Effects of 3FTx Protein Fraction from Naja ashei Venom on the Model and Native Membranes: Recognition and Implications for the Mechanisms of Toxicity
Abstract
1. Introduction
2. Results
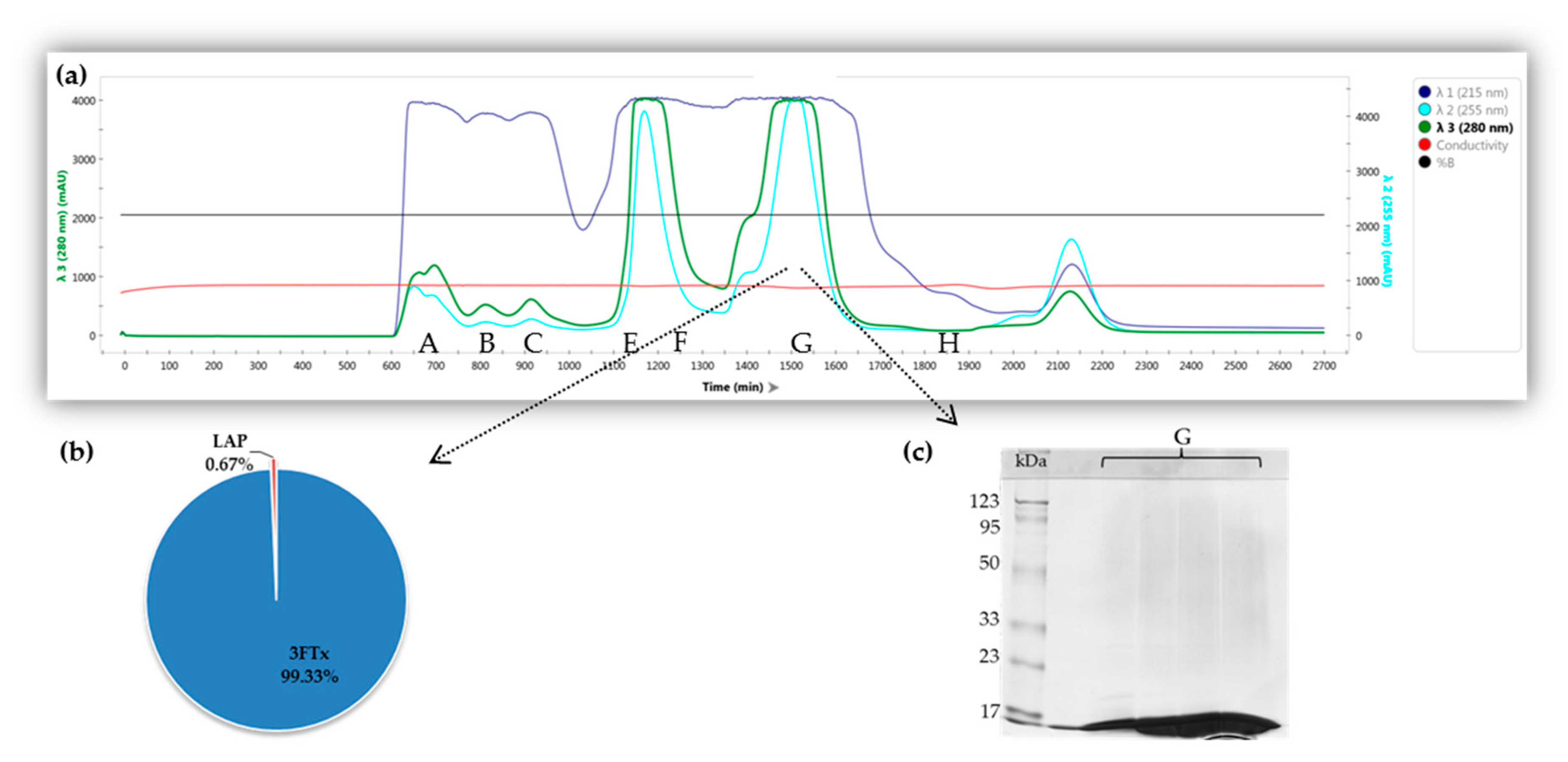
2.1. Fractionation of Crude Naja ashei Venom and Identification of Proteins in the Obtained Fractions
2.2. Characterization of Model Membranes and Their Physio-Chemical Parameters
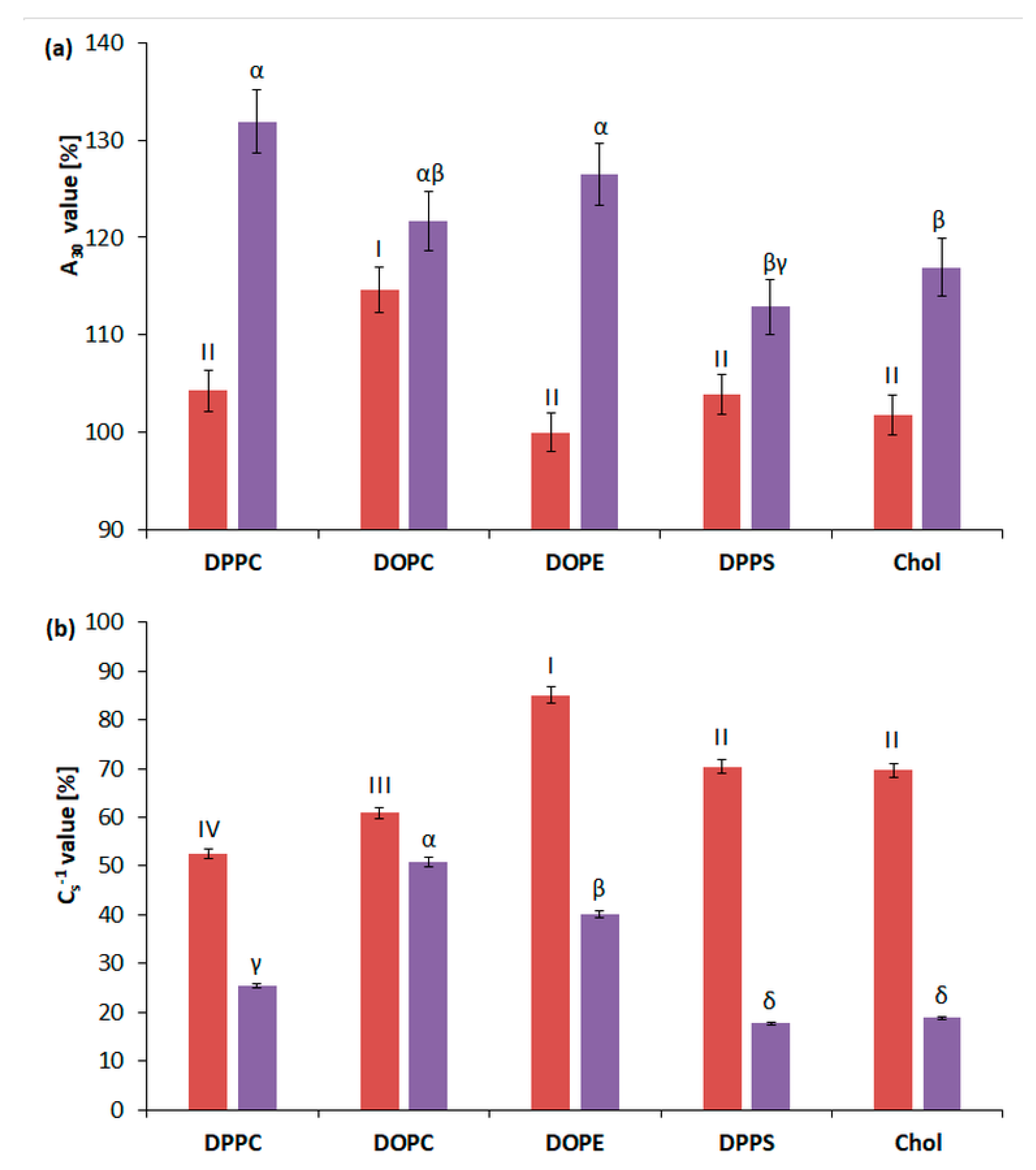
2.2.1. 3FTx Interaction with Multicomponent Monolayers
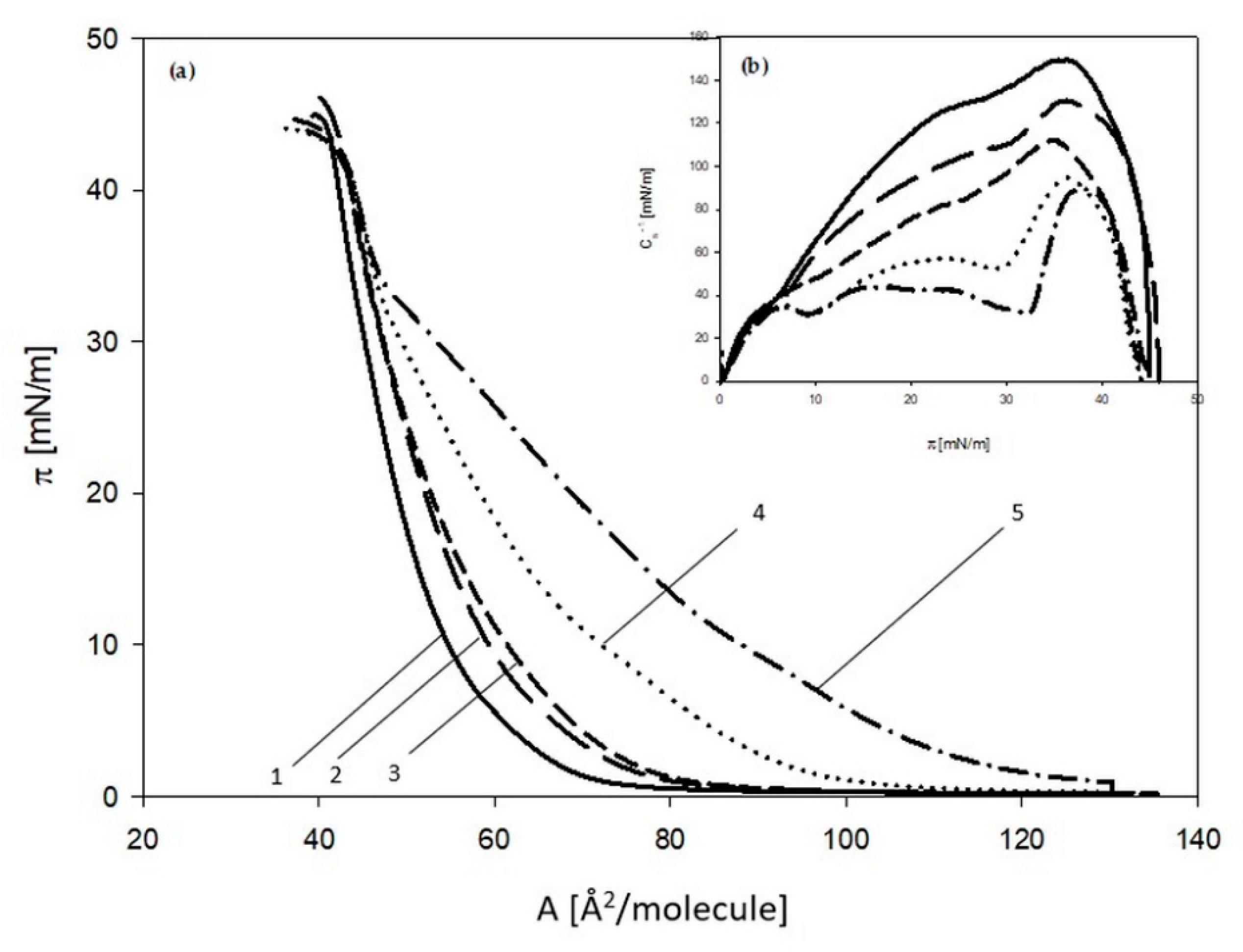
2.2.2. 3FTx Interaction with Single-Component Monolayers
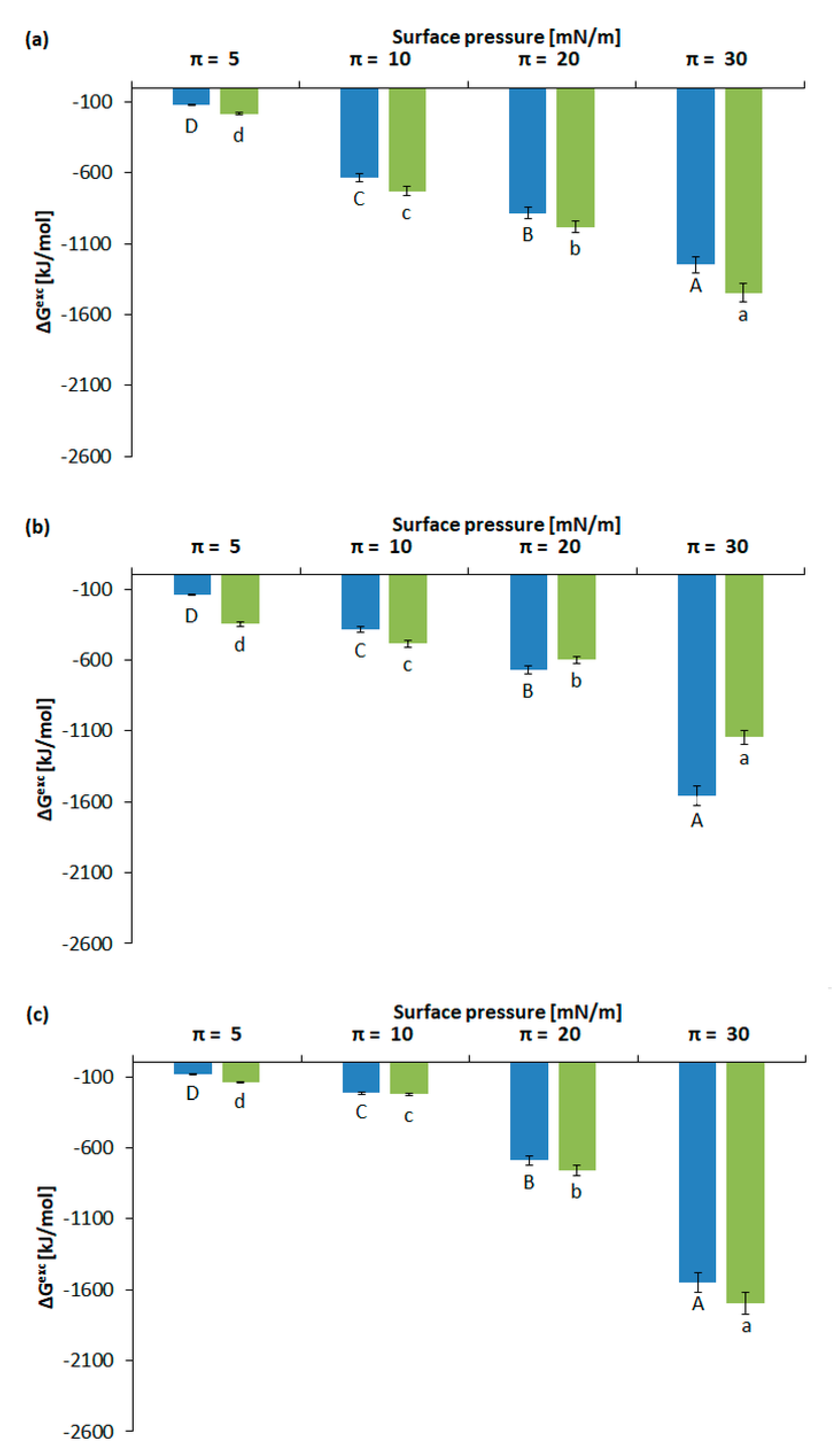
2.2.3. Calculation of Gibbs Excess Free Energy of Mixing
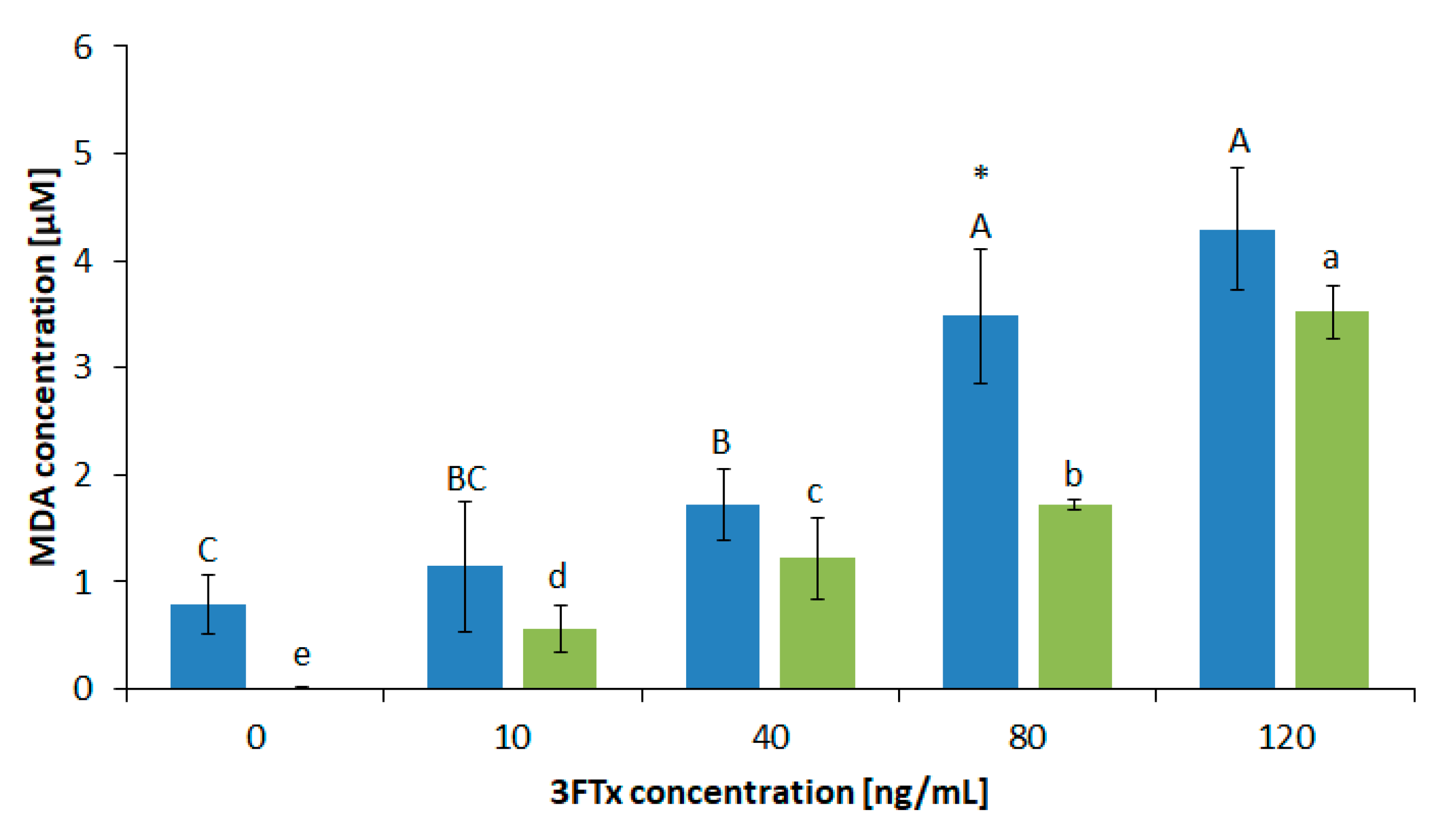
2.3. Biochemical Characterization of Membrane Properties Described by Lipid Peroxidation (MDA Concentration) and Membrane Damage (Lactate Dehydrogenase (LDH) Assay)
