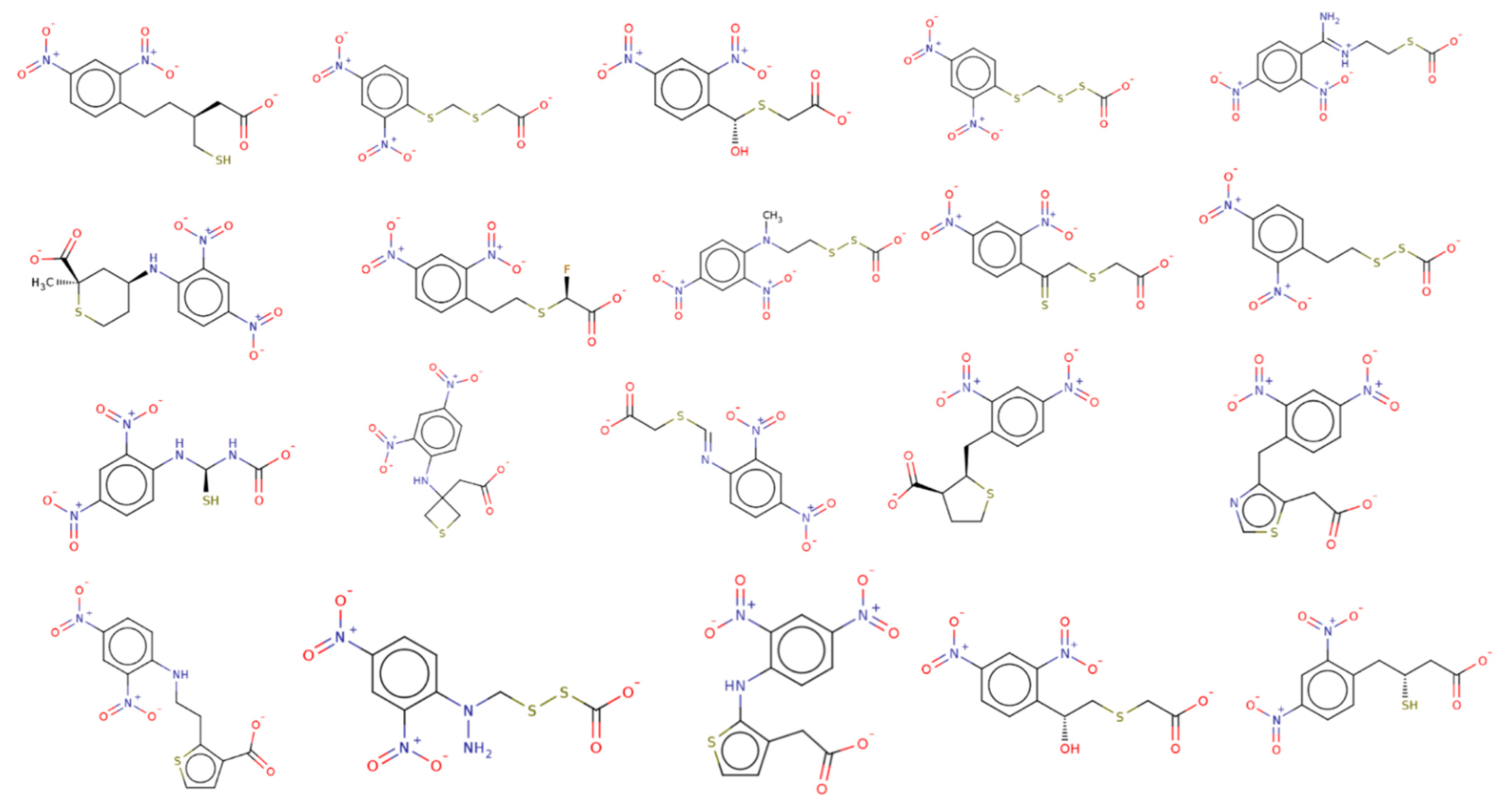
2.5.2. Novel Chemotypes Identification for Compound RJC02337 Based on Watermap, Electrostatic Complementarity and Bioisosteric Replacement
3. Discussion
4. Materials and Methods
4.1. Conserved Domain, Evolutionary and Interlog Analysis
4.2. Generation of PfDegP 3D Structure and Its MD Simulation
4.3. Binding Site Analysis
4.4. Virtual Screening and MD Trajectory Analysis
4.5. Absolute Binding Free Energy Calculation
4.6. Hydration Energy Calculations
4.7. Electrostatic Complementarity Analysis and Bio-Isosteric Replacements in Anti-Malarial Drug Design
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
Appendix A
Appendix A.1. Binding Energy Calculation Using AMBER
Appendix A.2. Parameters Used during WaterSwap Analysis
Appendix A.3. WaterMap Parameters
References
- Groll, M.; Bochtler, M.; Brandstetter, H.; Clausen, T.; Huber, R. Molecular Machines for Protein Degradation. ChemBioChem 2005, 6, 222–256. [Google Scholar] [CrossRef] [PubMed]
- Wickner, S. Posttranslational Quality Control: Folding, Refolding, and Degrading Proteins. Science 1999, 286, 1888–1893. [Google Scholar] [CrossRef] [PubMed]
- Gottesman, S.; Wickner, S.; Maurizi, M.R. Protein quality control: Triage by chaperones and proteases. Genes Dev. 1997, 11, 815–823. [Google Scholar] [CrossRef] [PubMed]
- Lipinska, B.; Zylicz, M.; Georgopoulos, C. The HtrA (DegP) protein, essential for Escherichia coli survival at high temperatures, is an endopeptidase. J. Bacteriol. 1990, 172, 1791–1797. [Google Scholar] [CrossRef]
- Spiess, C.; Beil, A.; Ehrmann, M. A Temperature-Dependent Switch from Chaperone to Protease in a Widely Conserved Heat Shock Protein. Cell 1999, 97, 339–347. [Google Scholar] [CrossRef]
- Sharma, D.; Soni, R.; Patel, S.; Joshi, D.; Bhatt, T.K. In-silico studies on DegP protein of Plasmodium falciparum in search of anti-malarials. J. Mol. Model. 2016, 22, 201. [Google Scholar] [CrossRef]
- Clausen, T.; Kaiser, M.; Huber, R.; Ehrmann, M. HTRA proteases: Regulated proteolysis in protein quality control. Nat. Rev. Mol. Cell Biol. 2011, 12, 152–162. [Google Scholar] [CrossRef]
- Jones, C.H.; Bolken, T.C.; Jones, K.F.; Zeller, G.O.; Hruby, D.E. Conserved DegP Protease in Gram-Positive Bacteria Is Essential for Thermal and Oxidative Tolerance and Full Virulence inStreptococcus pyogenes. Infect. Immun. 2001, 69, 5538–5545. [Google Scholar] [CrossRef]
- Sklar, J.G.; Wu, T.; Kahne, D.; Silhavy, T.J. Defining the roles of the periplasmic chaperones SurA, Skp, and DegP in Escherichia coli. Genes Dev. 2007, 21, 2473–2484. [Google Scholar] [CrossRef]
- Wessler, S.; Schneider, G.; Backert, S. Bacterial serine protease HtrA as a promising new target for antimicrobial therapy? Cell Commun. Signal. 2017, 15, 1–5. [Google Scholar] [CrossRef]
