ROS Dependent Antifungal and Anticancer Modulations of Piper colubrinum Osmotin
Abstract
1. Introduction
2. Results
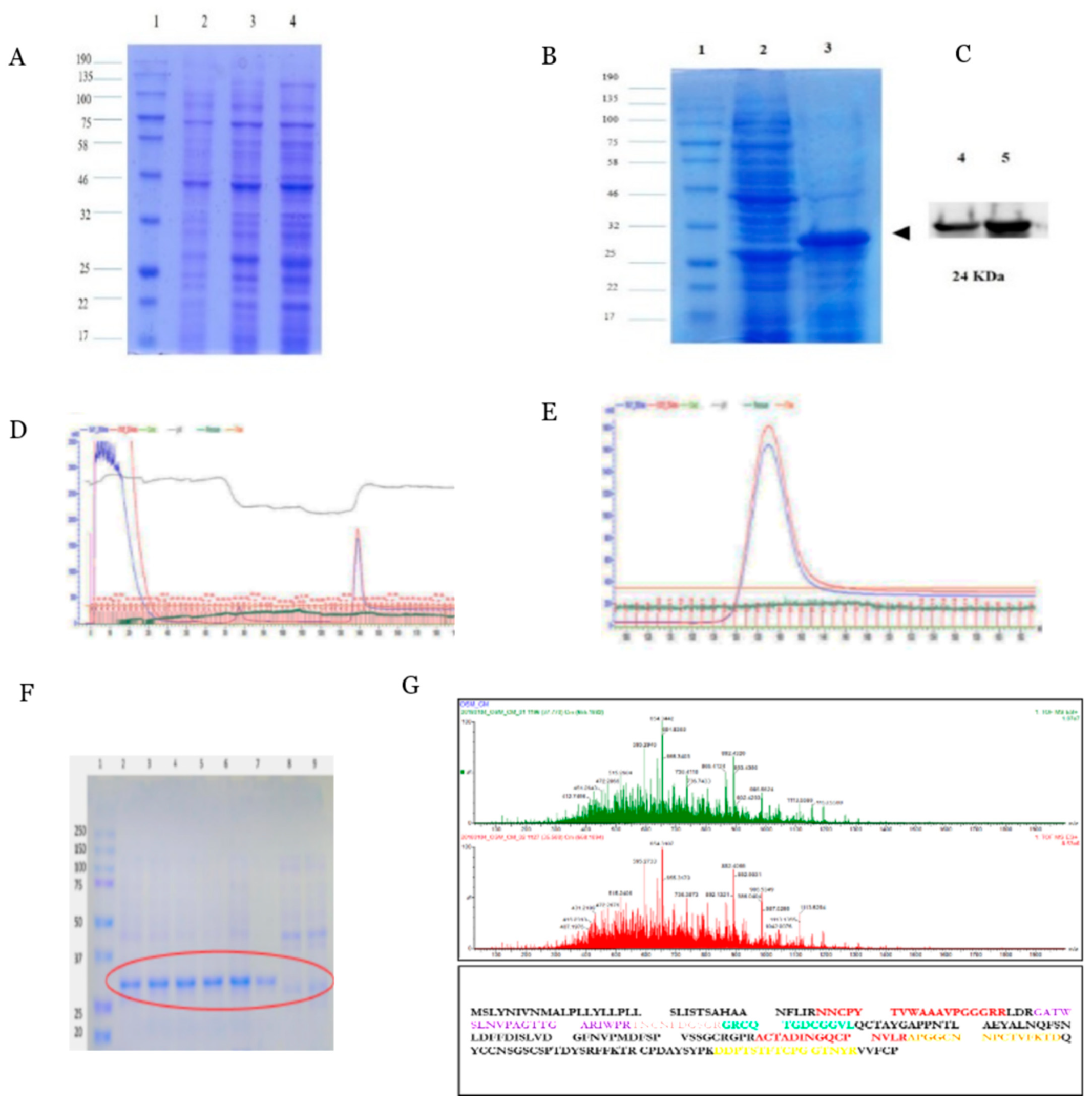
2.1. Expression and Purification of Recombinant PcOSM
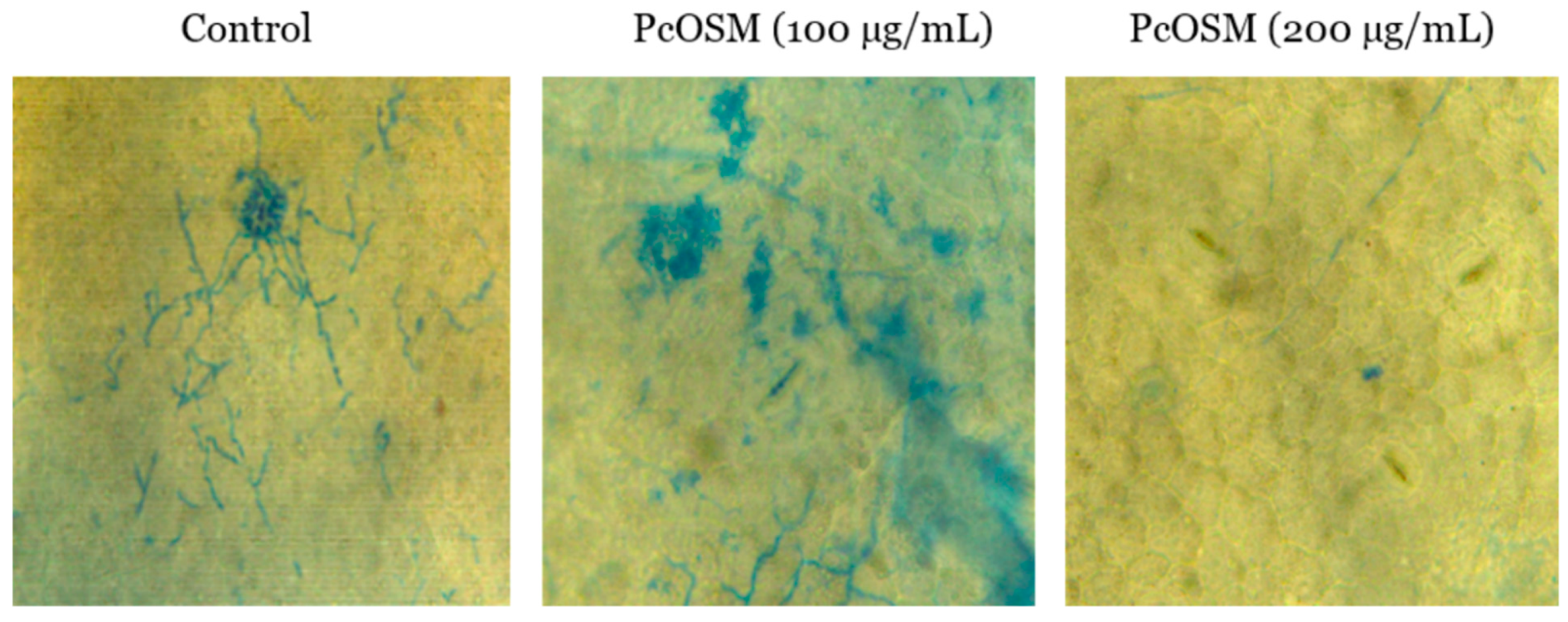
2.2. Inhibitory Activity of PcOSM on Hyphal Growth of Phytophthora Capsici
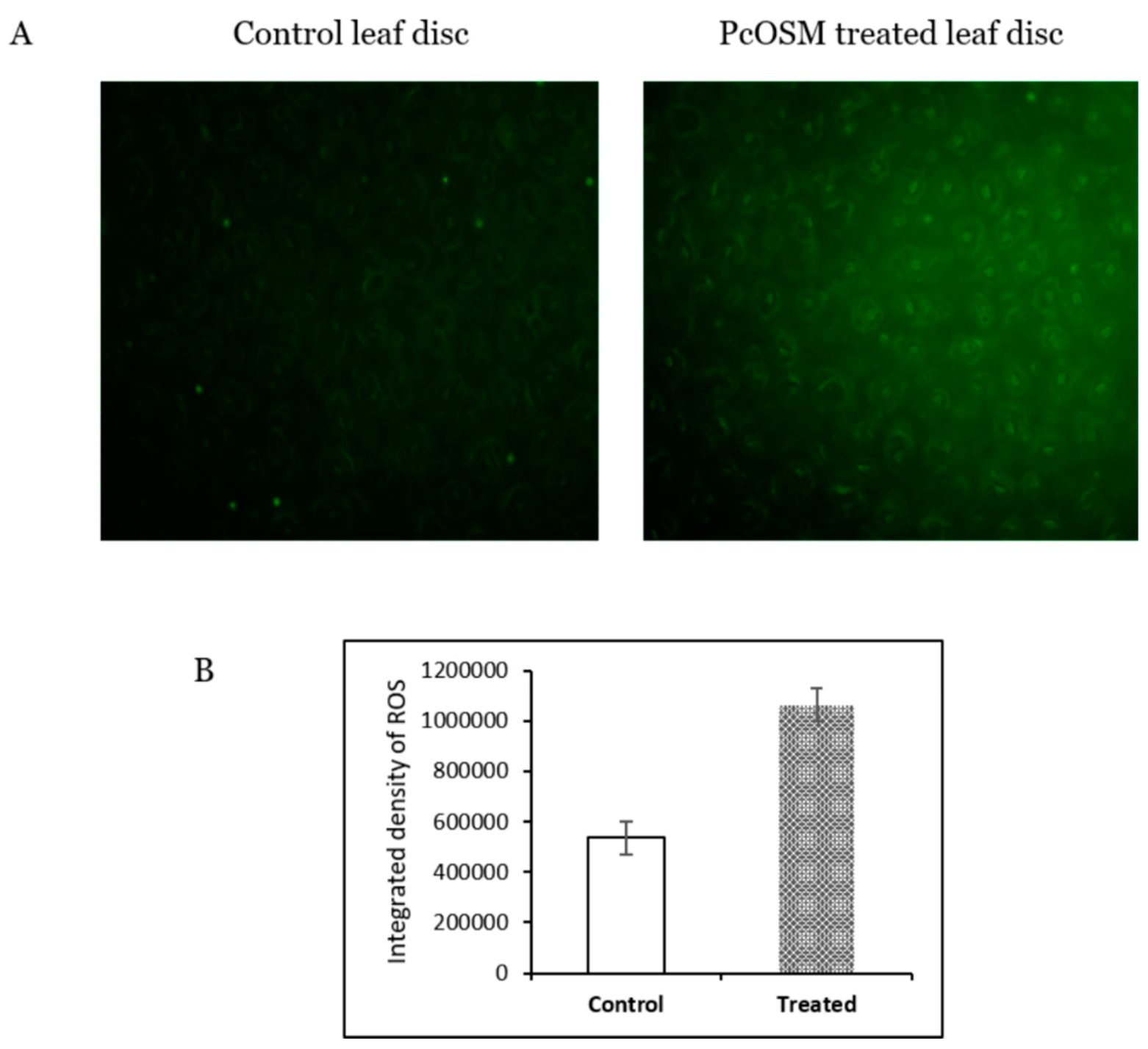
2.3. PcOSM Induced Intracellular ROS Accumulation in Leaves
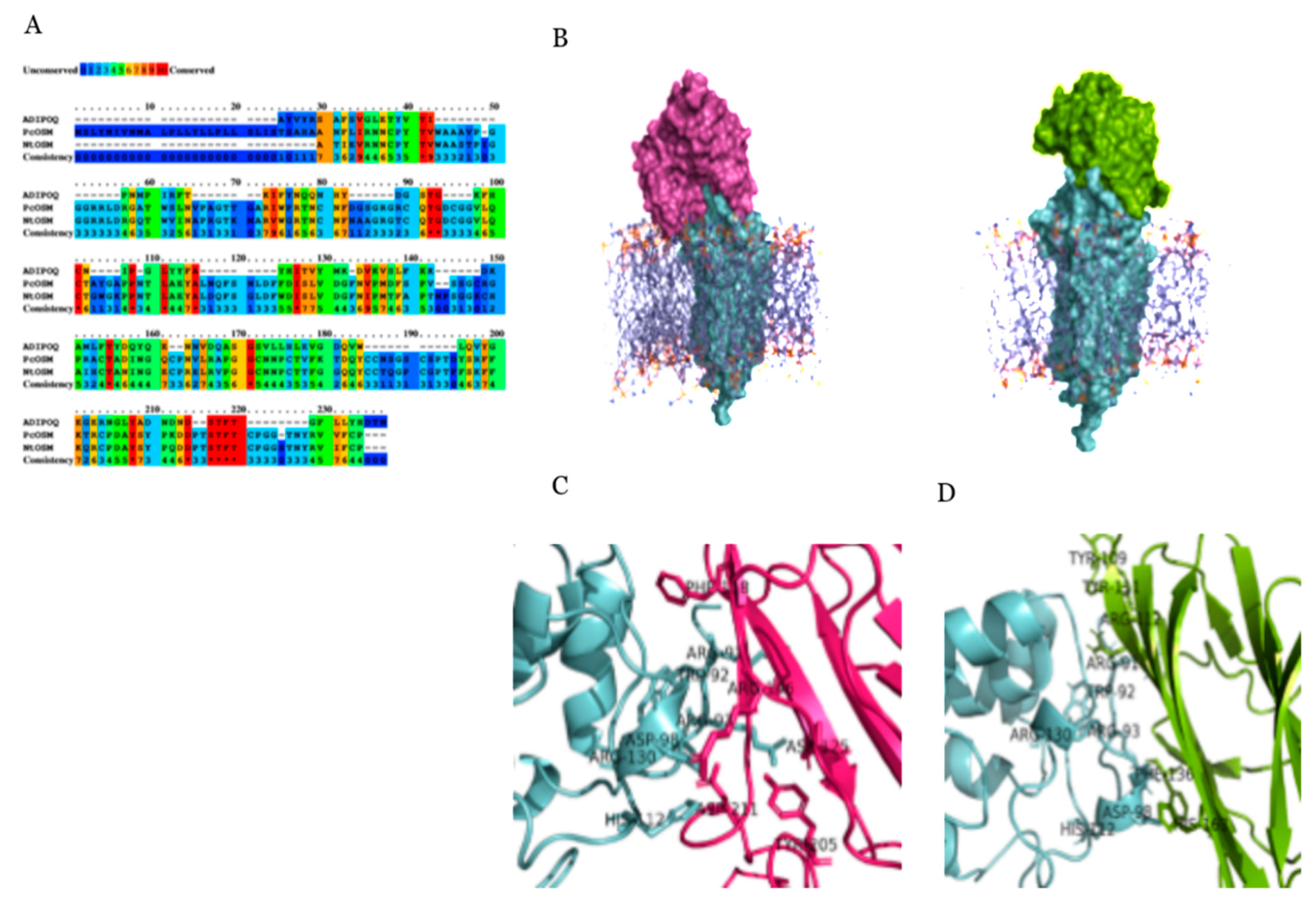
2.4. Molecular Docking and Simulation
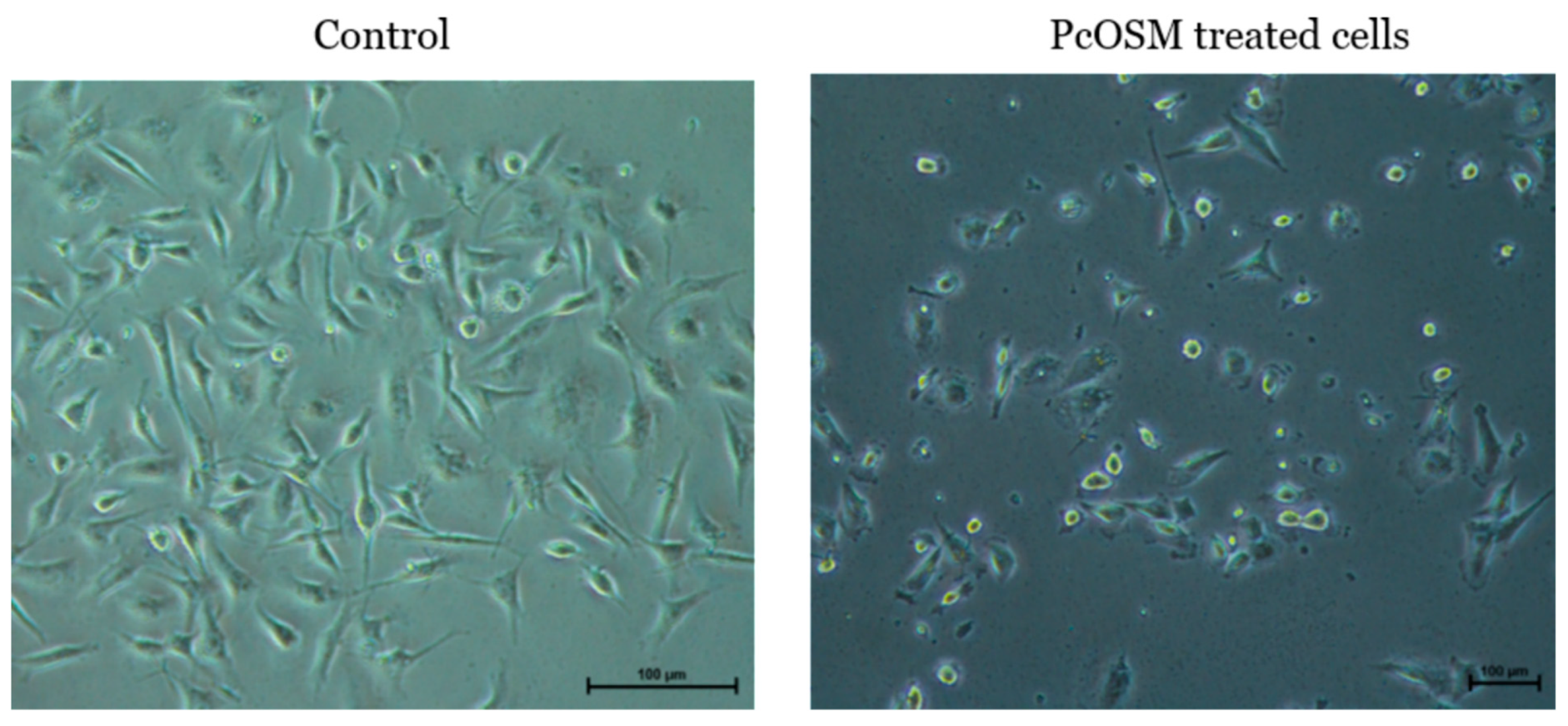
2.5. PcOSM Showed no Cytotoxicity in MDA MB231
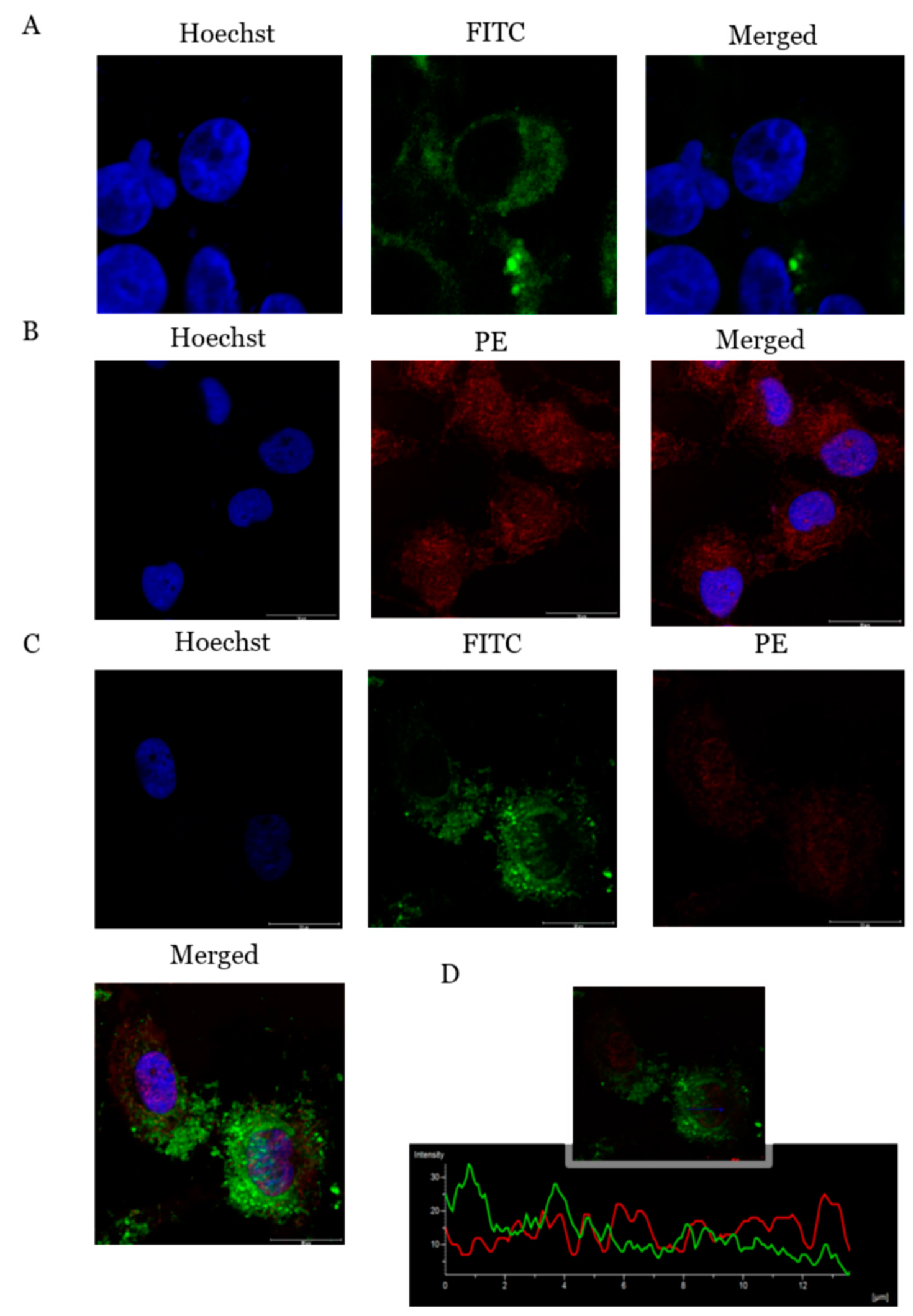
2.6. ADIPOR1 Localized on Cell Membrane and Showed Co-Localization with PcOSM–ADIPOR1
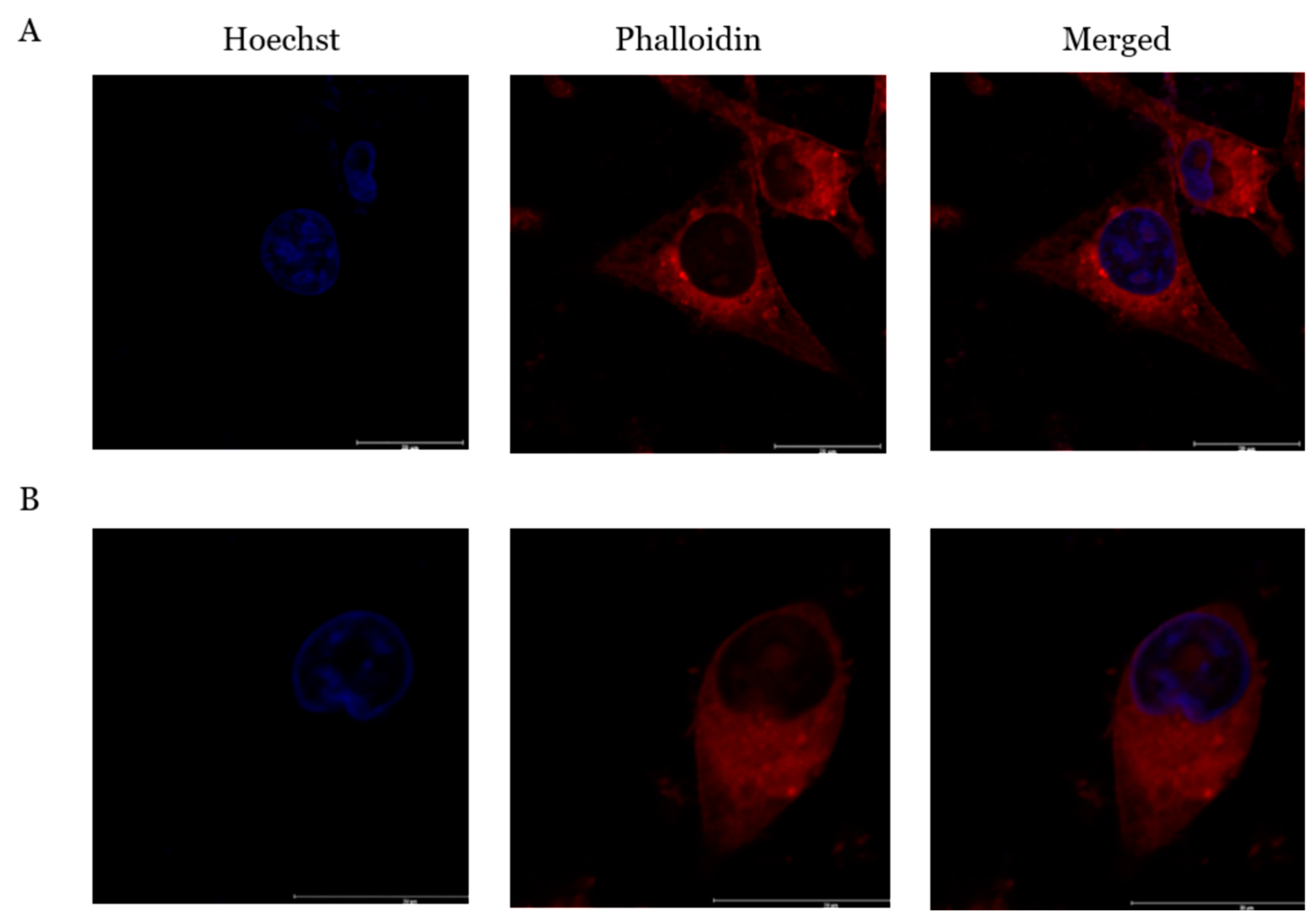
2.7. PcOSM Induced Disintegration of Cytoskeletal Elements
2.8. PcOSM Increased Intracellular Reactive Oxygen Species (ROS) Levels
2.9. PcOSM Induced Senescence in MDAMB231 Cells
2.10. PcOSM Induced Cell Cycle Arrest in MDAMB231 Cells
3. Discussion
4. Materials and Methods
4.1. Plants and Maintenance
4.2. Cloning and Expression of P. colubrinum Osmotin in E. coli
4.3. IPTG Induction and Purification of Recombinant PcOSM
4.4. SDS PAGE and Western Blot Analysis
4.5. Protein Purification
4.6. Liquid Chromatography Tandem Mass Spectrometry
4.7. LC-MS/MS Data Analysis
4.8. Fungal Growth Inhibition Bioassays
4.8.1. Infiltration Method
4.8.2. Infection Assay and Trypan Blue Staining
4.9. Measurement of ROS Production
4.10. Docking Studies
4.11. MDA MB231Proliferation Assay
4.12. Immunofluorescence Staining
4.13. ROS Detection Assay–DCFDA Fluorescent Microscopy and Flow Cytometry Assay
4.14. Senescence Assay-Beta Galactosidase Detection
4.15. Cell Cycle Analysis
4.16. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
Abbreviations
| PcOSM | Full length Piper colubrinum Osmotin (693 bop) |
| ROS | Reactive oxygen species |
| TBHP | Tert-butyl hydroperoxide |
| ADIPOR 1 | Adiponectin Receptor-1 |
| IPTG | Isopropyl β-d-1-thiogalactopyranoside |
| DCFDA | 2′,7′-Dichlorofluorescein Diacetate |