2.4. Human Cell Culture Tests under 3FTx Treatment
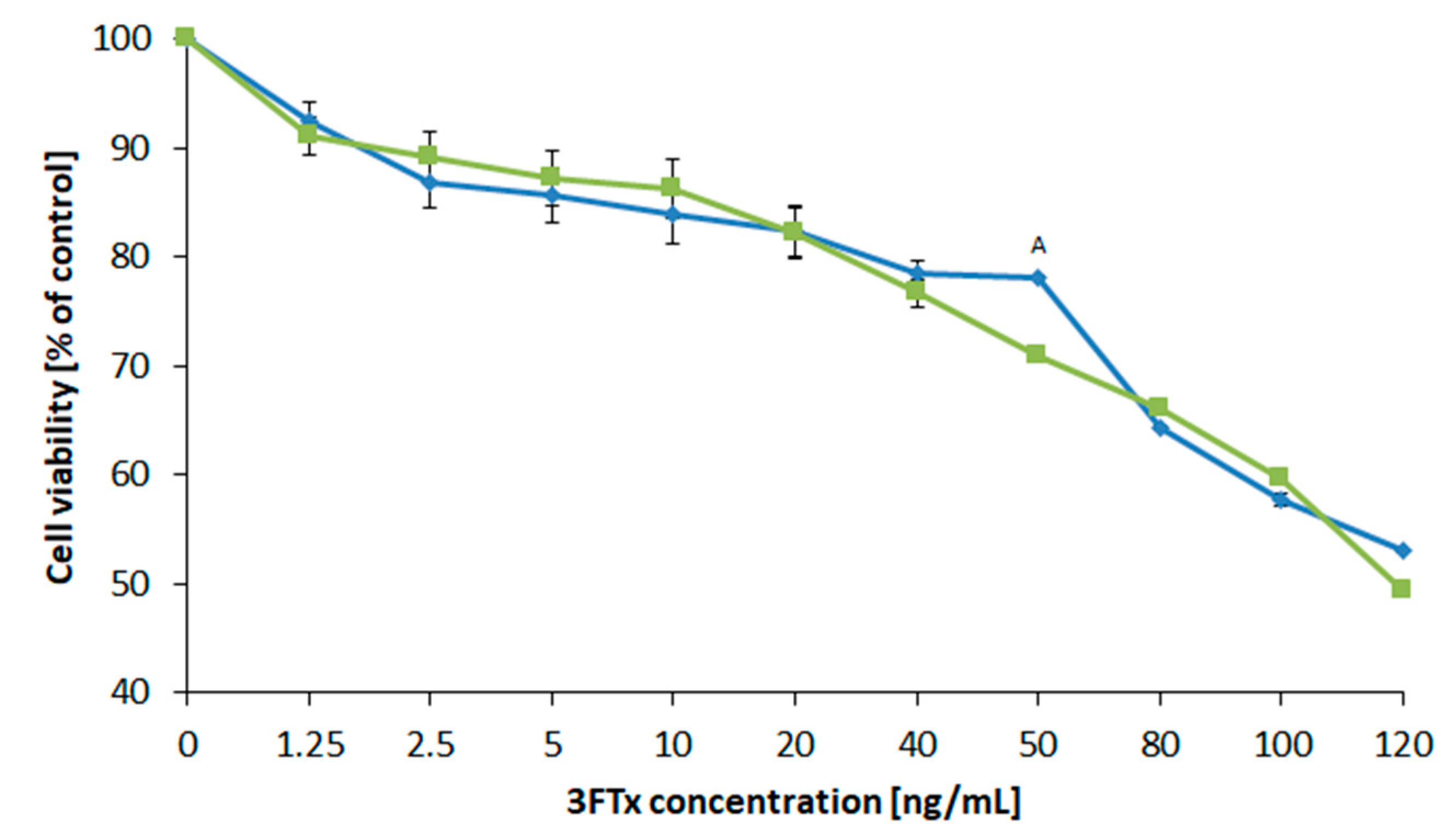
2.4.1. Determination of the Cell Viability (XTT Test)
2.4.2. Calculation of the Medial Lethal Dose (LD50)
3. Discussion
4. Materials and Methods
4.1. Venom Collection
4.2. Fractionation of Naja ashei Venom with Size-Exclusion Chromatography
4.3. Identification of Proteins in Obtained Fractions Using Shotgun LC-MS/MS Analysis
4.4. Model Membranes Composition
4.5. Langmuir Technique
4.6. Cell Cultures
4.7. Cell Viability Assay
4.8. Membrane Damage Assay (LDH Assay)
4.9. Determination of Membrane Lipid Peroxidation (MDA Concentration)
4.10. Nitric Oxide Production
4.11. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Sample Availability
Abbreviations
| Alim | the limiting area per molecule representing the maximal density of a layer |