- Altindis, E.; Fu, Y.; Mekalanos, J.J. Proteomic analysis of Vibrio cholerae outer membrane vesicles. Proc. Natl. Acad. Sci. USA 2014, 111, E1548–E1556. [Google Scholar] [CrossRef]
- Chien, J.; Ota, T.; Aletti, G.; Shridhar, R.; Boccellino, M.; Quagliuolo, L.; Baldi, A.; Shridhar, V. Serine Protease HtrA1 Associates with Microtubules and Inhibits Cell Migration. Mol. Cell Biol. 2009, 29, 4177–4187. [Google Scholar] [CrossRef]
- Sheng, M.; Sala, C. PDZ Domains and the Organization of Supramolecular Complexes. Annu. Rev. Neurosci. 2001, 24, 1–29. [Google Scholar] [CrossRef]
- Meltzer, M.; Hasenbein, S.; Hauske, P.; Kucz, N.; Merdanovic, M.; Grau, S.; Beil, A.; Jones, D.; Krojer, T.; Clausen, T.; et al. Allosteric Activation of HtrA Protease DegP by Stress Signals during Bacterial Protein Quality Control. Angew. Chem. Int. Ed. 2008, 47, 1332–1334. [Google Scholar] [CrossRef]
- Perona, J.J.; Craik, C.S. Structural basis of substrate specificity in the serine proteases. Protein Sci. 2008, 4, 337–360. [Google Scholar] [CrossRef]
- Krojer, T.; Garrido-Franco, M.; Huber, R.; Ehrmann, M.; Clausen, T. Crystal structure of DegP (HtrA) reveals a new protease-chaperone machine. Nat. Cell Biol. 2002, 416, 455–459. [Google Scholar] [CrossRef]
- Liao, I.D.; Qian, J.; Chisholm, A.D.; Jordan, D.B.; Diner, A.B. Crystal structures of the photosystem II D1 C-terminal processing protease. Nat. Genet. 2000, 7, 749–753. [Google Scholar]
- Cabral, J.H.M.; Petosa, C.; Sutcliffe, M.J.; Raza, S.; Byron, O.; Poy, F.; Marfatia, S.M.; Chishti, A.H.; Liddington, R.C. Crystal structure of a PDZ domain. Nature 1996, 382, 649–652. [Google Scholar] [CrossRef]
- Krojer, T.; Pangerl, K.; Kurt, J.; Sawa, J.; Stingl, C.; Mechtler, K.; Huber, R.; Ehrmann, M.; Clausen, T. Interplay of PDZ and protease domain of DegP ensures efficient elimination of misfolded proteins. Proc. Natl. Acad. Sci. USA 2008, 105, 7702–7707. [Google Scholar] [CrossRef]
- Merdanovic, M.; Burston, S.G.; Schmitz, A.L.; Köcher, S.; Knapp, S.; Clausen, T.; Kaiser, M.; Huber, R.; Ehrmann, M. Activation by substoichiometric inhibition. Proc. Natl. Acad. Sci. USA 2020, 117, 1414–1418. [Google Scholar] [CrossRef]
- Merdanovic, M.; Mamant, N.; Meltzer, M.; Poepsel, S.; Auckenthaler, A.; Melgaard, R.; Hauske, P.; Nagel-Steger, L.; Clarke, A.R.; Kaiser, M.; et al. Determinants of structural and functional plasticity of a widely conserved protease chaperone complex. Nat. Struct. Mol. Biol. 2010, 17, 837–843. [Google Scholar] [CrossRef]
- Aurrecoechea, C.; Brestelli, J.; Brunk, B.P.; Dommer, J.; Fischer, S.; Gajria, B.; Gao, X.; Gingle, A.; Grant, G.; Harb, O.S.; et al. PlasmoDB: A functional genomic database for malaria parasites. Nucleic Acids Res. 2008, 37, D539–D543. [Google Scholar] [CrossRef]
