Downfalls of Chemical Probes Acting at the Kinase ATP-Site: CK2 as a Case Study
Abstract
1. Introduction
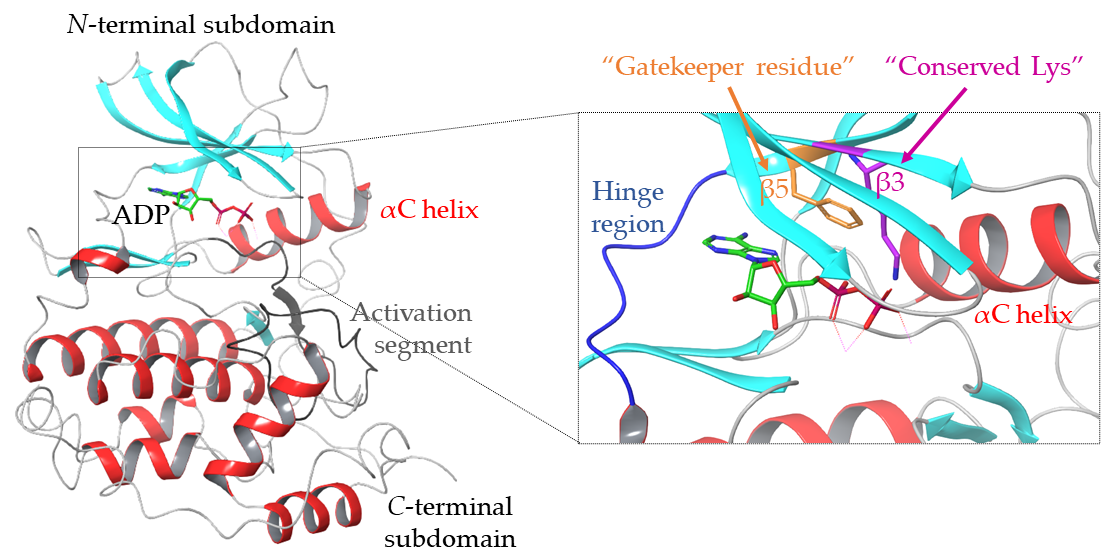
1.1. Structure of the ATP Binding Site of Protein Kinases