| A30 | the area per molecule representing the density of a layer at π = 30 mN/m |
| 3FTx | three-finger toxins |
| Cs−1 | static compression modulus |
| ∆Gexc | the excess free energy of mixing |
| DPPC | 1,2-dipalmitoyl-sn-glycero-3-phosphocholine (16:0) |
| DOPC | 1,2-dioleoyl-sn-glycero-3-phosphocholine (18:1) |
| DOPE | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine (18:1) |
| DPPS | 1,2-dipalmitoyl-sn-glycero-3-phospho-L-serine (16:0) |
| Chol | cholesterol |
| MDA | malondialdehyde |
| LDH | lactate dehydrogenase |
| LD50 | medial lethal dose |
| NO | nitric oxide |
References
- Ohsaka, A. Fractionation of Habu Snake Venom by Chromatography on Cm-Cellulose with Special Reference to Biological Activities. Jpn. J. Med. Sci. Biol. 1960, 13, 199–205. [Google Scholar] [CrossRef] [PubMed]
- Fox, J.W.; Serrano, S.M.T. Exploring Snake Venom Proteomes: Multifaceted Analyses for Complex Toxin Mixtures. Proteomics 2008, 8, 909–920. [Google Scholar] [CrossRef]
- Kumar, T.K.S.; Jayaraman, G.; Lee, C.S.; Arunkumar, A.I.; Sivaraman, T.; Samuel, D.; Yu, C. Snake Venom Cardiotoxins-Structure, Dynamics, Function and Folding. J. Biomol. Struct. Dyn. 1997, 15, 431–463. [Google Scholar] [CrossRef]
- Kini, R.M.; Doley, R. Structure, Function and Evolution of Three-Finger Toxins: Mini Proteins with Multiple Targets. Toxicon 2010, 56, 855–867. [Google Scholar] [CrossRef]
- Dufton, M.J. Classification of Elapid Snake Neurotoxins and Cytotoxins According to Chain Length: Evolutionary Implications. J. Mol. Evol. 1984, 20, 128–134. [Google Scholar] [CrossRef]
- Structural Classification of Proteins. Available online: http://supfam.org/SUPERFAMILY/cgi-bin/scop.cgi?sunid=57303#section_scop_classification (accessed on 15 February 2021).