- Lu, S.; Wang, J.; Chitsaz, F.; Derbyshire, M.K.; Geer, R.C.; Gonzales, N.R.; Gwadz, M.; Hurwitz, D.I.; Marchler, G.H.; Song, J.S.; et al. CDD/SPARCLE: The conserved domain database in 2020. Nucleic Acids Res. 2020, 48, D265–D268. [Google Scholar] [CrossRef]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; Mcgettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R.; et al. Clustal W and Clustal X version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef]
- Li, L.; Stoeckert, C.J.; Roos, D.S. OrthoMCL: Identification of Ortholog Groups for Eukaryotic Genomes. Genome Res. 2003, 13, 2178–2189. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A Software Environment for Integrated Models of Biomolecular Interaction Networks. Genome Res. 2013, 13, 2498–2504. [Google Scholar] [CrossRef]
- Jones, D.T. Protein secondary structure prediction based on position-specific scoring matrices. J. Mol. Biol. 1999, 292, 195–202. [Google Scholar] [CrossRef]
- Zheng, W.; Wuyun, Q.; Li, Y.; Mortuza, S.M.; Zhang, C.; Pearce, R.; Ruan, J.; Zhang, Y. Detecting distant-homology protein structures by aligning deep neural-network based contact maps. PLoS Comput. Biol. 2019, 15, e100741. [Google Scholar] [CrossRef]
- Sapay, N.; Tieleman, D.P. Combination of the CHARMM27 force field with united-atom lipid force fields. J. Comput. Chem. 2010, 32, 1400–1410. [Google Scholar] [CrossRef]
- Van Der Spoel, D.; Lindahl, E.; Hess, B.; Groenhof, G.; Mark, A.E.; Berendsen, H.J.C. GROMACS: Fast, flexible, and free. J. Comput. Chem. 2005, 26, 1701–1718. [Google Scholar] [CrossRef]
- Colovos, C.; Yeates, T.O. Verification of protein structures: Patterns of nonbonded atomic interactions. Protein Sci. 1993, 2, 1511–1519. [Google Scholar] [CrossRef]
- Hauske, P.; Meltzer, M.; Ottmann, C.C.; Krojer, T.; Clausen, T.; Ehrmann, M.; Kaiser, M. Selectivity profiling of DegP substrates and inhibitors. Bioorganic. Med. Chem. 2009, 17, 2920–2924. [Google Scholar] [CrossRef]
- Zurawa-Janicka, D.; Wenta, T.; Jarzab, M.; Skorko-Glonek, J.; Glaza, P.; Gieldon, A.; Ciarkowski, J.; Lipinska, B. Structural insights into the activation mechanisms of human HtrA serine proteases. Arch. Biochem. Biophys. 2017, 621, 6–23. [Google Scholar] [CrossRef]
- Perona, J.J.; Craik, C.S. Evolutionary Divergence of Substrate Specificity within the Chymotrypsin-like Serine Protease Fold. J. Biol. Chem. 1997, 272, 29987–29990. [Google Scholar] [CrossRef]
- Krojer, T.; Sawa, J.; Huber, R.; Clausen, T. HtrA proteases have a conserved activation mechanism that can be triggered by distinct molecular cues. Nat. Struct. Mol. Biol. 2010, 17, 844–852. [Google Scholar] [CrossRef]
- Stroganov, O.V.; Novikov, F.N.; Stroylov, V.S.; Kulkov, V.; Chilov, G.G. Lead Finder: An Approach To Improve Accuracy of Protein−Ligand Docking, Binding Energy Estimation, and Virtual Screening. J. Chem. Inf. Model. 2008, 48, 2371–2385. [Google Scholar] [CrossRef]
- Cheeseright, T.; Mackey, M.; Rose, S.; Vinter, A. Molecular Field Extrema as Descriptors of Biological Activity: Definition and Validation. J. Chem. Inf. Model. 2006, 46, 665–676. [Google Scholar] [CrossRef]
