Multiscale Virtual Screening Optimization for Shotgun Drug Repurposing Using the CANDO Platform
Abstract
1. Introduction
1.1. Drug Repurposing
1.2. Computational Drug Repurposing Using Molecular Docking
1.3. Shotgun Multitarget Multi-Disease Drug Repurposing Using the CANDO Platform
2. Results
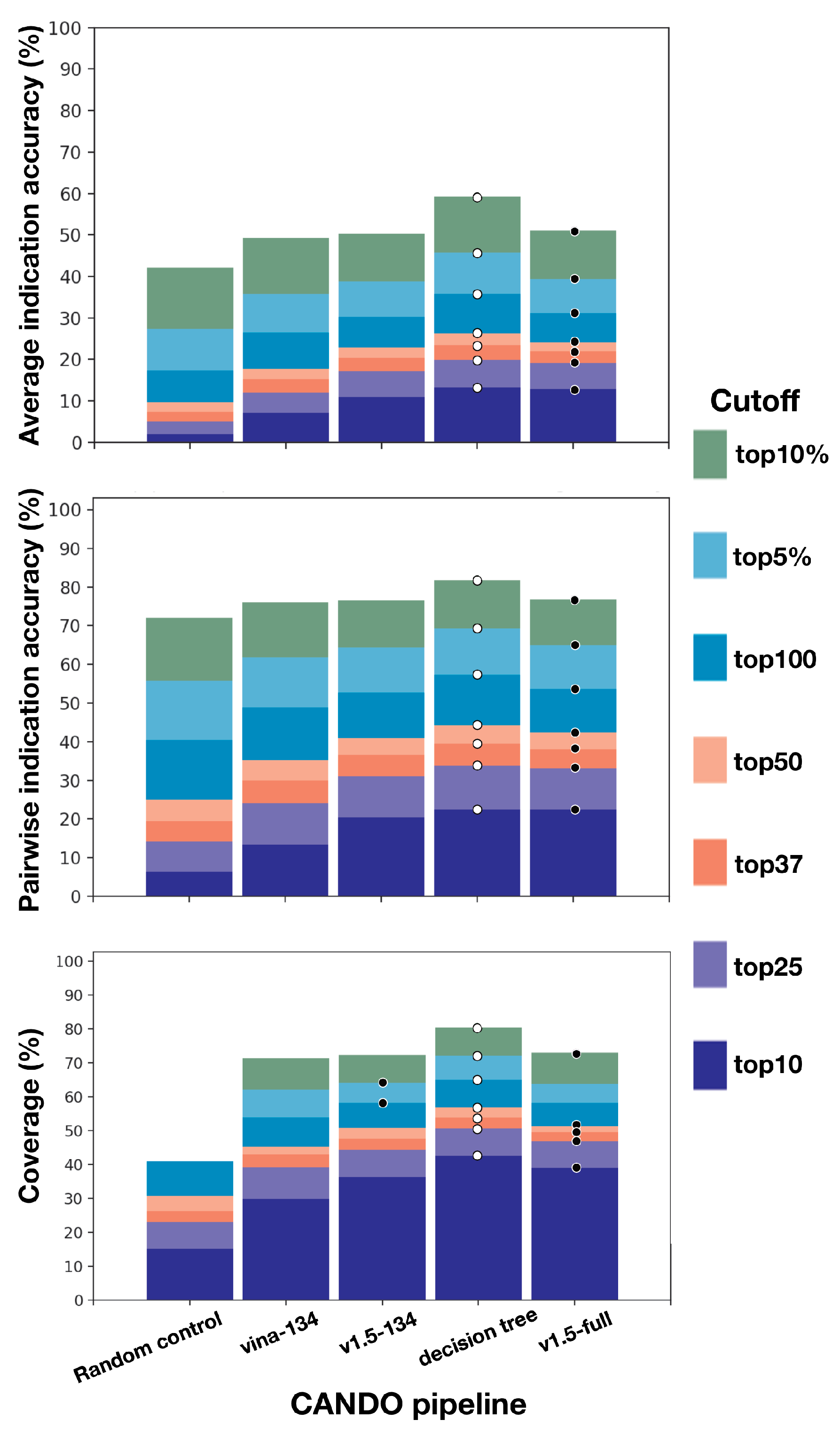
2.1. Benchmarking Performance of the Different Pipelines