- Hus, K.K.; Buczkowicz, J.; Petrilla, V.; Petrillová, M.; Łyskowski, A.; Legáth, J.; Bocian, A. First Look at the Venom of Naja Ashei. Molecules 2018, 23, 609. [Google Scholar] [CrossRef]
- Hus, K.K.; Marczak, Ł.; Petrilla, V.; Petrillová, M.; Legáth, J.; Bocian, A. Different Research Approaches in Unraveling the Venom Proteome of Naja Ashei. Biomolecules 2020, 10, 1282. [Google Scholar] [CrossRef]
- Wüster, W.; Broadley, D.G. Get an Eyeful of This: A New Species of Giant Spitting Cobra from Eastern and North-Eastern Africa (Squamata: Serpentes: Elapidae: Naja). Zootaxa 2007, 1532, 51–68. [Google Scholar] [CrossRef]
- Antolikova, N.R.; Kello, M.; Zigova, M.; Tischlerova, V.; Petrilla, V.; Pirnik, Z.; Mojzisova, G.; Mojzis, J. Naja Ashei Venom Induces Mitochondria-Mediated Apoptosis in Human Colorectal Cancer Cells. Acta Biochim. Pol. 2019, 66, 207–213. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Ming, W.; Wang, Y.; Liu, S.; Qiu, Y.; Xiang, Y.; Hu, L.; Fan, L.; Peng, X.; Wang, H.; et al. Cytotoxin 1 from Naja Atra Cantor Venom Induced Necroptosis of Leukemia Cells. Toxicon 2019, 165, 110–115. [Google Scholar] [CrossRef] [PubMed]
- Lewinska, A.; Bocian, A.; Petrilla, V.; Adamczyk-Grochala, J.; Szymura, K.; Hendzel, W.; Kaleniuk, E.; Hus, K.K.; Petrillova, M.; Wnuk, M. Snake Venoms Promote Stress-Induced Senescence in Human Fibroblasts. J. Cell. Physiol. 2019, 234, 6147–6160. [Google Scholar] [CrossRef]
- Bocian, A.; Ciszkowicz, E.; Hus, K.K.; Buczkowicz, J.; Lecka-Szlachta, K.; Pietrowska, M.; Petrilla, V.; Petrillova, M.; Legáth, Ľ.; Legáth, J. Antimicrobial Activity of Protein Fraction from Naja Ashei Venom against Staphylococcus Epidermidis. Molecules 2020, 25, 293. [Google Scholar] [CrossRef]
- Kuna, E.; Bocian, A.; Hus, K.K.; Petrilla, V.; Petrillova, M.; Legath, J.; Lewinska, A.; Wnuk, M. Evaluation of Antifungal Activity of Naja Pallida and Naja Mossambica Venoms against Three Candida Species. Toxins (Basel) 2020, 12, 500. [Google Scholar] [CrossRef] [PubMed]
- Konshina, A.G.; Boldyrev, I.A.; Utkin, Y.N.; Omel’kov, A.V.; Efremov, R.G. Snake Cytotoxins Bind to Membranes via Interactions with Phosphatidylserine Head Groups of Lipids. PLoS ONE 2011, 6, e19064. [Google Scholar] [CrossRef]
- Kessler, P.; Marchot, P.; Silva, M.; Servent, D. The Three-Finger Toxin Fold: A Multifunctional Structural Scaffold Able to Modulate Cholinergic Functions. J. Neurochem. 2017, 142, 7–18. [Google Scholar] [CrossRef] [PubMed]
- Dubovskii, P.V.; Lesovoy, D.M.; Dubinnyi, M.A.; Konshina, A.G.; Utkin, Y.N.; Efremov, R.G.; Arseniev, A.S. Interaction of Three-Finger Toxins with Phospholipid Membranes: Comparison of S- and P-Type Cytotoxins. Biochem. J. 2005, 387, 807–815. [Google Scholar] [CrossRef] [PubMed]
- Girish, V.M.; Kumar, S.; Joseph, L.; Jobichen, C.; Kini, R.M.; Sivaraman, J. Identification and Structural Characterization of a New Three-Finger Toxin Hemachatoxin from Hemachatus Haemachatus Venom. PLoS ONE 2012, 7, e48112. [Google Scholar] [CrossRef] [PubMed]
- Konshina, A.G.; Dubovskii, P.V.; Efremov, R.G. Structure and Dynamics of Cardiotoxins. Curr. Protein Pept. Sci. 2012, 13, 570–584. [Google Scholar] [CrossRef]
- Dubovskii, P.V.; Konshina, A.G.; Efremov, R.G. Cobra Cardiotoxins: Membrane Interactions and Pharmacological Potential. Curr. Med. Chem. 2014, 21, 270–287. [Google Scholar] [CrossRef]
- Das, D.; Sharma, M.; Kumar Das, H.; Pratim Sahu, P.; Doley, R. Purification and Characterization of Nk-3FTx: A Three Finger Toxin from the Venom of North East Indian Monocled Cobra. J. Biochem. Mol. Toxicol. 2016, 30, 59–70. [Google Scholar] [CrossRef]
- Song, J.K.; Jo, M.R.; Park, M.H.; Song, H.S.; An, B.J.; Song, M.J.; Han, S.B.; Hong, J.T. Cell Growth Inhibition and Induction of Apoptosis by Snake Venom Toxin in Ovarian Cancer Cell via Inactivation of Nuclear Factor ΚB and Signal Transducer and Activator of Transcription 3. Arch. Pharm. Res. 2012, 35, 867–876. [Google Scholar] [CrossRef] [PubMed]
- Conlon, J.M.; Attoub, S.; Musale, V.; Leprince, J.; Casewell, N.R.; Sanz, L.; Calvete, J.J. Isolation and Characterization of Cytotoxic and Insulin-Releasing Components from the Venom of the Black-Necked Spitting Cobra Naja Nigricollis (Elapidae). Toxicon: X 2020, 6, 100030. [Google Scholar] [CrossRef] [PubMed]
- Vyas, V.K.; Brahmbhatt, K.; Bhatt, H.; Parmar, U. Therapeutic Potential of Snake Venom in Cancer Therapy: Current Perspectives. Asian Pac. J. Trop. Biomed. 2013, 3, 156–162. [Google Scholar] [CrossRef]
- Williams, H.F.; Hayter, P.; Ravishankar, D.; Baines, A.; Layfield, H.J.; Croucher, L.; Wark, C.; Bicknell, A.B.; Trim, S.; Vaiyapuri, S. Impact of Naja Nigricollis Venom on the Production of Methaemoglobin. Toxins 2018, 10, 539. [Google Scholar] [CrossRef] [PubMed]