| ADIPOQ | Adiponectin |
| NtOSM | Nicotiana tabacum Osmotin |
| SA-β-gal | Senescence Associated Beta galactosidase |
References
- Roux, F.; Voisin, D.; Badet, T.; Balagué, C.; Barlet, X.; Huard-Chauveau, C.; Roby, D.; Raffaele, S. Resistance to phytopathogens e tutti quanti: Placing plant quantitative disease resistance on the map. Mol. Plant. Pathol. 2014, 15, 427–432. [Google Scholar] [CrossRef] [PubMed]
- Bowles, D.J. Defense-related proteins in higher plants. Annu. Rev. Biochem. 1990, 59, 873–907. [Google Scholar] [CrossRef] [PubMed]
- Sels, J.; Mathys, J.; De Coninck, B.M.; Cammue, B.P.; De Bolle, M.F. Plant pathogenesis-related (PR) proteins: A focus on PR peptides. Plant. Physiol. Biochem. 2008, 46, 941–950. [Google Scholar] [CrossRef] [PubMed]
- Miele, M.; Costantini, S.; Colonna, G. Structural and functional similarities between osmotin from Nicotiana tabacum seeds and human adiponectin. PLoS ONE 2011, 6, e16690. [Google Scholar] [CrossRef]
- Dicto, J.; Manjula, S. Identification of elicitor-induced PR5 gene homologue in Piper colubrinum Link by suppression subtractive hybridization. Curr. Sci. 2005, 25, 624–627. [Google Scholar]
- Mani, T.; Sivakumar, K.C.; Manjula, S. Expression and functional analysis of two osmotin (PR5) isoforms with differential anti-fungal activity from Piper colubrinum: Prediction of structure–function relationship by bioinformatics approach. Mol. Biotechnol. 2012, 52, 251–261. [Google Scholar] [CrossRef]
- Chowdhury, S.; Basu, A.; Kundu, S. Overexpression of a New Osmotin-Like Protein Gene (SindOLP) Confers Tolerance against Biotic and Abiotic Stresses in Sesame. Front. Plant. Sci. 2017, 8, 410. [Google Scholar] [CrossRef]
- Narasimhan, M.L.; Coca, M.A.; Jin, J.; Yamauchi, T.; Ito, Y.; Kadowaki, T.; Kim, K.K.; Pardo, J.M.; Damsz, B.; Hasegawa, P.M.; et al. Osmotin is a homolog of mammalian adiponectin and controls apoptosis in yeast through a homolog of mammalian adiponectin receptor. Mol. Cell. 2005, 17, 171–180. [Google Scholar] [CrossRef]