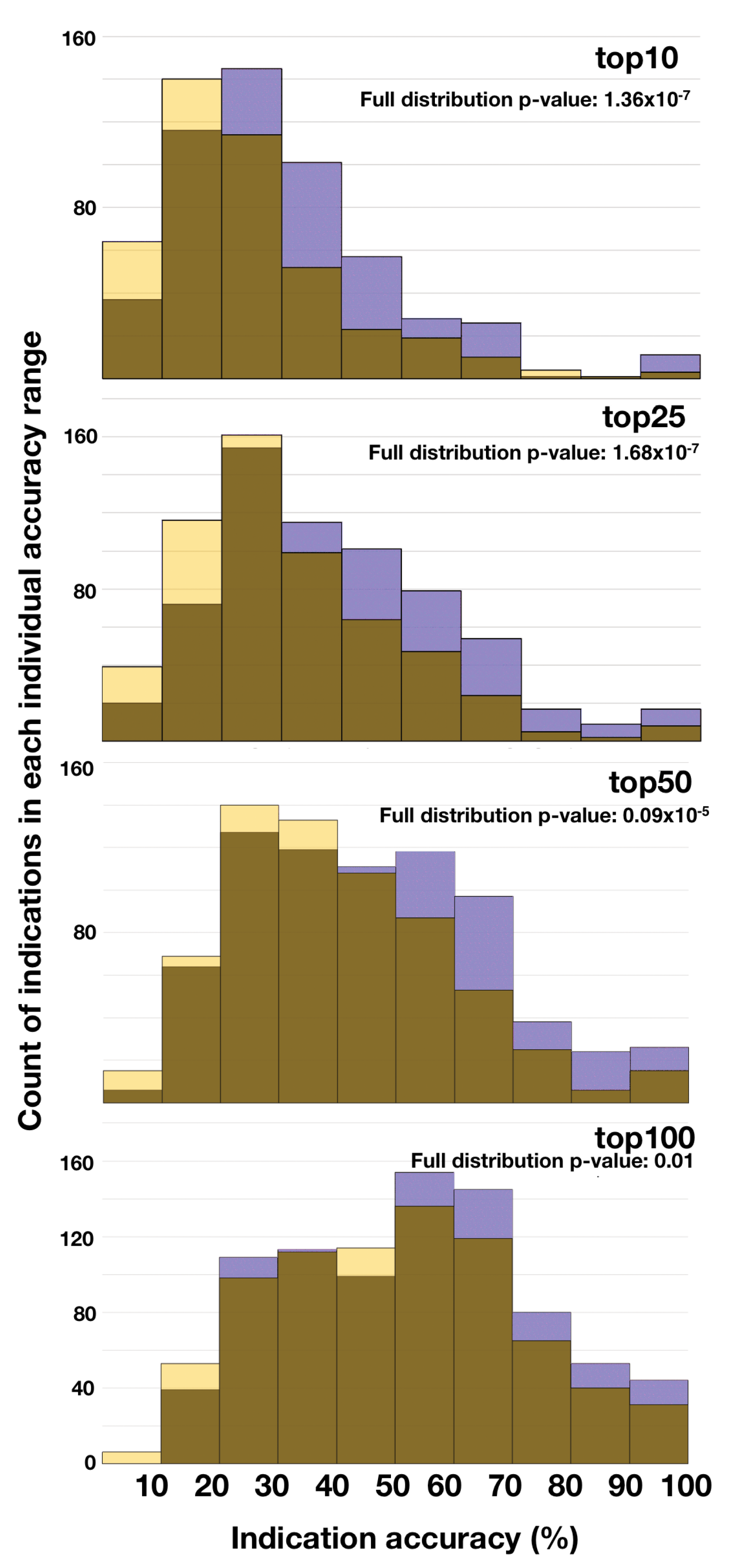
2.2. Divergence in Indication Accuracy at Various Thresholds
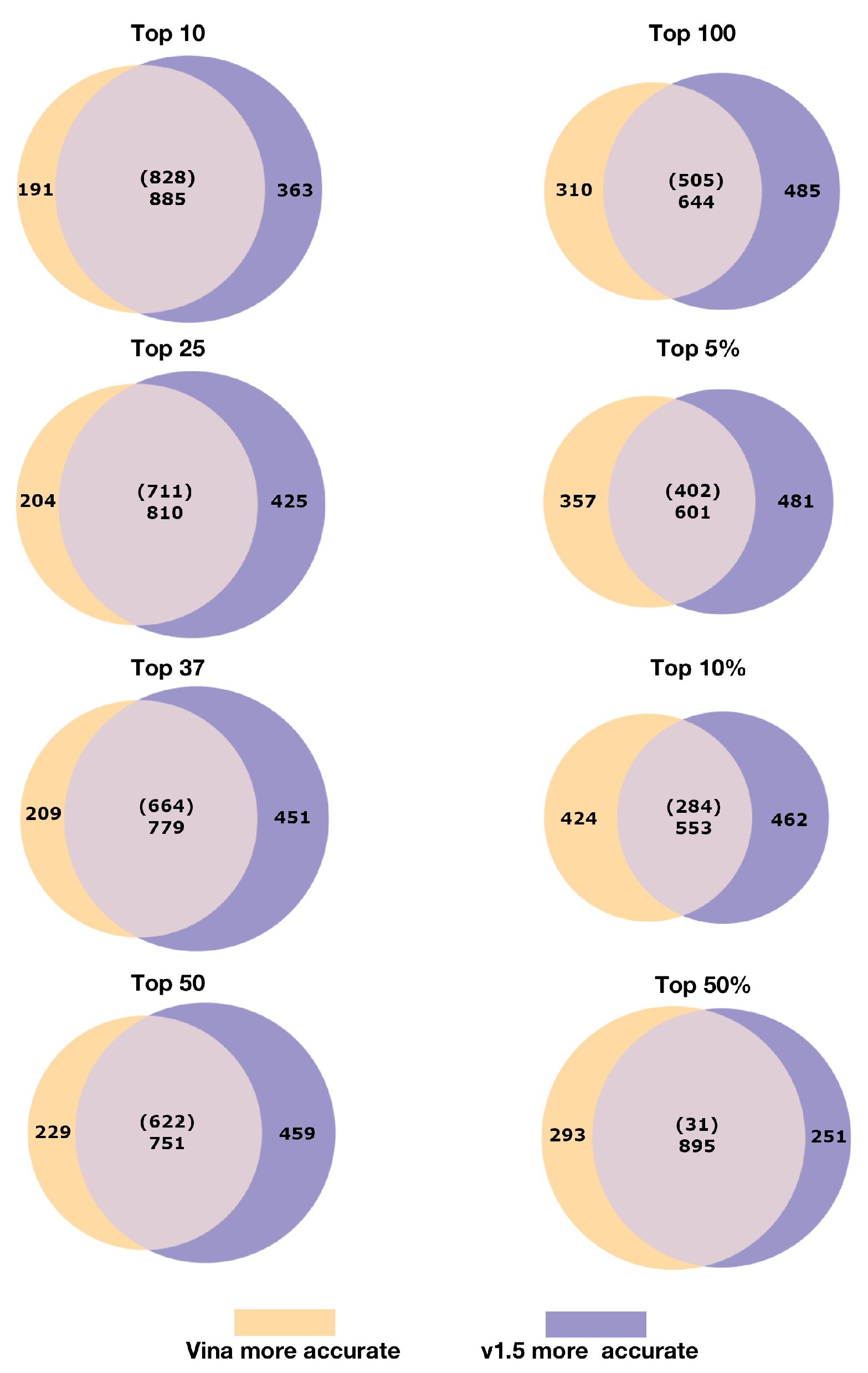
2.3. Net Differences in Indication Accuracy
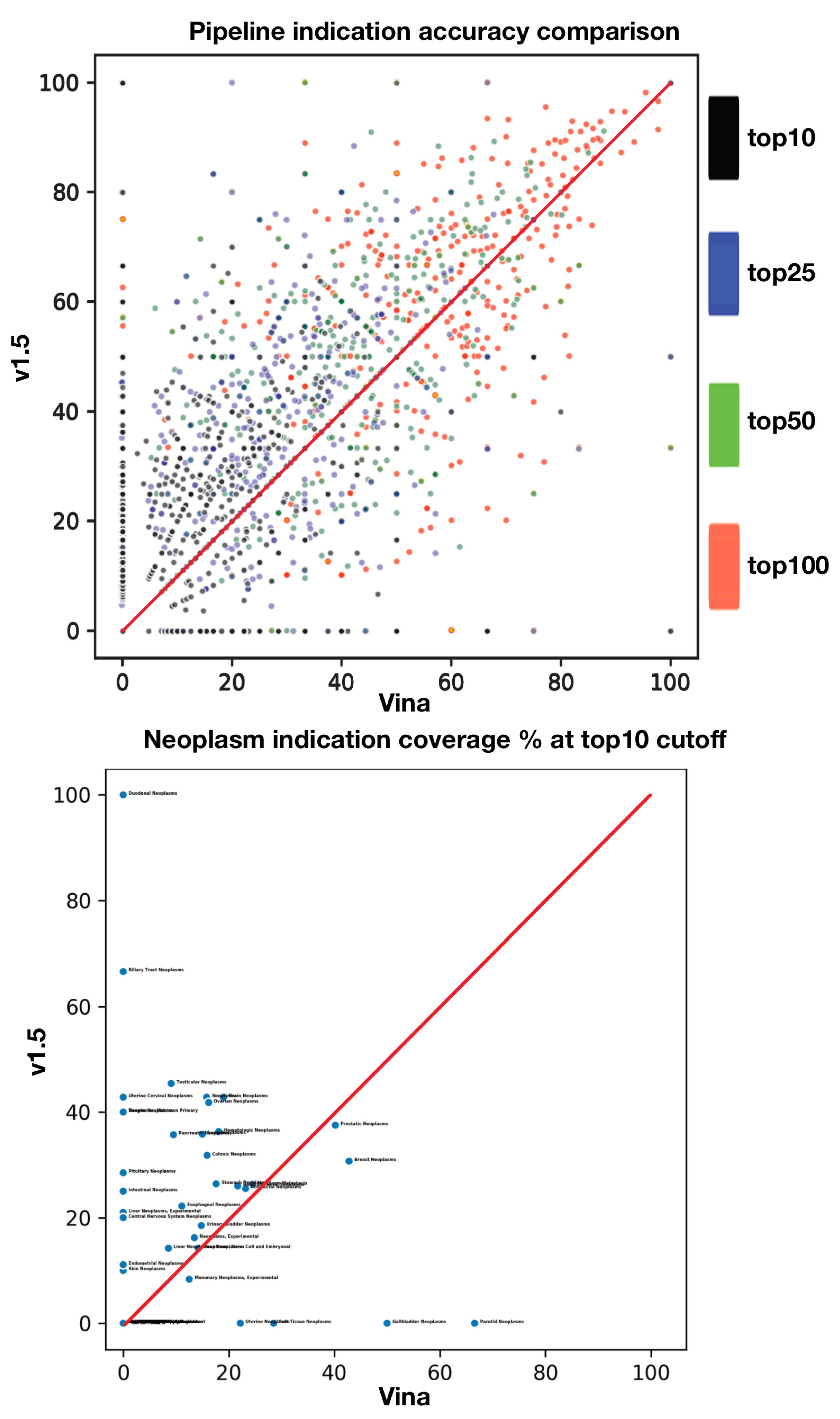
2.4. Relative Pipeline Indication Accuracy
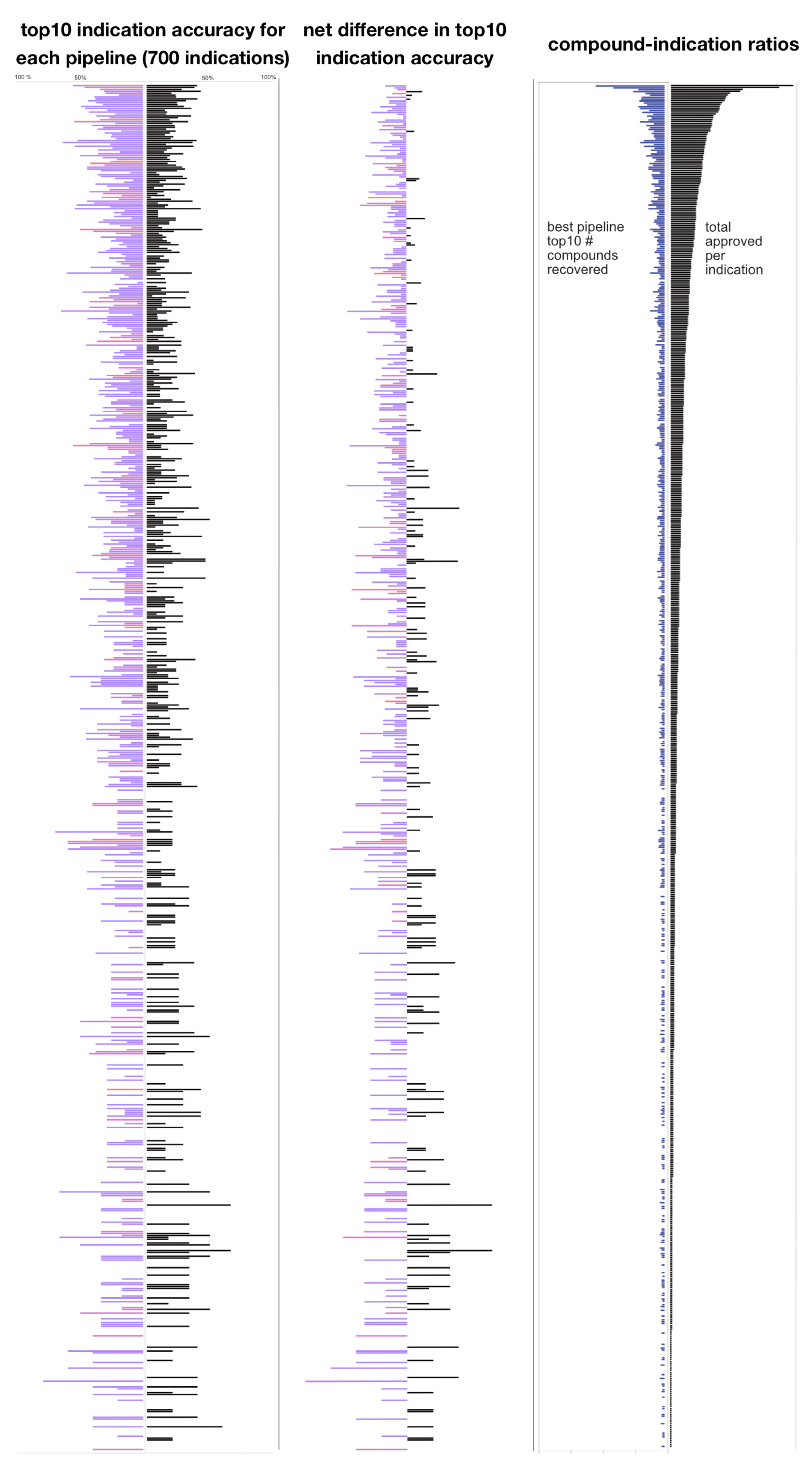
2.5. Comparison of the Pipeline Distribution of per Indication Accuracies