- Fakhri, A.; Omranipour, R.; Fakhri, S.; Mirshamsi, M.; Zangeneh, F.; Vatanpour, H.; Pourahmadjaktaji, J. Naja Naja Oxiana Venom Fraction Selectively Induces ROS-Mediated Apoptosis in Human Colorectal Tumor Cells by Directly Targeting Mitochondria. Asian Pac. J. Cancer Prev. 2017, 18, 2201–2208. [Google Scholar] [CrossRef]
- Wang, C.H.; Wu, W. Amphiphilic β-Sheet Cobra Cardiotoxin Targets Mitochondria and Disrupts Its Network. FEBS Lett. 2005, 579, 3169–3174. [Google Scholar] [CrossRef]
- Czyżowska, A.; Dyba, B.; Rudolphi-Szydło, E.; Barbasz, A. Structural and Biochemical Modifications of Model and Native Membranes of Human Immune Cells in Response to the Action of Zinc Oxide Nanoparticles. J. Appl. Toxicol. 2020, 41, 458–469. [Google Scholar] [CrossRef]
- de Carvalho, A.C.; Girola, N.; de Figueiredo, C.R.; Machado, A.C.; de Medeiros, L.S.; Guadagnin, R.C.; Caseli, L.; Veiga, T.A.M. Understanding the Cytotoxic Effects of New Isovanillin Derivatives through Phospholipid Langmuir Monolayers. Bioorg. Chem. 2019, 83, 205–213. [Google Scholar] [CrossRef]
- Ładniak, A.; Jurak, M.; Wiącek, A.E. Langmuir Monolayer Study of Phospholipid DPPC on the Titanium Dioxide–Chitosan–Hyaluronic Acid Subphases. Adsorption 2019, 25, 469–476. [Google Scholar] [CrossRef]
- Das, D. Studies on the Crude Venom and Purified Three Finger Toxin of Naja Kaouthia from North East India. Doctoral Thesis, Department of Molecular Biology and Biotechnology School of Sciences, Tezpur University, Tezpur, India, 2015. [Google Scholar]
- Zbinden, G.; Flury-Roversi, M. Significance of the LD50-Test for the Toxicological Evaluation of Chemical Substances. Arch. Toxicol. 1981, 47, 77–99. [Google Scholar] [CrossRef]
- Condrea, E. Membrane-Active Polypeptides from Snake Venom: Cardiotoxins and Haemocytotoxins. Experientia 1974, 30, 121–129. [Google Scholar] [CrossRef]
- Peetla, C.; Stine, A.; Labhasetwar, V. Biophysical Interactions with Model Lipid Membranes: Applications in Drug Discovery and Drug Delivery. Mol. Pharm. 2009, 6, 1264–1276. [Google Scholar] [CrossRef]
- Brockman, H. Lipid Monolayers: Why Use Half a Membrane to Characterize Protein-Membrane Interactions? Curr. Opin. Struct. Biol. 1999, 9, 438–443. [Google Scholar] [CrossRef]
- Pastor, R.W. Molecular Dynamics and Monte Carlo Simulations of Lipid Bilayers. Curr. Opin. Struct. Biol. 1994, 4, 486–492. [Google Scholar] [CrossRef]
- White, S.H.; Wiener, M.C. The Liquid-Crystallographic Structure of Fluid Lipid Bilayer Membranes. In Biological Membranes: A Molecular Perspective from Computation and Experiment; Merz, K.M., Roux, B., Eds.; Birkhäuser: Boston, MA, USA, 1996; pp. 127–144. [Google Scholar]
- Konshina, A.G.; Boldyrev, I.A.; Omelkov, A.V.; Utkin, Y.; Efremov, R. Anionic Lipids: Determinants of Binding Cytotoxins from Snake Venom on the Surface of Cell Membranes. Acta Nat. 2010, 2, 88–96. [Google Scholar] [CrossRef][Green Version]
- Huang, W.N.; Sue, S.C.; Wang, D.S.; Wu, P.L.; Wu, W. Peripheral Binding Mode and Penetration Depth of Cobra Cardiotoxin on Phospholipid Membranes as Studied by a Combined FTIR and Computer Simulation Approach. Biochemistry 2003, 42, 7457–7466. [Google Scholar] [CrossRef]
- Levtsova, O.V.; Antonov, M.Y.; Mordvintsev, D.Y.; Utkin, Y.N.; Shaitan, K.V.; Kirpichnikov, M.P. Steered Molecular Dynamics Simulations of Cobra Cytotoxin Interaction with Zwitterionic Lipid Bilayer: No Penetration of Loop Tips into Membranes. Comput. Biol. Chem. 2009, 33, 29–32. [Google Scholar] [CrossRef] [PubMed]
- Dutta, S.; Sinha, A.; Dasgupta, S.; Mukherjee, A.K. Binding of a Naja Naja Venom Acidic Phospholipase A2 Cognate Complex to Membrane-Bound Vimentin of Rat L6 Cells: Implications in Cobra Venom-Induced Cytotoxicity. Biochim. Biophys. Acta (BBA) Biomembr. 2019, 1861, 958–977. [Google Scholar] [CrossRef] [PubMed]
- Dutta, S.; Chanda, A.; Kalita, B.; Islam, T.; Patra, A.; Mukherjee, A.K. Proteomic Analysis to Unravel the Complex Venom Proteome of Eastern India Naja Naja: Correlation of Venom Composition with Its Biochemical and Pharmacological Properties. J. Proteom. 2017, 156, 29–39. [Google Scholar] [CrossRef] [PubMed]
- Ownby, C.L.; Fletcher, J.E.; Colberg, T.R. Cardiotoxin 1 from Cobra (Naja Naja Atra) Venom Causes Necrosis of Skeletal Muscle in Vivo. Toxicon 1993, 31, 697–709. [Google Scholar] [CrossRef]