- Narasimhan, M.L.; Damsz, B.; Coca, M.A.; Ibeas, J.I.; Yun, D.-J.; Pardo, J.M.; Hasegawa, P.M.; Bressan, R.A. A Plant Defense Response Effector Induces Microbial Apoptosis. Mol. Cell 2001, 8, 921–930. [Google Scholar] [CrossRef]
- Nigro, E.; Scudiero, O.; Monaco, M.L.; Palmieri, A.; Mazzarella, G.; Costagliola, C.; Bianco, A.; Daniele, A. New Insight into Adiponectin Role in Obesity and Obesity-Related Diseases. Biomed. Res. Int. 2014, 2014, 1–14. [Google Scholar] [CrossRef]
- Peng, Y.-J.; Shen, T.-L.; Chen, Y.-S.; Mersmann, H.J.; Liu, B.-H.; Ding, S.-T. Adiponectin and adiponectin receptor 1 overexpression enhance inflammatory bowel disease. J. Biomed. Sci. 2018, 25, 24. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, A.; Ali, T.; Kim, M.W.; Khan, A.; Jo, M.H.; Rehman, S.U.; Khan, M.S.; Abid, N.B.; Khan, M.; Ullah, R.; et al. Adiponectin homolog novel osmotin protects obesity/diabetes-induced NAFLD by upregulating AdipoRs/PPARα signaling in ob/ob and db/db transgenic mouse models. Metabolism 2019, 90, 31–43. [Google Scholar] [CrossRef]
- Liu, J.; Sui, H.; Zhao, J.; Wang, Y. Osmotin protects H9c2 cells from simulated ischemia-reperfusion injury through Adi-poR1/PI3K/AKT signaling pathway. Front. Physiol. 2017, 8, 611. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, Y.; Watanabe, R.; Sato, Y.; Ozawa, N.; Kojima, M.; Watanabe-Kominato, K.; Shirai, R.; Sato, K.; Hirano, T.; Watanabe, T. Novel phytopeptide osmotin mimics preventive effects of adiponectin on vascular inflammation and atherosclerosis. Metabolism 2018, 83, 128–138. [Google Scholar] [CrossRef] [PubMed]
- Shah, S.A.; Lee, H.Y.; Bressan, R.A.; Yun, D.J.; Kim, M.O. Novel osmotin attenuates glutamate-induced synaptic dysfunction and neurodegeneration via the JNK/PI3K/Akt pathway in postnatal rat brain. Cell Death Dis. 2014, 5, e1026. [Google Scholar] [CrossRef]
- Kang, J.H.; Lee, Y.Y.; Yu, B.Y.; Yang, B.-S.; Cho, K.-H.; Yoon, D.K.; Roh, Y.K. Adiponectin induces growth arrest and apoptosis of MDA-MB-231 breast cancer cell. Arch. Pharmacal Res. 2005, 28, 1263–1269. [Google Scholar] [CrossRef] [PubMed]
