Genome-Wide Analysis of PL7 Alginate Lyases in the Genus Zobellia
Abstract
1. Introduction
2. Results and Discussion
2.1. Genomic Comparison of Zobellia Species
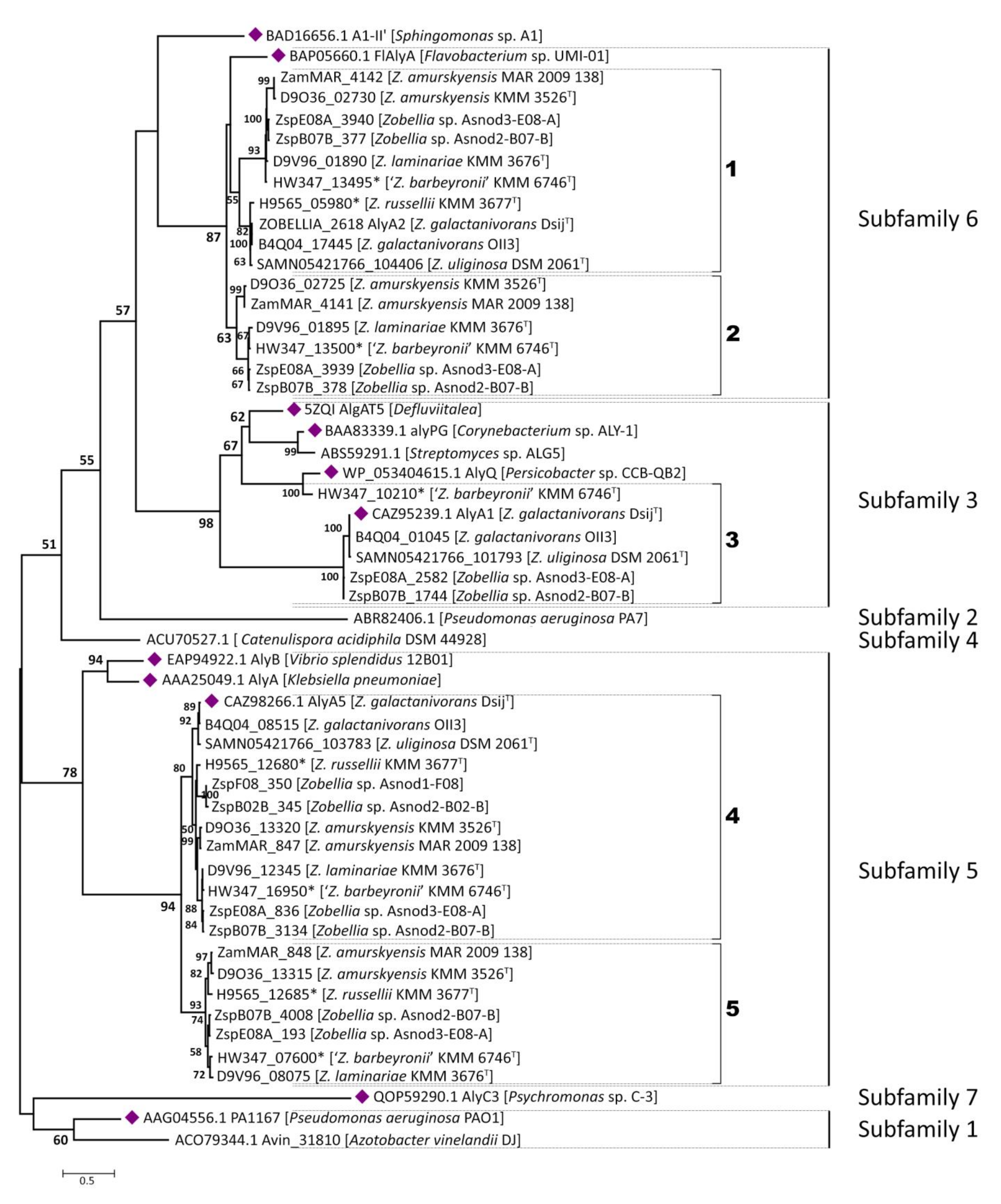
2.2. PL7 Phylogenic and Structural Analyses
2.3. Comparative Analysis of PL7-Containing Loci between Zobellia Genomes
3. Materials and Methods
3.1. Phylogenomic Analysis
3.2. Annotation of Carbohydrate-Active Enzymes
3.3. Sequence Analyses and Homology Modelling
3.4. Comparative Analysis of PL7-Containing Loci
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kirchman, D.L. The ecology of Cytophaga-Flavobacteria in aquatic environments. FEMS Microbiol. Ecol. 2002, 39, 91–100. [Google Scholar]
- Thomas, F.; Hehemann, J.-H.; Rebuffet, E.; Czjzek, M.; Michel, G. Environmental and gut Bacteroidetes: The food connection. Front. Microbiol. 2011, 2, 93. [Google Scholar] [CrossRef] [PubMed]
- Thomas, F.; Barbeyron, T.; Tonon, T.; Génicot, S.; Czjzek, M.; Michel, G. Characterization of the first alginolytic operons in a marine bacterium: From their emergence in marine Flavobacteriia to their independent transfers to marine Proteobacteria and human gut Bacteroides. Environ. Microbiol. 2012, 14, 2379–2394. [Google Scholar] [CrossRef] [PubMed]
- Sun, C.; Fu, G.; Zhang, C.; Hu, J.; Xu, L.; Wang, R.; Su, Y.; Han, S.; Yu, X.; Cheng, H.; et al. Isolation and complete genome sequence of Algibacter alginolytica sp. nov., a novel seaweed-degrading Bacteroidetes bacterium with diverse putative polysaccharide utilization loci. Appl. Environ. Microbiol. 2016, 82, 2975–2987. [Google Scholar]
- Mann, A.J.; Hahnke, R.L.; Huang, S.; Werner, J.; Xing, P.; Barbeyron, T.; Huettel, B.; Stüber, K.; Reinhardt, R.; Harder, J.; et al. The genome of the alga-associated marine flavobacterium Formosa agariphila KMM 3901T reveals a broad potential for degradation of algal polysaccharides. Appl. Environ. Microbiol. 2013, 79, 6813–6822. [Google Scholar]
- Kabisch, A.; Otto, A.; Konig, S.; Becher, D.; Albrecht, D.; Schuler, M.; Teeling, H.; Amann, R.I.; Scheweder, T. Functional characterization of polysaccharide utilization loci in the marine Bacteroidetes ‘Gramella forsetii’ KT0803. ISME J. 2014, 8, 1492–1502. [Google Scholar]
- Chaq, Q.Q.; Wang, X.J.; Ren, X.B.; Li, D.; Wang, P.; Li, P.Y.; Fu, H.H.; Zhang, X.Y.; Chen, X.L.; Zhang, Y.Z.; et al. Comparison of alginate utilization pathways in culturable bacteria isolated from Arctic and Antarctic marine environments. Front. Microbiol. 2021, 12, 609393. [Google Scholar]