- Su, Z.Y.; Wang, Y.T. Coarse-Grained Molecular Dynamics Simulations of Cobra Cytotoxin A3 Interactions with a Lipid Bilayer: Penetration of Loops into Membranes. J. Phys. Chem. B. 2011, 115, 796–802. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.C.; Lin, C.C.; Wang, C.H.; Wu, P.L.; Huang, H.W.; Chang, C.I.; Wu, W. Endocytotic Routes of Cobra Cardiotoxins Depend on Spatial Distribution of Positively Charged and Hydrophobic Domains to Target Distinct Types of Sulfated Glycoconjugates on Cell Surface. J. Biol. Chem. 2014, 289, 20170–20181. [Google Scholar] [CrossRef] [PubMed]
- Hanzal-Bayer, M.F.; Hancock, J.F. Lipid Rafts and Membrane Traffic. FEBS Lett. 2007, 581, 2098–2104. [Google Scholar] [CrossRef]
- Wang, C.-H.; Liu, J.-H.; Lee, S.-C.; Hsiao, C.-D.; Wu, W. Glycosphingolipid-Facilitated Membrane Insertion and Internalization of Cobra Cardiotoxin: The sulfatide·cardiotoxin complex structure in a membrane-like environment suggests a lipid-dependent cell-penetrating mechanism for membrane binding polypeptides*. J. Biol. Chem. 2006, 281, 656–667. [Google Scholar] [CrossRef] [PubMed]
- Kuo, P.; Weinfeld, M.; Rudd, M.A.; Amarante, P.; Loscalzo, J. Plasma Membrane Enrichment with Cis-Unsaturated Fatty Acids Enhances LDL Metabolism in U937 Monocytes. Arteriosclerosis 1990, 10, 111–118. [Google Scholar] [CrossRef]
- Ip, S.; Cooper, R. Decreased Membrane Fluidity during Differentiation of Human Promyelocytic Leukemia Cells in Culture. Blood 1980, 56, 227–232. [Google Scholar] [CrossRef]
- Bilwes, A.; Rees, B.; Moras, D.; Ménez, R.; Ménez, A. X-Ray Structure at 1.55 A of Toxin Gamma, a Cardiotoxin from Naja Nigricollis Venom. Crystal Packing Reveals a Model for Insertion into Membranes. J. Mol. Biol. 1994, 239, 122–136. [Google Scholar] [CrossRef] [PubMed]
- Wu, W.G.; Tjong, S.C.; Wu, P.L.; Kuo, J.H.; Wu, K. Role of Heparan Sulfates and Glycosphingolipids in the Pore Formation of Basic Polypeptides of Cobra Cardiotoxin. Adv. Exp. Med. Biol. 2010, 677, 143–149. [Google Scholar] [CrossRef] [PubMed]
- Feofanov, A.V.; Sharonov, G.V.; Dubinnyi, M.A.; Astapova, M.V.; Kudelina, I.A.; Dubovskii, P.V.; Rodionov, D.I.; Utkin, Y.N.; Arseniev, A.S. Comparative Study of Structure and Activity of Cytotoxins from Venom of the Cobras Naja Oxiana, NajaKaouthia, and NajaHaje. Biochemistry (Mosc) 2004, 69, 1148–1157. [Google Scholar] [CrossRef] [PubMed]
- Gasanov, S.E.; Dagda, R.K.; Rael, E.D. Snake Venom Cytotoxins, Phospholipase A2s, and Zn2+-Dependent Metalloproteinases: Mechanisms of Action and Pharmacological Relevance. J. Clin. Toxicol. 2014, 4, 1000181. [Google Scholar] [CrossRef] [PubMed]
- Feofanov, A.V.; Sharonov, G.V.; Astapova, M.V.; Rodionov, D.I.; Utkin, Y.N.; Arseniev, A.S. Cancer Cell Injury by Cytotoxins from Cobra Venom Is Mediated through Lysosomal Damage. Biochem. J. 2005, 390, 11–18. [Google Scholar] [CrossRef]
- Chiou, S.H.; Raynor, R.L.; Zheng, B.; Chambers, T.C.; Kuo, J.F. Cobra Venom Cardiotoxin (Cytotoxin) Isoforms and Neurotoxin: Comparative Potency of Protein Kinase C Inhibition and Cancer Cell Cytotoxicity and Modes of Enzyme Inhibition. Biochemistry 1993, 32, 2062–2067. [Google Scholar] [CrossRef] [PubMed]
- Chiou, J.-T.; Shi, Y.-J.; Wang, L.-J.; Huang, C.-H.; Lee, Y.-C.; Chang, L.-S. Naja Atra Cardiotoxin 3 Elicits Autophagy and Apoptosis in U937 Human Leukemia Cells through the Ca2+/PP2A/AMPK Axis. Toxins (Basel) 2019, 11, 527. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Li, F.; Chen, Z.; Shrivastava, I.H.; Gasanoff, E.S.; Dagda, R.K. Naja Mossambica Mossambica Cobra Cardiotoxin Targets Mitochondria to Disrupt Mitochondrial Membrane Structure and Function. Toxins 2019, 11, 152. [Google Scholar] [CrossRef]
- Tambourgi, D.V.; dos Santos, M.C.; Furtado, M.D.F.; de Freitas, M.C.; da Silva, W.D.; Kipnis, T.L. Pro-Inflammatory Activities in Elapid Snake Venoms. Br. J. Pharmacol. 1994, 112, 723–727. [Google Scholar] [CrossRef] [PubMed]
- Deka, A.; Sharma, M.; Mukhopadhyay, R.; Devi, A.; Doley, R. Naja Kaouthia Venom Protein, Nk-CRISP, Upregulates Inflammatory Gene Expression in Human Macrophages. Int. J. Biol. Macromol. 2020, 160, 602–611. [Google Scholar] [CrossRef] [PubMed]
- Okumu, M.O.; Mbaria, J.M.; Gikunju, J.K.; Mbuthia, P.G.; Madadi, V.O.; Ochola, F.O. Enzymatic Activity and Brine Shrimp Lethality of Venom from the Large Brown Spitting Cobra (Naja Ashei) and Its Neutralization by Antivenom. BMC Res. Notes 2020, 13, 325. [Google Scholar] [CrossRef] [PubMed]
- Clarkson, C.; Maharaj, V.J.; Crouch, N.R.; Grace, O.M.; Pillay, P.; Matsabisa, M.G.; Bhagwandin, N.; Smith, P.J.; Folb, P.I. In Vitro Antiplasmodial Activity of Medicinal Plants Native to or Naturalised in South Africa. J. Ethnopharmacol. 2004, 92, 177–191. [Google Scholar] [CrossRef]
- Meyer, B.N.; Ferrigni, N.R.; Putnam, J.E.; Jacobsen, L.B.; Nichols, D.E.; McLaughlin, J.L. Brine Shrimp: A Convenient General Bioassay for Active Plant Constituents. Planta Med. 1982, 45, 31–34. [Google Scholar] [CrossRef] [PubMed]