- Chung, S.J.; Nagaraju, G.P.; Nagalingam, A.; Muniraj, N.; Kuppusamy, P.; Walker, A.; Woo, J.; Győrffy, B.; Gabrielson, E.; Saxena, N.K.; et al. ADIPOQ/adiponectin induces cytotoxic autophagy in breast cancer cells through STK11/LKB1-mediated activation of the AMPK-ULK1 axis. Autophagy 2017, 13, 1386–1403. [Google Scholar] [CrossRef]
- Dieudonne, M.-N.; Bussiere, M.; Dos Santos, E.; Leneveu, M.-C.; Giudicelli, Y.; Pecquery, R. Adiponectin mediates antiproliferative and apoptotic responses in human MCF7 breast cancer cells. Biochem. Biophys. Res. Commun. 2006, 345, 271–279. [Google Scholar] [CrossRef] [PubMed]
- Anil Kumar, S.; Hima Kumari, P.; Shravan Kumar, G.; Mohanalatha, C.; Kavi Kishor, P.B. Osmotin: A plant sentinel and a possible agonist of mammalian adiponectin. Front. Plant Sci. 2015, 6, 163. [Google Scholar] [CrossRef]
- Ali, T.; Yoon, G.H.; Shah, S.A.; Lee, H.Y.; Kim, M.O. Osmotin attenuates amyloid beta-induced memory impairment, tau phosphor-ylation and neurodegeneration in the mouse hippocampus. Sci. Rep. 2015, 5, 1–7. [Google Scholar] [CrossRef]
- Trivedi, V.R.; Chorawala, M.R.; Shah, G.B. Antiatherosclerotic activity of osmotin, an adiponectin agonist in atherogenic diet in-duced hypertriglyceridemia and hypercholesterolemia in wistar rats. Adv. Res. Pharm. Biol. 2012, 2, 196–207. [Google Scholar]
- Watanabe, T. Therapeutic properties of the new phytochemical osmotin for preventing atherosclerosis. Vessel Plus 2020, 4. [Google Scholar] [CrossRef]
- Arsenescu, V.; Narasimhan, M.L.; Halide, T.; Bressan, R.A.; Barisione, C.; Cohen, N.A.; De Villiers, W.J.S.; Arsenescu, R. Adiponectin and Plant-Derived Mammalian Adiponectin Homolog Exert a Protective Effect in Murine Colitis. Dig. Dis. Sci. 2011, 56, 2818. [Google Scholar] [CrossRef]
- Bashir, M.A.; Silvestri, C.; Ahmad, T.; Hafiz, I.A.; Abbasi, N.A.; Manzoor, A.; Cristofori, V.; Rugini, E. Osmotin: A Cationic Protein Leads to Improve Biotic and Abiotic Stress Tolerance in Plants. Plants 2020, 9, 992. [Google Scholar] [CrossRef]