- Wolter, L.A.; Mitulla, M.; Kalem, J.; Daniel, R.; Simon, M.; Wietz, M. CAZymes in Maribacter dokdonensis 62-1 from the Patagonian shelf: Genomics and physiology compared to related flavobacteria and a co-occurring Alteromonas strain. Front. Microbiol. 2020, 12, 717. [Google Scholar]
- Reintjes, G.; Arnosti, C.; Fuchs, B.M.; Amann, R. An alternative polysaccharide uptake mechanism of marine bacteria. ISME J. 2017, 11, 1640–1650. [Google Scholar] [CrossRef]
- Matos, M.N.; Lozada, M.; Anselmino, L.E.; Musumeci, M.A.; Henrissat, B.; Jansson, J.K.; Mac Cormack, W.P.; Carroll, J.; Sjöling, S.; Lundgren, L.; et al. Metagenomics unveils the attributes of the alginolytic guilds of sediments from four distant cold coastal environments. Environ. Microbiol. 2016, 18, 4471–4484. [Google Scholar]
- Barbeyron, T.; L’Haridon, S.; Corre, E.; Kloareg, B.; Potin, P. Zobellia galactanovorans gen. nov., sp. nov., a marine species of Flavobacteriaceae isolated from a red alga, and classification of [Cytophaga] uliginosa (ZoBell and Upham 1944) Reichenbach 1989 as Zobellia uliginosa gen. nov., comb. nov. Int. J. Syst. Evol. Microbiol. 2001, 51, 985–997. [Google Scholar]
- Nedashkovskaya, O.I.; Suzuki, M.; Vancanneyt, M.; Cleenwerck, I.; Lysenko, A.M.; Mikhailov, V.V.; Swings, J. Zobellia amurskyensis sp. nov., Zobellia laminariae sp. nov. and Zobellia russellii sp. nov., novel marine bacteria of the family Flavobacteriaceae. Int. J. Syst. Evol. Microbiol. 2004, 54, 1643–1648. [Google Scholar]
- Martin, M.; Portetelle, D.; Michel, G.; Vandenbol, M. Microorganisms living on macroalgae: Diversity, interactions, and biotechnological applications. Appl. Microbiol. Biotechnol. 2014, 98, 2917–2935. [Google Scholar]
- Barbeyron, T.; Thomas, F.; Barbe, V.; Teeling, H.; Schenowitz, C.; Dossat, C.; Goesmann, A.; Leblanc, C.; Glöckner, F.O.; Czjzek, M.; et al. Habitat and taxon as driving forces of carbohydrate catabolism in marine heterotrophic bacteria: Example of the model algae-associated bacterium Zobellia galactanivorans Dsij. Environ. Microbiol. 2016, 18, 4610–4627. [Google Scholar] [CrossRef]
- Hehemann, J.H.; Correc, G.; Thomas, F.; Bernard, T.; Barbeyron, T.; Jam, M.; Helbert, W.; Michel, G.; Czjzek, M. Biochemical and structural characterization of the complex agarolytic enzyme system from the marine bacterium Zobellia galactanivorans. J. Biol. Chem. 2012, 287, 30571–30584. [Google Scholar]
- Barbeyron, T.; Gerard, A.; Potin, P. The kappa-carrageenase of the marine bacterium Cytophaga drobachiensis. Structural and phylogenetic relationships within family-16 glycoside hydrolases. Mol. Biol. Evol. 1998, 15, 528–537. [Google Scholar]