1.2. Problems with ATP-Competitive Inhibitors
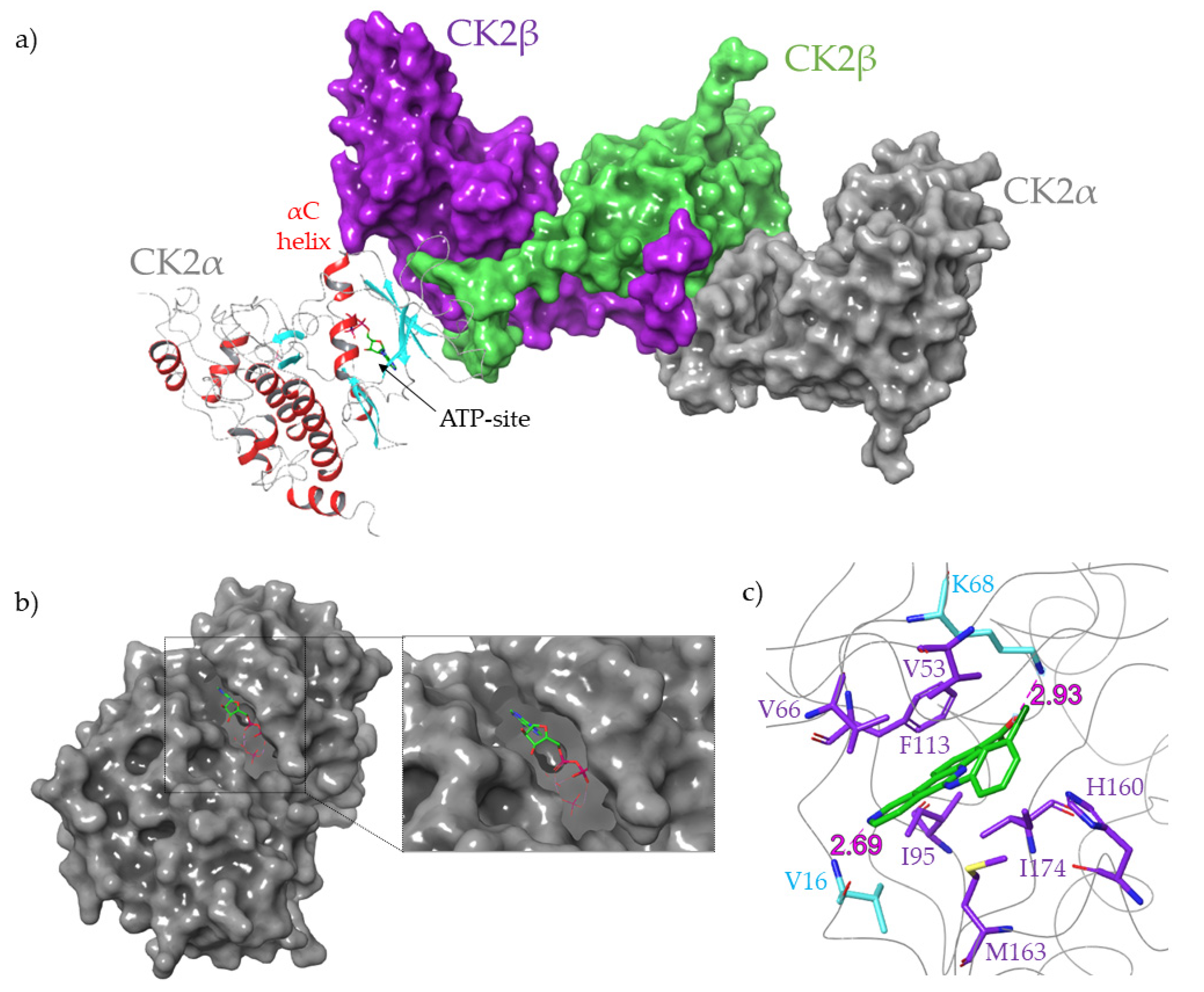
1.3. Protein Kinase CK2
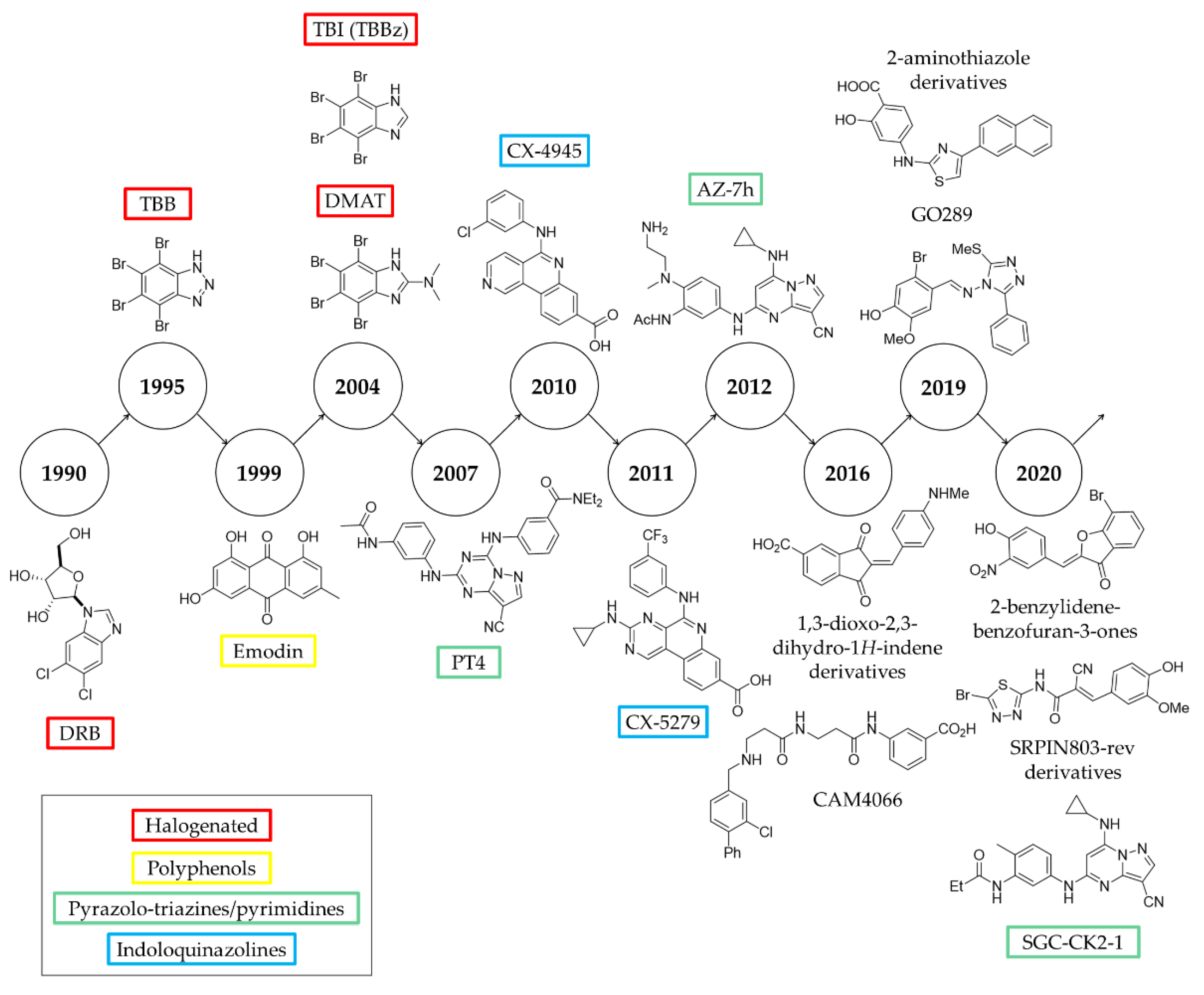
2. History of ATP-Competitive Inhibitors of CK2