- Qu, D.; Ma, W.; Ye, Y.; Han, J. Effect of dinitolmide intercalated into Montmorillonite on E. tenella infection in chickens. Parasitol. Res. 2014, 113, 1233–1238. [Google Scholar] [CrossRef]
- Khedr, L.; Rahmo, R.M.; Farag, D.B.; Schaalan, M.F.; El Magdoub, H.M. Crocin attenuates cisplatin-induced hepatotoxicity via TLR4/NF-κBp50 signaling and BAMBI modulation of TGF-β activity: Involvement of miRNA-9 and miRNA-29. Food Chem. Toxicol. 2020, 140, 111307. [Google Scholar] [CrossRef]
- Cui, Y.; Han, J.; Ren, J.; Chen, H.; Xu, B.; Song, N.; Li, H.; Liang, A.; Shen, G. Untargeted LC-MS-based metabonomics revealed that aristolochic acid I induces testicular toxicity by inhibiting amino acids metabolism, glucose metabolism, β-oxidation of fatty acids and the TCA cycle in male mice. Toxicol. Appl. Pharmacol. 2019, 373, 26–38. [Google Scholar] [CrossRef]
- Kavasi, R.-M.; Berdiaki, A.; Spyridaki, I.; Papoutsidakis, A.; Corsini, E.; Tsatsakis, A.; Tzanakakis, G.N.; Nikitovic, D. Contact allergen (PPD and DNCB)-induced keratinocyte sensitization is partly mediated through a low molecular weight hyaluronan (LMWHA)/TLR4/NF-κB signaling axis. Toxicol. Appl. Pharmacol. 2019, 377, 114632. [Google Scholar] [CrossRef]
- Li, C.; Wang, X.; Bi, Y.; Yu, H.; Wei, J.; Zhang, Y.; Han, L.; Zhang, Y. Potent Inhibitors of Organic Anion Transporters 1 and 3 From Natural Compounds and Their Protective Effect on Aristolochic Acid Nephropathy. Toxicol. Sci. 2020, 175, 279–291. [Google Scholar] [CrossRef]
- Sborchia, M.; De Prez, E.G.; Antoine, M.-H.; Bienfait, L.; Indra, R.; Valbuena, G.; Phillips, D.H.; Nortier, J.L.; Stiborová, M.; Keun, H.C.; et al. The impact of p53 on aristolochic acid I-induced nephrotoxicity. Arch. Toxicol. 2019, 93, 3345–3366. [Google Scholar] [CrossRef]
- Ye, J.; Qian, Z.; Xue, M.; Liu, Y.; Zhu, S.; Li, Y.; Liu, X.; Cai, D.; Rui, J.; Zhang, L. Aristolochic acid I aggravates renal injury by activating the C3a/C3aR complement system. Toxicol. Lett. 2019, 312, 118–124. [Google Scholar] [CrossRef]
- Asari, Y.; Kageyama, K.; Sugiyama, A.; Kogawa, H.; Niioka, K.; Daimon, M. Lapatinib decreases the ACTH production and proliferation of corticotroph tumor cells. Endocr. J. 2019, 66, 515–522. [Google Scholar] [CrossRef]
- Coker, S.A.; Hurwitz, H.I.; Sharma, S.; Wang, D.; Jordaan, P.; Zarate, J.P.; Lewis, L.D. The effects of lapatinib on cardiac repolarization: Results from a placebo controlled, single sequence, crossover study in patients with advanced solid tumors. Cancer Chemother. Pharmacol. 2019, 84, 383–392. [Google Scholar] [CrossRef]
- Breccia, M.; Cannella, L.; Nanni, M.; Stefanizzi, C.; Alimena, G. Nilotinib Can Override Dasatinib Resistance in Chronic Myeloid Leukemia Patients with Secondary Resistance to Imatinib First-Line Therapy. Acta Haematol. 2007, 118, 162–164. [Google Scholar] [CrossRef]