- Rebuffet, E.; Barbeyron, T.; Jeudy, A.; Jam, M.; Czjzek, M.; Michel, G. Identification of catalytic residues and mechanistic analysis of family GH82 iota-carrageenases. Biochemistry 2010, 49, 7590–7599. [Google Scholar] [CrossRef]
- Harms, H.; Poehlein, A.; Thürmer, A.; König, G.M.; Schäberle, T.F. Draft Genome Sequence of Zobellia sp. Strain OII3, Isolated from the Coastal Zone of the Baltic Sea. Genome Announc. 2017, 5, e00737-17. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Thomas, F.; Larocque, R.; Li, N.; Duffieux, D.; Cladière, L.; Souchaud, F.; Michel, G.; McBride, M.J. Genetic analyses unravel the crucial role of a horizontally acquired alginate lyase for brown algal biomass degradation by Zobellia galactanivorans. Environ. Microbiol. 2017, 19, 2164–2181. [Google Scholar] [CrossRef]
- Labourel, A.; Jam, M. The beta-glucanase ZgLamA from Zobellia galactanivorans evolved a bent active site adapted for efficient degradation of algal laminarin. J. Biol. Chem. 2014, 289, 2027–2042. [Google Scholar]
- Thomas, F.; Lundqvist, L.C.E.; Jam, M.; Jeudy, A.; Barbeyron, T.; Sandström, C.; Michel, G.; Czjzek, M. Comparative Characterization of Two Marine Alginate Lyases from Zobellia galactanivorans Reveals Distinct Modes of Action and Exquisite Adaptation to Their Natural Substrate. J. Biol. Chem. 2013, 288, 23021–23037. [Google Scholar] [CrossRef]
- Asnicar, F.; Thomas, A.M.; Beghini, F.; Mengoni, C.; Manara, S.; Manghi, P.; Zhu, Q.; Bolzan, M.; Cumbo, F.; May, U.; et al. Precise phylogenetic analysis of microbial isolates and genomes from metagenomes using PhyloPhlAn 3.0. Nat. Commun. 2020, 11, 2500. [Google Scholar] [CrossRef] [PubMed]
- Chernysheva, N.; Bystritskaya, E.; Stenkova, A.; Golovkin, I.; Nedashkovskaya, O.; Isaeva, M. Comparative Genomics and CAZyme Genome Repertoires of Marine Zobellia Amurskyensis KMM 3526T and Zobellia Laminariae KMM 3676T. Mar. Drugs 2019, 17, 661. [Google Scholar]
- Zhang, H.; Yohe, T.; Huang, L.; Entwistle, S.; Wu, P.; Yang, Z.; Busk, P.K.; Xu, Y.; Yin, Y. dbCAN2: A meta server for automated carbohydrate-active enzyme annotation. Nucleic Acids Res. 2018, 46, W95–W101. [Google Scholar] [CrossRef] [PubMed]
- Thomas, F.; Barbeyron, T.; Michel, G. Evaluation of reference genes for real-time quantitative PCR in the marine flavobacterium Zobellia galactanivorans. J. Microbiol. Methods 2011, 84, 61–66. [Google Scholar]
- Lombard, V.; Bernard, T.; Rancurel, C.; Brumer, H.; Coutinho, P.M.; Henrissat, B. A hierarchical classification of polysaccharide lyases for glycogenomics. Biochem. J. 2010, 432, 437–444. [Google Scholar] [CrossRef] [PubMed]