2.6. Indication Accuracy Distribution
2.7. Distribution of Individual Drug-Indication Pair Rankings
3. Discussion
3.1. Multiple Large-Scale Virtual Screening Pipelines
3.2. Limitations and Future Work
4. Materials and Methods
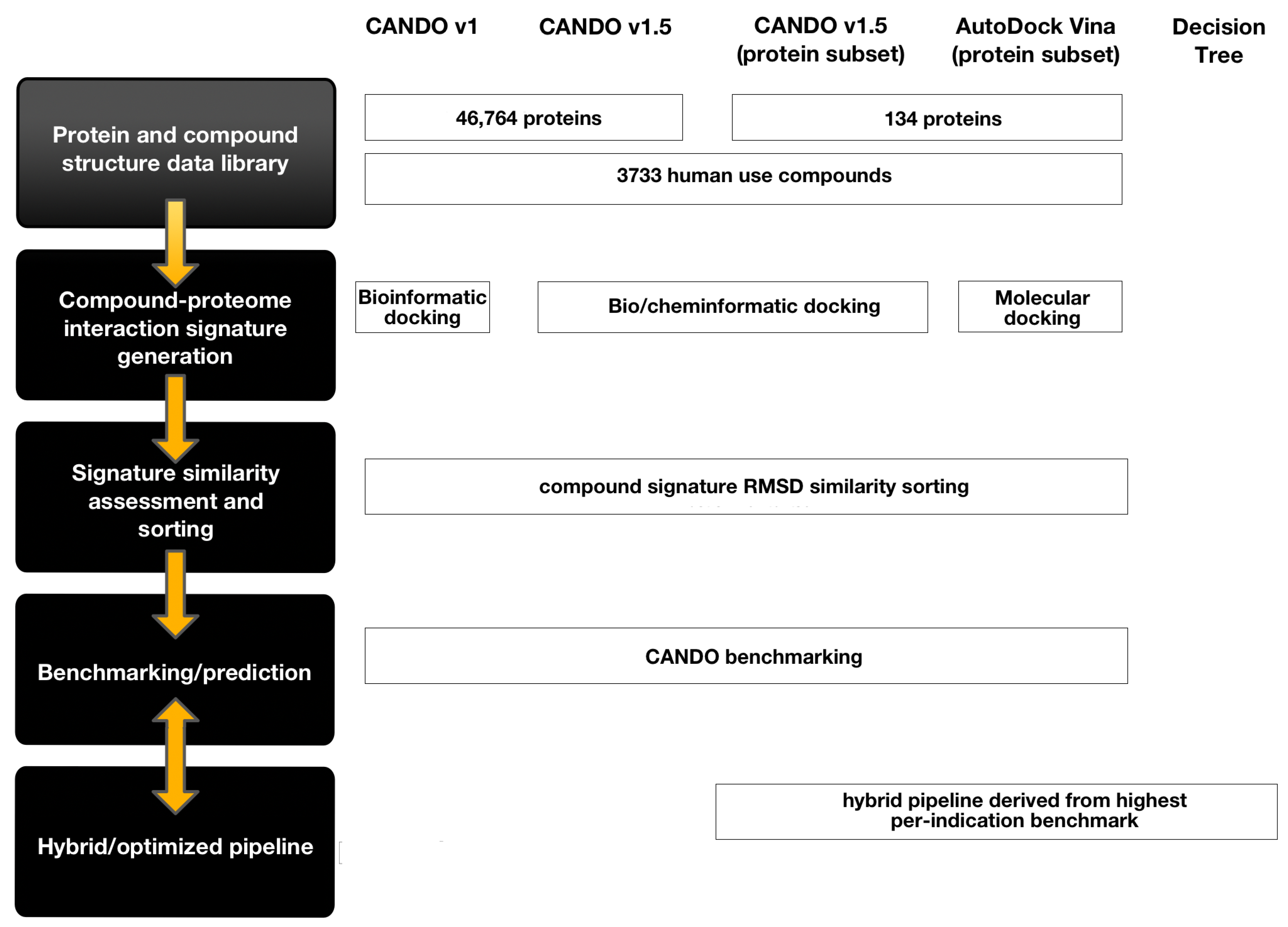
4.1. CANDO Platform and Pipeline Implementation
4.1.1. Drug/Compound, Protein Structure, and Indication Library Curation
4.1.2. Drug- and Compound-Proteome Interaction Signature Generation
4.1.3. Drug- and Compound-Proteome Signature Similarity Calculation and Sorting
4.2. Benchmarking CANDO Platform Pipelines
4.3. New and Hybrid Pipelines
4.3.1. Virtual Screening Pipeline Using Autodock Vina
4.3.2. Decision Tree Pipeline
4.4. Controls
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| CANDO | Computational Analysis of Novel Drug Opportunities |
| FDA | Food and Drug Administration |
| NMR | Nuclear Magnetic Resonance |
References
- Lichtenberg, F.R. Pharmaceutical Innovation, Mortality Reduction, and Economic Growth; Technical Report; National Bureau of Economic Research: Cambridge, MA, USA, 1998. [Google Scholar]
- FDA. U.S. New Drug Development & Approval Process. Available online: fda.gov/drugs (accessed on 24 April 2021).