- Kantarjian, H.; Giles, F.; Wunderle, L.; Bhalla, K.; O’Brien, S.; Wassmann, B.; Tanaka, C.; Manley, P.; Rae, P.; Mietlowski, W.; et al. Nilotinib in Imatinib-Resistant CML and Philadelphia Chromosome–Positive ALL. N. Engl. J. Med. 2006, 354, 2542–2551. [Google Scholar] [CrossRef]
- Kantarjian, H.M.; Giles, F.; Gattermann, N.; Bhalla, K.; Alimena, G.; Palandri, F.; Ossenkoppele, G.J.; Nicolini, F.-E.; O’Brien, S.G.; Litzow, M.; et al. Nilotinib (formerly AMN107), a highly selective BCR-ABL tyrosine kinase inhibitor, is effective in patients with Philadelphia chromosome–positive chronic myelogenous leukemia in chronic phase following imatinib resistance and intolerance. Blood 2007, 110, 3540–3546. [Google Scholar] [CrossRef]
- Maekawa, T.; Ashihara, E.; Kimura, S. The Bcr-Abl tyrosine kinase inhibitor imatinib and promising new agents against Philadelphia chromosome-positive leukemias. Int. J. Clin. Oncol. 2007, 12, 327–340. [Google Scholar] [CrossRef]
- Kim, S.-H.; Menon, H.; Jootar, S.; Saikia, T.; Kwak, J.-Y.; Sohn, S.-K.; Park, J.S.; Jeong, S.H.; Kim, H.J.; Kim, Y.-K.; et al. Efficacy and safety of radotinib in chronic phase chronic myeloid leukemia patients with resistance or intolerance to BCR-ABL1 tyrosine kinase inhibitors. Haematologica 2014, 99, 1191–1196. [Google Scholar] [CrossRef] [PubMed]
- O’Hare, T.; Zabriskie, M.S.; Eiring, A.M.; Deininger, M.W. Pushing the limits of targeted therapy in chronic myeloid leukaemia. Nat. Rev. Cancer 2012, 12, 513–526. [Google Scholar] [CrossRef] [PubMed]
- Zabriskie, M.S.; Vellore, N.A.; Gantz, K.C.; Deininger, M.W.; O’Hare, T. Radotinib is an effective inhibitor of native and kinase domain-mutant BCR-ABL1. Leukemia 2015, 29, 1939–1942. [Google Scholar] [CrossRef] [PubMed]
- Salomon-Ferrer, R.; Case, D.A.; Walker, R.C. An overview of the Amber biomolecular simulation package. Wiley Interdiscip. Rev. Comput. Mol. Sci. 2013, 3, 198–210. [Google Scholar] [CrossRef]
- Wang, J.; Wolf, R.M.; Caldwell, J.W.; Kollman, P.A.; Case, D.A. Development and testing of a general amber force field. J. Comput. Chem. 2004, 25, 1157–1174. [Google Scholar] [CrossRef]
- Jakalian, A.; Jack, D.B.; Bayly, C.I. Fast, efficient generation of high-quality atomic charges. AM1-BCC model: II. Parameterization and validation. J. Comput. Chem. 2002, 23, 1623–1641. [Google Scholar] [CrossRef]
- Kollman, P.A.; Massova, I.; Reyes, C.; Kuhn, B.; Huo, S.; Chong, L.; Lee, M.; Lee, T.; Duan, Y.; Wang, W.; et al. Calculating Structures and Free Energies of Complex Molecules: Combining Molecular Mechanics and Continuum Models. Accounts Chem. Res. 2000, 33, 889–897. [Google Scholar] [CrossRef]
- Hou, T.; Wang, J.; Li, Y.; Wang, W. Assessing the performance of the molecular mechanics/Poisson Boltzmann surface area and molecular mechanics/generalized Born surface area methods. II. The accuracy of ranking poses generated from docking. J. Comput. Chem. 2010, 32, 866–877. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar] [CrossRef]