- González-Bosch, C. Priming plant resistance by activation of redox-sensitive genes. Free Radic. Biol. Med. 2018, 122, 171–180. [Google Scholar] [CrossRef]
- Kupchak, B.R.; Villa, N.Y.; Kulemina, L.V.; Lyons, T.J. Dissecting the regulation of yeast genes by the osmotin receptor. Biochem. Biophys. Res. Commun. 2008, 374, 210–213. [Google Scholar] [CrossRef][Green Version]
- Jabs, T. Reactive oxygen intermediates as mediators of programmed cell death in plants and animals. Biochem. Pharmacol. 1999, 57, 231–245. [Google Scholar] [CrossRef]
- Madeo, F.; Fröhlich, E.; Ligr, M.; Grey, M.; Sigrist, S.J.; Wolf, D.H.; Fröhlich, K.-U. Oxygen Stress: A Regulator of Apoptosis in Yeast. J. Cell Biol. 1999, 145, 757–767. [Google Scholar] [CrossRef]
- Naseer, M.I.; Ullah, I.; Narasimhan, M.L.; Lee, H.Y.; Bressan, R.A.; Yoon, G.H.; Yun, D.J.; Kim, M.O. Neuroprotective effect of osmotin against ethanol-induced apoptotic neurodegeneration in the developing rat brain. Cell Death Dis. 2014, 5, e1150. [Google Scholar] [CrossRef]
- Oh, S.W.; Park, C.-Y.; Lee, E.S.; Yoon, Y.S.; Park, S.S.; Kim, Y.; Sung, N.J.; Yun, Y.H.; Lee, K.S.; Kang, H.S.; et al. Adipokines, insulin resistance, metabolic syndrome, and breast cancer recurrence: A cohort study. Breast Cancer Res. 2011, 13, 1–10. [Google Scholar] [CrossRef]
- Viktorova, J.; Klcova, B.; Rehorova, K.; Vlcko, T.; Stankova, L.; Jelenova, N.; Cejnar, P.; Kundu, J.K.; Ohnoutkova, L.; Macek, T. Recom-binant expression of osmotin in barley improves stress resistance and food safety during adverse growing conditions. PLoS ONE 2019, 14, e0212718. [Google Scholar] [CrossRef] [PubMed]
- Abad, L.R.; D’Urzo, M.P.; Liu, D.; Narasimhan, M.L.; Reuveni, M.; Zhu, J.K.; Niu, X.; Singh, N.K.; Hasegawa, P.M.; Bressan, R.A. Antifungal activity of tobacco osmotin has specificity and involves plasma membrane permeabilization. Plant. Sci. 1996, 118, 11–23. [Google Scholar] [CrossRef]
- Yun, D.-J.; Zhao, Y.; Pardo, J.M.; Narasimhan, M.L.; Damsz, B.; Lee, H.; Abad, L.R.; D’Urzo, M.P.; Hasegawa, P.M.; Bressan, R.A. Stress proteins on the yeast cell surface determine resistance to osmotin, a plant antifungal protein. Proc. Natl. Acad. Sci. USA 1997, 94, 7082–7087. [Google Scholar] [CrossRef]