- Pei, X.; Chang, Y.; Shen, J. Cloning, expression and characterization of an endo-acting bifunctional alginate lyase of marine bacterium Wenyingzhuangia fucanilytica. Protein Expr. Purif. 2019, 154, 44–51. [Google Scholar] [CrossRef]
- Xu, F.; Chen, X.L.; Sun, X.H.; Dong, F.; Li, C.Y.; Li, P.Y.; Ding, H.; Chen, Y.; Zhang, Y.Z.; Wang, P. Structural and molecular basis for the substrate positioning mechanism of a new PL7 subfamily alginate lyase from the arctic. J. Biol. Chem. 2020, 295, 16380–16392. [Google Scholar] [CrossRef]
- Sim, P.F.; Furusawa, G.; Teh, A.H. Functional and structural studies of a multidomain alginate lyase from Persicobacter sp. CCB-QB2. Sci. Rep. 2017, 7, 13656. [Google Scholar] [CrossRef]
- Inoue, A.; Takadono, K.; Nishiyama, R.; Tajima, K.; Kobayashi, T.; Ojima, T. Characterization of an Alginate Lyase, FlAlyA, from Flavobacterium sp. Strain UMI-01 and Its Expression in Escherichia coli. Mar. Drugs 2014, 12, 4693–4712. [Google Scholar] [CrossRef] [PubMed]
- Qin, H.M.; Miyakawa, T.; Inoue, A.; Nishiyama, R.; Nakamura, A.; Asano, A.; Ojima, T.; Tanokura, M. Structural basis for controlling the enzymatic properties of polymannuronate preferred alginate lyase FlAlyA from the PL-7 family. Chem. Commun. (Camb.) 2018, 54, 555–558. [Google Scholar] [CrossRef]
- Thomas, F.; Bordron, P.; Eveillard, D.; Michel, G. Gene expression analysis of Zobellia galactanivorans during the degradation of algal polysaccharides reveals both substrate-specific and shared transcriptome-wide responses. Front. Microbiol. 2017, 8, 1808. [Google Scholar] [CrossRef] [PubMed]
- Dudek, M.; Dieudonné, A.; Jouanneau, D.; Rochat, T.; Michel, G.; Sarels, B.; Thomas, F. Regulation of alginate catabolism involves a GntR family repressor in the marine flavobacterium Zobellia galactanivorans Dsij. Nucleic Acids Res. 2020, 48, 7786–7800. [Google Scholar] [CrossRef] [PubMed]
- Stamatakis, A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef]
- Wickham, H. ggplot2. Wiley Interdiscip. Rev. Comput. Stat. 2011, 3, 180–185. [Google Scholar] [CrossRef]
- Kolde, R.; Kolde, M.R. Package ‘pheatmap’. R Package 2015, 1, 790. [Google Scholar]
- Blum, M.; Chang, H.-Y.; Chuguransky, S.; Grego, T.; Kandasaamy, S.; Mitchell, A.; Nuka, G.; Paysan-Lafosse, T.; Qureshi, M.; Raj, S. The InterPro protein families and domains database: 20 years on. Nucleic Acids Res. 2021, 49, D344–D354. [Google Scholar] [CrossRef]