- Felsenstein, J. Confidence Limits on Phylogenies: An Approach Using the Bootstrap. Evolution 1985, 39, 783. [Google Scholar] [CrossRef]
- Jones, D.T.; Taylor, W.R.; Thornton, J.M. The rapid generation of mutation data matrices from protein sequences. Bioinformatics 1992, 8, 275–282. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree Of Life (iTOL): An online tool for phylogenetic tree display and annotation. Bioinformatics 2006, 23, 127–128. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Franceschini, A.; Wyder, S.; Forslund, K.; Heller, D.; Huerta-Cepas, J.; Simonovic, M.; Roth, A.; Santos, A.; Tsafou, K.P.; et al. STRING v10: Protein–protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015, 43, D447–D452. [Google Scholar] [CrossRef]
- Le Guilloux, V.; Schmidtke, P.; Tuffery, P. Fpocket: An open source platform for ligand pocket detection. BMC Bioinform. 2009, 10, 168. [Google Scholar] [CrossRef]
- Morris, G.M.; Huey, R.; Lindstrom, W.; Sanner, M.F.; Belew, R.K.; Goodsell, D.S.; Olson, A.J. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J. Comput. Chem. 2009, 30, 2785–2791. [Google Scholar] [CrossRef]
- Friesner, R.A.; Banks, J.L.; Murphy, R.B.; Halgren, T.A.; Klicic, J.J.; Mainz, D.T.; Repasky, M.P.; Knoll, E.H.; Shelley, M.; Perry, J.K.; et al. Glide: A New Approach for Rapid, Accurate Docking and Scoring. 1. Method and Assessment of Docking Accuracy. J. Med. Chem. 2004, 47, 1739–1749. [Google Scholar] [CrossRef]
- Woods, C.J.; Malaisree, M.; Michel, J.; Long, B.; McIntosh-Smith, S.; Mulholland, A.J. Rapid decomposition and visualisation of protein–ligand binding free energies by residue and by water. Faraday Discuss. 2014, 169, 477–499. [Google Scholar] [CrossRef]
- Abel, R.; Young, T.; Farid, R.; Berne, B.J.; Friesner, R.A. Role of the Active-Site Solvent in the Thermodynamics of Factor Xa Ligand Binding. J. Am. Chem. Soc. 2008, 130, 2817–2831. [Google Scholar] [CrossRef]
- Lazaridis, T. Inhomogeneous Fluid Approach to Solvation Thermodynamics. 1. Theory. J. Phys. Chem. B 1998, 102, 3531–3541. [Google Scholar] [CrossRef]
- Young, T.; Abel, R.; Kim, B.; Berne, B.J.; Friesner, R.A. Motifs for molecular recognition exploiting hydrophobic enclosure in protein–ligand binding. Proc. Natl. Acad. Sci. USA 2007, 104, 808–813. [Google Scholar] [CrossRef] [PubMed]
- Kaminski, G.A.; Friesner, R.A.; Tirado-Rives, J.; Jorgensen, W.L. Evaluation and Reparametrization of the OPLS-AA Force Field for Proteins via Comparison with Accurate Quantum Chemical Calculations on Peptides. J. Phys. Chem. B 2001, 105, 6474–6487. [Google Scholar] [CrossRef]
- Jorgensen, W.L.; Maxwell, D.S.; Tirado-Rives, J. Development and Testing of the OPLS All-Atom Force Field on Conformational Energetics and Properties of Organic Liquids. J. Am. Chem. Soc. 1996, 118, 11225–11236. [Google Scholar] [CrossRef]