- Tan, L.T.-H.; Chan, K.-G.; Pusparajah, P.; Lee, W.-L.; Chuah, L.-H.; Khan, T.M.; Lee, L.-H.; Goh, B.-H. Targeting Membrane Lipid a Potential Cancer Cure? Front. Pharmacol. 2017, 8, 12. [Google Scholar] [CrossRef]
- Evans, J.; Wang, Y.D.; Shaw, K.P.; Vernon, L.P. Cellular responses to Pyrularia thionin are mediated by Ca2+ influx and phospho-lipase A2 activation and are inhibited by thionin tyrosine iodination. Proc. Natl. Acad. Sci. USA 1989, 86, 5849–5853. [Google Scholar] [CrossRef]
- Mulder, K.; Lima, L.A.; Miranda, V.; Dias, S.C.; Franco, O.L. Current scenario of peptide-based drugs: The key roles of cationic anti-tumor and antiviral peptides. Front. Microbiol. 2013, 4, 321. [Google Scholar] [CrossRef] [PubMed]
- D’Angeli, S.; Altamura, M.M. Osmotin induces cold protection in olive trees by affecting programmed cell death and cytoskeleton organization. Planta 2007, 225, 1147–1163. [Google Scholar] [CrossRef]
- Vallejo, M.J.; Salazar, L.; Grijalva, M. Oxidative Stress Modulation and ROS-Mediated Toxicity in Cancer: A Review on In Vitro Models for Plant-Derived Compounds. Oxidative Med. Cell. Longev. 2017, 2017, 1–9. [Google Scholar] [CrossRef]
- Trachootham, D.; Alexandre, J.; Huang, P. Targeting cancer cells by ROS-mediated mechanisms: A radical therapeutic approach? Nat. Rev. Drug Discov. 2009, 8, 579–591. [Google Scholar] [CrossRef]
- Redza-Dutordoir, M.; Averill-Bates, D.A. Activation of apoptosis signalling pathways by reactive oxygen species. Biochim. Biophys. Acta BBA Mol. Cell Res. 2016, 1863, 2977–2992. [Google Scholar] [CrossRef] [PubMed]
- Liou, G.-Y.; Storz, P. Reactive oxygen species in cancer. Free Radic. Res. 2010, 44, 479–496. [Google Scholar] [CrossRef]
- Li, M.; You, L.; Xue, J.; Lu, Y. Ionizing Radiation-Induced Cellular Senescence in Normal, Non-transformed Cells and the Involved DNA Damage Response: A Mini Review. Front. Pharmacol. 2018, 9, 522. [Google Scholar] [CrossRef]