- DiMasi, J.A. New drug development in the United States from 1963 to 1999. Clin. Pharmacol. Ther. 2001, 69, 286–296. [Google Scholar] [CrossRef] [PubMed]
- DiMasi, J.A.; Hansen, R.W.; Grabowski, H.G. The price of innovation: New estimates of drug development costs. J. Health Econ. 2003, 22, 151–185. [Google Scholar] [CrossRef]
- DiMasi, J.A.; Grabowski, H.G.; Hansen, R.W. Innovation in the pharmaceutical industry: New estimates of R&D costs. J. Health Econ. 2016, 47, 20–33. [Google Scholar]
- Scannell, J.W.; Blanckley, A.; Boldon, H.; Warrington, B. Diagnosing the decline in pharmaceutical R&D efficiency. Nat. Rev. Drug Discov. 2012, 11, 191. [Google Scholar]
- Broach, J.R.; Thorner, J. High-throughput screening for drug discovery. Nature 1996, 384, 14–16. [Google Scholar]
- Michelini, E.; Cevenini, L.; Mezzanotte, L.; Coppa, A.; Roda, A. Cell-based assays: Fuelling drug discovery. Anal. Bioanal. Chem. 2010, 398, 227–238. [Google Scholar] [CrossRef]
- Macalino, S.J.Y.; Gosu, V.; Hong, S.; Choi, S. Role of computer-aided drug design in modern drug discovery. Arch. Pharm. Res. 2015, 38, 1686–1701. [Google Scholar] [CrossRef]
- Lionta, E.; Spyrou, G.; Vassilatis, D.K.; Cournia, Z. Structure-based virtual screening for drug discovery: Principles, applications and recent advances. Curr. Top. Med. Chem. 2014, 14, 1923–1938. [Google Scholar] [CrossRef] [PubMed]
- Patel, C.N.; Georrge, J.J.; Modi, K.M.; Narechania, M.B.; Patel, D.P.; Gonzalez, F.J.; Pandya, H.A. Pharmacophore-based virtual screening of catechol-o-methyltransferase (COMT) inhibitors to combat Alzheimer’s disease. J. Biomol. Struct. Dyn. 2018, 36, 3938–3957. [Google Scholar] [CrossRef] [PubMed]
- Medina-Franco, J.L.; Giulianotti, M.A.; Welmaker, G.S.; Houghten, R.A. Shifting from the single to the multitarget paradigm in drug discovery. Drug Discov. Today 2013, 18, 495–501. [Google Scholar] [CrossRef] [PubMed]
- Bolognesi, M.L. Polypharmacology in a single drug: Multitarget drugs. Curr. Med. Chem. 2013, 20, 1639–1645. [Google Scholar] [CrossRef]
- Hu, Y.; Bajorath, J. Monitoring drug promiscuity over time. F1000Research 2014, 3, 218. [Google Scholar] [CrossRef]
- Ashburn, T.T.; Thor, K.B. Drug repositioning: Identifying and developing new uses for existing drugs. Nat. Rev. Drug Discov. 2004, 3, 673–683. [Google Scholar] [CrossRef] [PubMed]
- Langedijk, J.; Mantel-Teeuwisse, A.K.; Slijkerman, D.S.; Schutjens, M.H.D. Drug repositioning and repurposing: Terminology and definitions in literature. Drug Discov. Today 2015, 20, 1027–1034. [Google Scholar] [CrossRef] [PubMed]
- Palumbo, A.; Facon, T.; Sonneveld, P.; Blade, J.; Offidani, M.; Gay, F.; Moreau, P.; Waage, A.; Spencer, A.; Ludwig, H.; et al. Thalidomide for treatment of multiple myeloma: 10 years later. Blood J. Am. Soc. Hematol. 2008, 111, 3968–3977. [Google Scholar] [CrossRef] [PubMed]
- Roth, B.L.; Sheffler, D.J.; Kroeze, W.K. Magic shotguns versus magic bullets: Selectively non-selective drugs for mood disorders and schizophrenia. Nat. Rev. Drug Discov. 2004, 3, 353–359. [Google Scholar] [CrossRef] [PubMed]
- de Lera, A.R.; Ganesan, A. Epigenetic polypharmacology: From combination therapy to multitargeted drugs. Clin. Epigenet. 2016, 8, 105. [Google Scholar] [CrossRef]
- Arts, E.J.; Hazuda, D.J. HIV-1 antiretroviral drug therapy. Cold Spring Harb. Perspect. Med. 2012, 2, a007161. [Google Scholar] [CrossRef]
- Sardana, D.; Zhu, C.; Zhang, M.; Gudivada, R.C.; Yang, L.; Jegga, A.G. Drug repositioning for orphan diseases. Brief. Bioinform. 2011, 12, 346–356. [Google Scholar] [CrossRef]
- Minie, M.; Chopra, G.; Sethi, G.; Horst, J.; White, G.; Roy, A.; Hatti, K.; Samudrala, R. CANDO and the infinite drug discovery frontier. Drug Discov. Today 2014, 19, 1353–1363. [Google Scholar] [CrossRef]
- Mangione, W.; Falls, Z.; Chopra, G.; Samudrala, R. Cando. py: Open source software for analyzing large scale drug-protein-disease data. J. Chem. Inf. Model. 2020, 60, 4131–4136. [Google Scholar] [CrossRef]