- Tran, Q.-T.; Williams, S.; Farid, R.; Erdemli, G.; Pearlstein, R. The translocation kinetics of antibiotics through porin OmpC: Insights from structure-based solvation mapping using WaterMap. Proteins Struct. Funct. Bioinform. 2013, 81, 291–299. [Google Scholar] [CrossRef]
- Bauer, M.R.; Mackey, M.D. Electrostatic Complementarity as a Fast and Effective Tool to Optimize Binding and Selectivity of Protein–Ligand Complexes. J. Med. Chem. 2019, 62, 3036–3050. [Google Scholar] [CrossRef]
- Dick, A.; Cocklin, S. Bioisosteric Replacement as a Tool in Anti-HIV Drug Design. Pharmaceuticals 2020, 13, 36. [Google Scholar] [CrossRef]
- Huang, Y.; Harris, R.C.; Shen, J. Generalized Born Based Continuous Constant pH Molecular Dynamics in Amber: Implementation, Benchmarking and Analysis. J. Chem. Inf. Model. 2018, 58, 1372–1383. [Google Scholar] [CrossRef]
- Salomon-Ferrer, R.; Götz, A.W.; Poole, D.; Le Grand, S.; Walker, R.C. Routine Microsecond Molecular Dynamics Simulations with AMBER on GPUs. 2. Explicit Solvent Particle Mesh Ewald. J. Chem. Theory Comput. 2013, 9, 3878–3888. [Google Scholar] [CrossRef]
- Case, D.A.; Cheatham, T.E., III; Darden, T.; Gohlke, H.; Luo, R.; Merz, K.M., Jr.; Onufriev, A.; Simmerling, C.; Wang, B.; Woods, R.J. The Amber biomolecular simulation programs. J. Comput. Chem. 2005, 26, 1668–1688. [Google Scholar] [CrossRef]
| Structure | 2D Structure | Title | LF Rank Score 1 | LF dG 2 | LF vs. Score 3 | MW 4 | Atoms | SlogP 5 | TPSA 6 | RB 7 | Rof5 8 |
|---|---|---|---|---|---|---|---|---|---|---|---|
| [O-][N+](=O)c1cc([N+]([O-])=O)ccc1NCCCC([O-])=O | 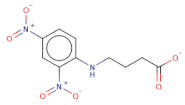 | RJC02337 | −16.742 | −8.149 | −9.451 | 268.2 | 19 | 1.6 | 143.8 | 7 | 0 |
| O=C(N)c1cc([N+]([O-])=O)cc([N+]([O-])=O)c1C | 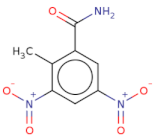 | T0873 | −15.539 | −7.431 | −9.133 | 225.2 | 16 | 0.7 | 134.7 | 3 | 0 |
| O[C@@H]1[C@H](O[C@@H]([C@H](O)[C@@H]1O)CO)OC[C@@H]2[C@H](O)[C@H](O)[C@H](O)[C@H](O2)OC(=O)/C(=C/C=C/C(C)=C/C=C/C=C(\C=C\C=C(\C(O[C@@H]3[C@@H](O)[C@@H](O)[C@@H](O)[C@H](O3)CO[C@@H]4[C@@H](O)[C@@H](O)[C@@H](O)[C@H](O4)CO)=O)C)C)C | 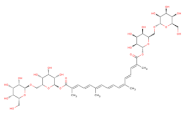 | T2823 | −15.515 | −13.277 | −19.591 | 977 | 68 | −0.6 | 391.2 | 32 | 3 |
| [O-][N+](=O)c1cc2c(OC)cccc2c3c4c(OCO4)cc(c13)C([O-])=O | 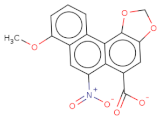 | T2801 | −15.35 | −8.138 | −10.132 | 340.3 | 25 | 3.2 | 113.6 | 3 | 0 |
| [O-][N+](=O)c1c(cc(N/N=C/c2ccco2)c([N+]([O-])=O)c1)C | 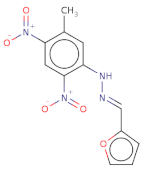 | CD00811 | −15.321 | −7.022 | −9.005 | 290.2 | 21 | 2.1 | 129.2 | 5 | 0 |
| OP(OC(C)C)OC(C)C | 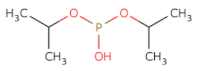 | DFP | −9.916 | −5.191 | −5.268 | 165.1 | 10 | 2.5 | 35.5 | 4 | 0 |
| Structure | 2D Structure | Title | LF Rank Score | LF dG | LF vs. Score | MW | Atoms | SlogP | TPSA | RB | Rof5 |