- Ji, S.; Zheng, Z.; Liu, S.; Ren, G.; Gao, J.; Zhang, Y.; Li, G. Resveratrol promotes oxidative stress to drive DLC1 mediated cellular se-nescence in cancer cells. Exp. Cell Res. 2018, 370, 292–302. [Google Scholar] [CrossRef]
- Ewald, J.A.; Desotelle, J.A.; Wilding, G.; Jarrard, D.F. Therapy-Induced Senescence in Cancer. J. Natl. Cancer Inst. 2010, 102, 1536–1546. [Google Scholar] [CrossRef]
- Rioja, C.; Van Wees, S.C.; Charlton, K.A.; Pieterse, C.M.J.; Lorenzo, O.; García-Sánchez, S. Wide Screening of Phage-Displayed Libraries Identifies Immune Targets in Planta. PLoS ONE 2013, 8, e54654. [Google Scholar] [CrossRef]
- Beihaghi, M.; Marashi, H.; Bagheri, A.; Sankian, M. Transient expression of CCL21as recombinant protein in tomato. Biotechnol. Rep. 2018, 17, 10–15. [Google Scholar] [CrossRef] [PubMed]
- De Campos, M.A.; Silva, M.S.; Magalhães, C.P.; Ribeiro, S.G.; Sarto, R.P.; Vieira, E.A.; Grossi-De-Sa, M.F. Expression in Escherichia coli, purification, refolding and antifungal activity of an osmotin from Solanum nigrum. Microb. Cell Factories 2008, 7, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Anu, K.; Jessymol, K.K.; Chidambareswaren, M.; Gayathri, G.S.; Manjula, S. Down-regulation of osmotin (PR5) gene by virus-induced gene silencing (VIGS) leads to susceptibility of resistant Piper colubrinum Link. to the oomycete pathogen Phytophthora capsici Leonian. Indian J. Exp. Biol. 2015, 53, 329–334. [Google Scholar]
- Rossi, F.R.; Krapp, A.R.; Bisaro, F.; Maiale, S.J.; Pieckenstain, F.L.; Carrillo, N. Reactive oxygen species generated in chloroplasts con-tribute to tobacco leaf infection by the necrotrophic fungus Botrytis cinerea. Plant. J. 2017, 92, 761–773. [Google Scholar] [CrossRef] [PubMed]
- Simossis, V.A.; Heringa, J. PRALINE: A multiple sequence alignment toolbox that integrates homology-extended and secondary structure information. Nucleic Acids Res. 2005, 33 (Suppl. 2), W289–W294. [Google Scholar] [CrossRef]
- Schneidman-Duhovny, D.; Inbar, Y.; Nussinov, R.; Wolfson, H.J. PatchDock and SymmDock: Servers for rigid and symmetric docking. Nucleic Acids Res. 2005, 33 (Suppl. 2), W363–W367. [Google Scholar] [CrossRef]