- Xu, M.; Lee, E.M.; Wen, Z.; Cheng, Y.; Huang, W.K.; Qian, X.; Julia, T.; Kouznetsova, J.; Ogden, S.C.; Hammack, C.; et al. Identification of small-molecule inhibitors of Zika virus infection and induced neural cell death via a drug repurposing screen. Nat. Med. 2016, 22, 1101. [Google Scholar] [CrossRef] [PubMed]
- Schuler, J.; Hudson, M.L.; Schwartz, D.; Samudrala, R. A systematic review of computational drug discovery, development, and repurposing for Ebola virus disease treatment. Molecules 2017, 22, 1777. [Google Scholar] [CrossRef] [PubMed]
- Roder, C.; Thomson, M.J. Auranofin: Repurposing an old drug for a golden new age. Drugs R D 2015, 15, 13–20. [Google Scholar] [CrossRef] [PubMed]
- Ou-Yang, S.S.; Lu, J.Y.; Kong, X.Q.; Liang, Z.J.; Luo, C.; Jiang, H. Computational drug discovery. Acta Pharmacol. Sin. 2012, 33, 1131–1140. [Google Scholar] [CrossRef] [PubMed]
- Taylor, R.D.; Jewsbury, P.J.; Essex, J.W. A review of protein-small molecule docking methods. J. Comput. Aided Mol. Des. 2002, 16, 151–166. [Google Scholar] [CrossRef]
- Pagadala, N.S.; Syed, K.; Tuszynski, J. Software for molecular docking: A review. Biophys. Rev. 2017, 9, 91–102. [Google Scholar] [CrossRef]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef] [PubMed]
- Fine, J.; Konc, J.; Samudrala, R.; Chopra, G. CANDOCK: Chemical atomic network based hierarchical flexible docking algorithm using generalized statistical potentials. J. Chem. Inf. Model. 2020, 60, 1509–1527. [Google Scholar] [CrossRef]
- Yuriev, E.; Ramsland, P.A. Latest developments in molecular docking: 2010–2011 in review. J. Mol. Recognit. 2013, 26, 215–239. [Google Scholar] [CrossRef]
- Huang, N.; Jacobson, M.P. Physics-based methods for studying protein-ligand interactions. Curr. Opin. Drug Discov. Dev. 2007, 10, 325. [Google Scholar]
- Gohlke, H.; Klebe, G. Statistical potentials and scoring functions applied to protein–ligand binding. Curr. Opin. Struct. Biol. 2001, 11, 231–235. [Google Scholar] [CrossRef]
- Muegge, I. A knowledge-based scoring function for protein-ligand interactions: Probing the reference state. Perspect. Drug Discov. Des. 2000, 20, 99–114. [Google Scholar] [CrossRef]
- Lokhande, K.B.; Nagar, S.; Swamy, K.V. Molecular interaction studies of Deguelin and its derivatives with Cyclin D1 and Cyclin E in cancer cell signaling pathway: The computational approach. Sci. Rep. 2019, 9, 1–13. [Google Scholar] [CrossRef]
- Ma, D.L.; Chan, D.S.H.; Leung, C.H. Molecular docking for virtual screening of natural product databases. Chem. Sci. 2011, 2, 1656–1665. [Google Scholar] [CrossRef]
- Sethi, G.; Chopra, G.; Samudrala, R. Multiscale modeling of relationships between protein classes and drug behavior across all diseases using the CANDO platform. Mini Rev. Med. Chem. 2015, 15, 705–717. [Google Scholar] [CrossRef]
- Rodrigues, J.; Melquiond, A.; Karaca, E.; Trellet, M.; Van Dijk, M.; Van Zundert, G.; Schmitz, C.; De Vries, S.; Bordogna, A.; Bonati, L.; et al. Defining the limits of homology modeling in information-driven protein docking. Proteins Struct. Funct. Bioinform. 2013, 81, 2119–2128. [Google Scholar] [CrossRef]
- Yuriev, E.; Agostino, M.; Ramsland, P.A. Challenges and advances in computational docking: 2009 in review. J. Mol. Recognit. 2011, 24, 149–164. [Google Scholar] [CrossRef] [PubMed]
- Chopra, G.; Samudrala, R. Exploring polypharmacology in drug discovery and repurposing using the CANDO platform. Curr. Pharm. Des. 2016, 22, 3109–3123. [Google Scholar] [CrossRef]
- Chopra, G.; Kaushik, S.; Elkin, P.L.; Samudrala, R. Combating ebola with repurposed therapeutics using the CANDO platform. Molecules 2016, 21, 1537. [Google Scholar] [CrossRef] [PubMed]
- Schuler, J.; Samudrala, R. Fingerprinting CANDO: Increased Accuracy with Structure-and Ligand-Based Shotgun Drug Repurposing. ACS Omega 2019, 4, 17393–17403. [Google Scholar] [CrossRef] [PubMed]