- Berkovic, D.; Fleer, E.; Breass, J.; Pförtner, J.; Schleyer, E.; Hiddemann, W. The Influence of 1-β-D -Arabinofuranosylcytosine on the Metabolism of Phosphatidylcholine in Human Leukemic HL 60 and Raji Cells. Leukemia 1997, 11, 2079–2086. [Google Scholar] [CrossRef] [PubMed]
- Chabot, M.C.; Wykle, R.L.; Modest, E.J.; Daniel, L.W. Correlation of Ether Lipid Content of Human Leukemia Cell Lines and Their Susceptibility to 1-O-Octadecyl-2-O-Methyl-Rac-Glycero-3-Phosphocholine. Cancer Res. 1989, 49, 4441–4445. [Google Scholar] [PubMed]
- Ecker, J.; Liebisch, G.; Englmaier, M.; Grandl, M.; Schmitz, G. SREBP-1c Mediated Induction of Lipid Metabolism Is Crucial for Phagocytic Differentiation of Monocytes. Chem. Phys. Lipids 2009, 160, S20. [Google Scholar] [CrossRef]
- Lyberg, T.; Ivhed, I.; Prydz, H.; Nilsson, K. Thromboplastin as a Marker for Monocyte Differentiation. Br. J. Haematol. 1983, 53, 327–335. [Google Scholar] [CrossRef] [PubMed]
- Barbasz, A.; Kreczmer, B.; Dyba, B.; Filek, M.; Rudolphi-Szydło, E. The Direct Action of Hyaluronic Acid on Human U-937 and HL-60 Cells–Modification of Native and Model Membranes. Biologia 2016, 71, 1304–1314. [Google Scholar] [CrossRef]
- Rudolphi-Skórska, E.; Zembala, M.; Filek, M. Mechanical and Electrokinetic Effects of Polyamines/Phospholipid Interactions in Model Membranes. J. Membr. Biol. 2014, 247, 81–92. [Google Scholar] [CrossRef] [PubMed]
- Rudolphi-Szydło, E.; Filek, M.; Zembala, M. Physicochemical Aspects of Reaction of Ozone with Galactolipid and Galactolipid-Tocopherol Layers. J. Membr. Biol. 2014, 247, 639–649. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Barnes, G.; Gentle, I. Interfacial Science: An Introduction, 2nd ed.; Oxford University Press: New York, NY, USA, 2011. [Google Scholar]
- Costin, I.S.; Barnes, G.T. Two-Component Monolayers. II. Surface Pressure—Area Relations for the Octadecanol—Docosyl Sulphate System. J. Colloid Interface Sci. 1975, 51, 106–121. [Google Scholar] [CrossRef]
- Flasiński, M.; Hąc-Wydro, K.; Wydro, P.; Dynarowicz-Łątka, P. Influence of Platelet-Activating Factor, Lyso-Platelet-Activating Factor and Edelfosine on Langmuir Monolayers Imitating Plasma Membranes of Cell Lines Differing in Susceptibility to Anti-Cancer Treatment: The Effect of Plasmalogen Level. J. R. Soc. Interface 2014, 11, 20131103. [Google Scholar] [CrossRef] [PubMed]


| Experiment | Alim (Å2) | πcol (mN/m) | Cs−1max (mN/m) | |||
|---|---|---|---|---|---|---|
| U-937 | HL-60 | U-937 | HL-60 | U-937 | HL-60 | |
| Control | 54.2 ± 0.1 E,* | 52.9 ± 0.1 d | 44.0 ± 0.9 A | 45.9 ± 0.5 a | 149.4 ± 0.1 A | 170.6 ± 0.1 a,* |
| 5 ng/mL | 57.3 ± 0.2 D,* | 56.4 ± 0.1 c | 44.7 ± 0.8 A | 42.5 ± 0.8 b | 130.4 ± 0.2 B | 133.4 ± 0.1 b,* |
| 10 ng/mL | 58.6 ± 0.2 C | 58.6 ± 0.3 b | 42.3 ± 0.8 B | 42.3 ± 0.6 b | 112.4 ± 0.3 C | 113.0 ± 0.2 c,* |
| 20 ng/mL | 59.1 ± 0.1 B,* | 58.8 ± 0.1 b | 41.9 ± 0.6 B | 42.2 ± 0.5 b | 94.3 ± 0.3 D | 93.9 ± 0.2 d |
| 40 ng/mL | 60.2 ± 0.1 A | 61.1 ± 0.1 a,* | 44.5 ± 0.8 A | 42.4 ± 0.6 b | 89.9 ± 0.3 E | 89.6 ± 0.3 e |
| Cell line | LD50 [mg/L/1 × 106 cells] |
|---|---|
| U-937 | 126.80 ± 2.94 * |
| HL-60 | 121.29 ± 1.42 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dyba, B.; Rudolphi-Szydło, E.; Barbasz, A.; Czyżowska, A.; Hus, K.K.; Petrilla, V.; Petrillová, M.; Legáth, J.; Bocian, A. Effects of 3FTx Protein Fraction from Naja ashei Venom on the Model and Native Membranes: Recognition and Implications for the Mechanisms of Toxicity. Molecules 2021, 26, 2164. https://doi.org/10.3390/molecules26082164
Dyba B, Rudolphi-Szydło E, Barbasz A, Czyżowska A, Hus KK, Petrilla V, Petrillová M, Legáth J, Bocian A. Effects of 3FTx Protein Fraction from Naja ashei Venom on the Model and Native Membranes: Recognition and Implications for the Mechanisms of Toxicity. Molecules. 2021; 26(8):2164. https://doi.org/10.3390/molecules26082164
Chicago/Turabian StyleDyba, Barbara, Elżbieta Rudolphi-Szydło, Anna Barbasz, Agnieszka Czyżowska, Konrad Kamil Hus, Vladimír Petrilla, Monika Petrillová, Jaroslav Legáth, and Aleksandra Bocian. 2021. "Effects of 3FTx Protein Fraction from Naja ashei Venom on the Model and Native Membranes: Recognition and Implications for the Mechanisms of Toxicity" Molecules 26, no. 8: 2164. https://doi.org/10.3390/molecules26082164
APA StyleDyba, B., Rudolphi-Szydło, E., Barbasz, A., Czyżowska, A., Hus, K. K., Petrilla, V., Petrillová, M., Legáth, J., & Bocian, A. (2021). Effects of 3FTx Protein Fraction from Naja ashei Venom on the Model and Native Membranes: Recognition and Implications for the Mechanisms of Toxicity. Molecules, 26(8), 2164. https://doi.org/10.3390/molecules26082164