- Abraham, M.J.; Murtola, T.; Schulz, R.; Páll, S.; Smith, J.C.; Hess, B.; Lindahl, E. GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 2015, 1–2, 19–25. [Google Scholar] [CrossRef]
- Song, P.; Li, Y.; Dong, Y.; Liang, Y.; Qu, H.; Qi, D.; Lu, Y.; Jin, X.; Guo, Y.; Jia, Y.; et al. Estrogen receptor β inhibits breast cancer cells migration and invasion through CLDN6-mediated autophagy. J. Exp. Clin. Cancer Res. 2019, 38, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Jiang, M.; Wang, Z. TPM4 promotes cell migration by modulating F-actin formation in lung cancer. Oncotargets Ther. 2019, 12, 4055–4063. [Google Scholar] [CrossRef] [PubMed]
- Song, S.Y.; Jung, E.C.; Bae, C.H.; Choi, Y.S.; Kim, Y.D. Visfatin induces MUC8 and MUC5B expression via p38 MAPK/ROS/NF-κB in human airway epithelial cells. J. Biomed. Sci. 2014, 21, 1–9. [Google Scholar] [CrossRef] [PubMed]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Geetha, R.G.; Krishnankutty Nair Chandrika, S.; Saraswathy, G.G.; Nair Sivakumari, A.; Sakuntala, M. ROS Dependent Antifungal and Anticancer Modulations of Piper colubrinum Osmotin. Molecules 2021, 26, 2239. https://doi.org/10.3390/molecules26082239
Geetha RG, Krishnankutty Nair Chandrika S, Saraswathy GG, Nair Sivakumari A, Sakuntala M. ROS Dependent Antifungal and Anticancer Modulations of Piper colubrinum Osmotin. Molecules. 2021; 26(8):2239. https://doi.org/10.3390/molecules26082239
Chicago/Turabian StyleGeetha, Rajeswari Gopal, Sivakumar Krishnankutty Nair Chandrika, Gayathri G. Saraswathy, Asha Nair Sivakumari, and Manjula Sakuntala. 2021. "ROS Dependent Antifungal and Anticancer Modulations of Piper colubrinum Osmotin" Molecules 26, no. 8: 2239. https://doi.org/10.3390/molecules26082239
APA StyleGeetha, R. G., Krishnankutty Nair Chandrika, S., Saraswathy, G. G., Nair Sivakumari, A., & Sakuntala, M. (2021). ROS Dependent Antifungal and Anticancer Modulations of Piper colubrinum Osmotin. Molecules, 26(8), 2239. https://doi.org/10.3390/molecules26082239