|---|---|---|---|---|---|---|---|---|---|---|---|
| O[C@H]1[C@H]2C=CC=C[C@H]2[S@]3(N1C)[C@H]4CCC[C@H]5CCC[C@@H]([C@H]54)[C@@H](O3)O |  | BTB11534 | −21.512 | −9.444 | −10.711 | 351.5 | 24 | 3.4 | 52.9 | 2 | 0 |
| Clc1ccc(O)c([C@H]2C[C@H](NN2)[C@@H](O)Nc3ccc(NC(=O)C)cc3)c1 | 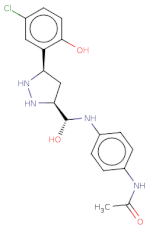 | MB_552691 | −19.348 | −12.373 | −13.444 | 376.8 | 26 | 2.7 | 105.6 | 7 | 1 |
| Clc1cc(Nc2c3c(ncn2)ccc(c4ccc(o4)C[NH2+]CCS(=O)(=O)C)c3)ccc1OCc5cc(F)ccc5 | 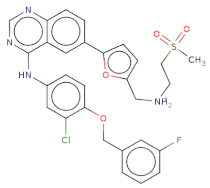 | T0078L | −18.51 | −11.313 | −14.248 | 582.1 | 40 | 5.9 | 110.9 | 8 | 1 |
| FC(F)(F)c1cc(NC(=O)c2ccc(c(Nc3nccc(n3)-c4cnccc4)c2)C)cc(-n5cnc(c5)C)c1 | 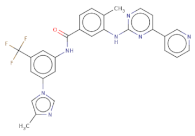 | T1524 | −17.871 | −10984 | −12.821 | 529.5 | 39 | 6.4 | 97.6 | 7 | 2 |
| FC(F)(F)c1cc(NC(=O)c2ccc(c(Nc3nccc(n3)-c4cnccn4)c2)C)cc(-n5cnc(c5)C)c1 | 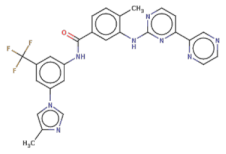 | T2328 | −17.553 | −10.541 | −12.76 | 530.5 | 39 | 5.8 | 110.5 | 7 | 2 |
| Structure | 2D Structure | MW | Atoms | Rof5 | Attachment Point 1 Type | Attachment Point 2 Type | LF dG | LF vs. Score | LF Rank Score |
|---|---|---|---|---|---|---|---|---|---|
| SC[COOH](CCc1ccc([N+]([O-])=O)cc1[N+]([O-])=O)CC([O-])=O | 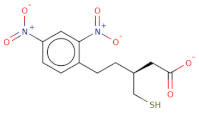 | 313.3 | 21 | 0 | Csp3 | Csp3 | −8.733 | −8.868 | −9.026 |
| [O-][N+](=O)c1cc([N+]([O-])=O)ccc1SCSCC([O-])=O | 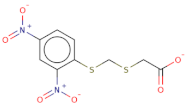 | 303.3 | 19 | 0 | S | Csp3 | −8.57 | −8.757 | −9.33 |
| O[COOH](SCC([O-])=O)c1ccc([N+]([O-])=O)cc1[N+]([O-])=O | 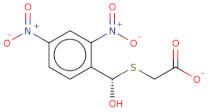 | 287.2 | 19 | 0 | Csp3 | Csp3 | −8.314 | −9.108 | −9.2 |
| [O-][N+](=O)c1cc([N+]([O-])=O)ccc1SCSSC([O-])=O | 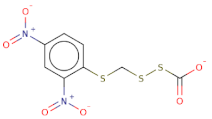 | 321.3 | 19 | 0 | S | S | −8.11 | −8.956 | −8.967 |
| [O-][N+](=O)c1cc([N+]([O-])=O)ccc1/C(=[NH+]/CCSC([O-])=O)N | 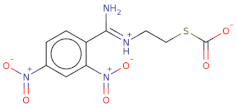 | 314.3 | 21 | 0 | Csp2 | S | −8.006 | −7.295 | −7.551 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shehzad, S.; Pandey, R.; Malhotra, P.; Gupta, D. Computational Design of Novel Allosteric Inhibitors for Plasmodium falciparum DegP. Molecules 2021, 26, 2742. https://doi.org/10.3390/molecules26092742
Shehzad S, Pandey R, Malhotra P, Gupta D. Computational Design of Novel Allosteric Inhibitors for Plasmodium falciparum DegP. Molecules. 2021; 26(9):2742. https://doi.org/10.3390/molecules26092742
Chicago/Turabian StyleShehzad, Sadaf, Rajan Pandey, Pawan Malhotra, and Dinesh Gupta. 2021. "Computational Design of Novel Allosteric Inhibitors for Plasmodium falciparum DegP" Molecules 26, no. 9: 2742. https://doi.org/10.3390/molecules26092742
APA StyleShehzad, S., Pandey, R., Malhotra, P., & Gupta, D. (2021). Computational Design of Novel Allosteric Inhibitors for Plasmodium falciparum DegP. Molecules, 26(9), 2742. https://doi.org/10.3390/molecules26092742