- Lombard, V.; Golaconda Ramulu, H.; Drula, E.; Coutinho, P.M.; Henrissat, B. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 2013, 42, D490–D495. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Nguyen, L.-T.; Schmidt, H.A.; von Haeseler, A.; Minh, B.Q. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef] [PubMed]
- Kalyaanamoorthy, S.; Minh, B.Q.; Wong, T.K.F.; von Haeseler, A.; Jermiin, L.S. Modelfinder: Fast model selection for accurate phylogenetic estimates. Nat. Methods 2017, 14, 587–589. [Google Scholar] [CrossRef]
- Madeira, F.; Park, Y.M.; Lee, J.; Buso, N.; Gur, T.; Madhusoodanan, N.; Basutkar, P.; Tivey, A.R.N.; Potter, S.C.; Finn, R.D.; et al. The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Res. 2019, 47, W636–W641. [Google Scholar] [CrossRef]
- Molecular Operating Environment (MOE); 2020.09; Chemical Computing Group ULC: Montreal, QC, Canada, 2020.
- Robert, X.; Gouet, P. Deciphering Key Features in Protein Structures with the New ENDscript Server. Nucleic Acids Res. 2014, 42, W320–W324. [Google Scholar] [CrossRef] [PubMed]
- Kearse, M.; Moir, R.; Wilson, A.; Stones-Havas, S.; Cheung, M.; Sturrock, S.; Buxton, S.; Cooper, A.; Markowitz, S.; Duran, C. Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 2012, 28, 1647–1649. [Google Scholar] [CrossRef]
- Aziz, R.K.; Bartels, D.; Best, A.A.; DeJongh, M.; Disz, T.; Edwards, R.A.; Formsma, K.; Gerdes, S.; Glass, E.M.; Kubal, M.; et al. The RAST Server: Rapid annotations using subsystems technology. BMC Genom. 2008, 9, 75. [Google Scholar] [CrossRef] [PubMed]
- Sullivan, M.J.; Petty, N.K.; Beatson, S.A. Easyfig: A genome comparison visualizer. Bioinformatics 2011, 27, 1009–1010. [Google Scholar] [CrossRef] [PubMed]




Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chernysheva, N.; Bystritskaya, E.; Likhatskaya, G.; Nedashkovskaya, O.; Isaeva, M. Genome-Wide Analysis of PL7 Alginate Lyases in the Genus Zobellia. Molecules 2021, 26, 2387. https://doi.org/10.3390/molecules26082387
Chernysheva N, Bystritskaya E, Likhatskaya G, Nedashkovskaya O, Isaeva M. Genome-Wide Analysis of PL7 Alginate Lyases in the Genus Zobellia. Molecules. 2021; 26(8):2387. https://doi.org/10.3390/molecules26082387
Chicago/Turabian StyleChernysheva, Nadezhda, Evgeniya Bystritskaya, Galina Likhatskaya, Olga Nedashkovskaya, and Marina Isaeva. 2021. "Genome-Wide Analysis of PL7 Alginate Lyases in the Genus Zobellia" Molecules 26, no. 8: 2387. https://doi.org/10.3390/molecules26082387
APA StyleChernysheva, N., Bystritskaya, E., Likhatskaya, G., Nedashkovskaya, O., & Isaeva, M. (2021). Genome-Wide Analysis of PL7 Alginate Lyases in the Genus Zobellia. Molecules, 26(8), 2387. https://doi.org/10.3390/molecules26082387