- Mangione, W.; Samudrala, R. Identifying protein features responsible for improved drug repurposing accuracies using the CANDO platform: Implications for drug design. Molecules 2019, 24, 167. [Google Scholar] [CrossRef] [PubMed]
- Falls, Z.; Mangione, W.; Schuler, J.; Samudrala, R. Exploration of interaction scoring criteria in the CANDO platform. BMC Res. Notes 2019, 12, 318. [Google Scholar] [CrossRef]
- Cavasotto, C.N.; Phatak, S.S. Homology modeling in drug discovery: Current trends and applications. Drug Discov. Today 2009, 14, 676–683. [Google Scholar] [CrossRef]
- Lipscomb, C.E. Medical subject headings (MeSH). Bull. Med. Libr. Assoc. 2000, 88, 265. [Google Scholar]
- Arp, R.; Smith, B.; Spear, A.D. Building Ontologies with Basic Formal Ontology; Mit Press: Cambridge, MA, USA, 2015. [Google Scholar]
- Schuler, J.; Mangione, W.; Samudrala, R.; Ceusters, W. Foundations for a Realism-based Drug Repurposing Ontology. In Proceedings of the 10th Annual International Conference on Biomedical Ontology, Buffalo, NY, USA, 30 July–2 August 2019. [Google Scholar]
- Schuler, J.; Falls, Z.; Mangione, W.; Hudson, M.; Bruggemann, L.; Samdurala, R. Evaluating performance of drug repurposing technologies. Drug Discov. Today 2020. [Google Scholar] [CrossRef]
- Mangione, W.; Falls, Z.; Melendy, T.; Chopra, G.; Samudrala, R. Shotgun drug repurposing biotechnology to tackle epidemics and pandemics. Drug Discov. Today 2020, 25, 1126–1128. [Google Scholar] [CrossRef] [PubMed]
- Berman, H.M.; Westbrook, J.; Feng, Z.; Gilliland, G.; Bhat, T.N.; Weissig, H.; Shindyalov, I.N.; Bourne, P.E. The protein data bank. Nucleic Acids Res. 2000, 28, 235–242. [Google Scholar] [CrossRef]
- Kim, S.; Thiessen, P.A.; Bolton, E.E.; Chen, J.; Fu, G.; Gindulyte, A.; Han, L.; He, J.; He, S.; Shoemaker, B.A.; et al. PubChem substance and compound databases. Nucleic Acids Res. 2016, 44, D1202–D1213. [Google Scholar] [CrossRef] [PubMed]
- Davis, A.P.; Murphy, C.G.; Johnson, R.; Lay, J.M.; Lennon-Hopkins, K.; Saraceni-Richards, C.; Sciaky, D.; King, B.L.; Rosenstein, M.C.; Wiegers, T.C.; et al. The comparative toxicogenomics database: Update 2013. Nucleic Acids Res. 2012, 41, D1104–D1114. [Google Scholar] [CrossRef]
- Knox, C.; Law, V.; Jewison, T.; Liu, P.; Ly, S.; Frolkis, A.; Pon, A.; Banco, K.; Mak, C.; Neveu, V.; et al. DrugBank 3.0: A comprehensive resource for ‘omics’ research on drugs. Nucleic Acids Res. 2010, 39, D1035–D1041. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y. I-TASSER server for protein 3D structure prediction. BMC Bioinform. 2008, 9, 40. [Google Scholar] [CrossRef] [PubMed]
- Roy, A.; Yang, J.; Zhang, Y. COFACTOR: An accurate comparative algorithm for structure-based protein function annotation. Nucleic Acids Res. 2012, 40, W471–W477. [Google Scholar] [CrossRef] [PubMed]
- Feinstein, W.P.; Brylinski, M. Calculating an optimal box size for ligand docking and virtual screening against experimental and predicted binding pockets. J. Cheminform. 2015, 7, 1–10. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hudson, M.L.; Samudrala, R. Multiscale Virtual Screening Optimization for Shotgun Drug Repurposing Using the CANDO Platform. Molecules 2021, 26, 2581. https://doi.org/10.3390/molecules26092581
Hudson ML, Samudrala R. Multiscale Virtual Screening Optimization for Shotgun Drug Repurposing Using the CANDO Platform. Molecules. 2021; 26(9):2581. https://doi.org/10.3390/molecules26092581
Chicago/Turabian StyleHudson, Matthew L., and Ram Samudrala. 2021. "Multiscale Virtual Screening Optimization for Shotgun Drug Repurposing Using the CANDO Platform" Molecules 26, no. 9: 2581. https://doi.org/10.3390/molecules26092581
APA StyleHudson, M. L., & Samudrala, R. (2021). Multiscale Virtual Screening Optimization for Shotgun Drug Repurposing Using the CANDO Platform. Molecules, 26(9), 2581. https://doi.org/10.3390/molecules26092581