- Indoloquinazolines such as CX-4945 [65]
2.1. DRB
2.2. TBB
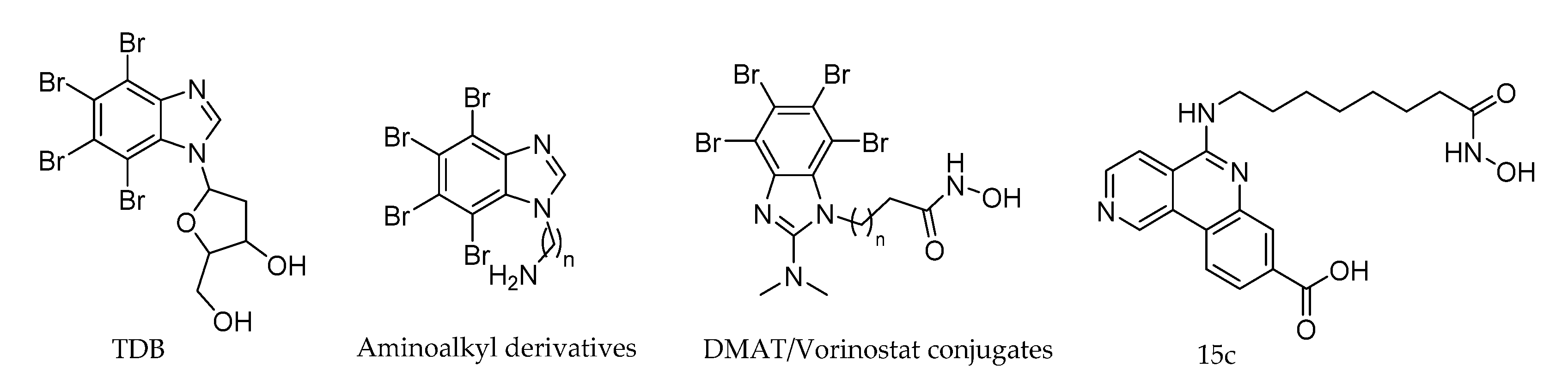
2.3. TBI (TBBz) and DMAT
2.4. Polyphenols
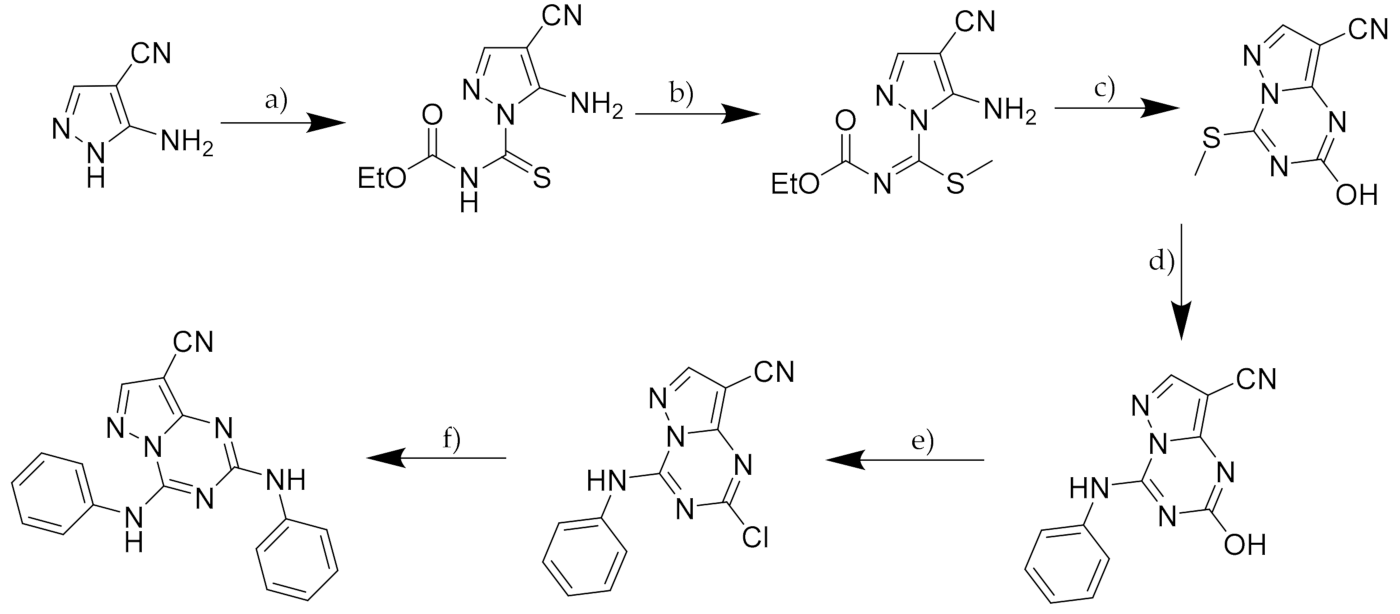
2.5. Pyrazolo-Triazines and Pyrazolo-Pyrimidines
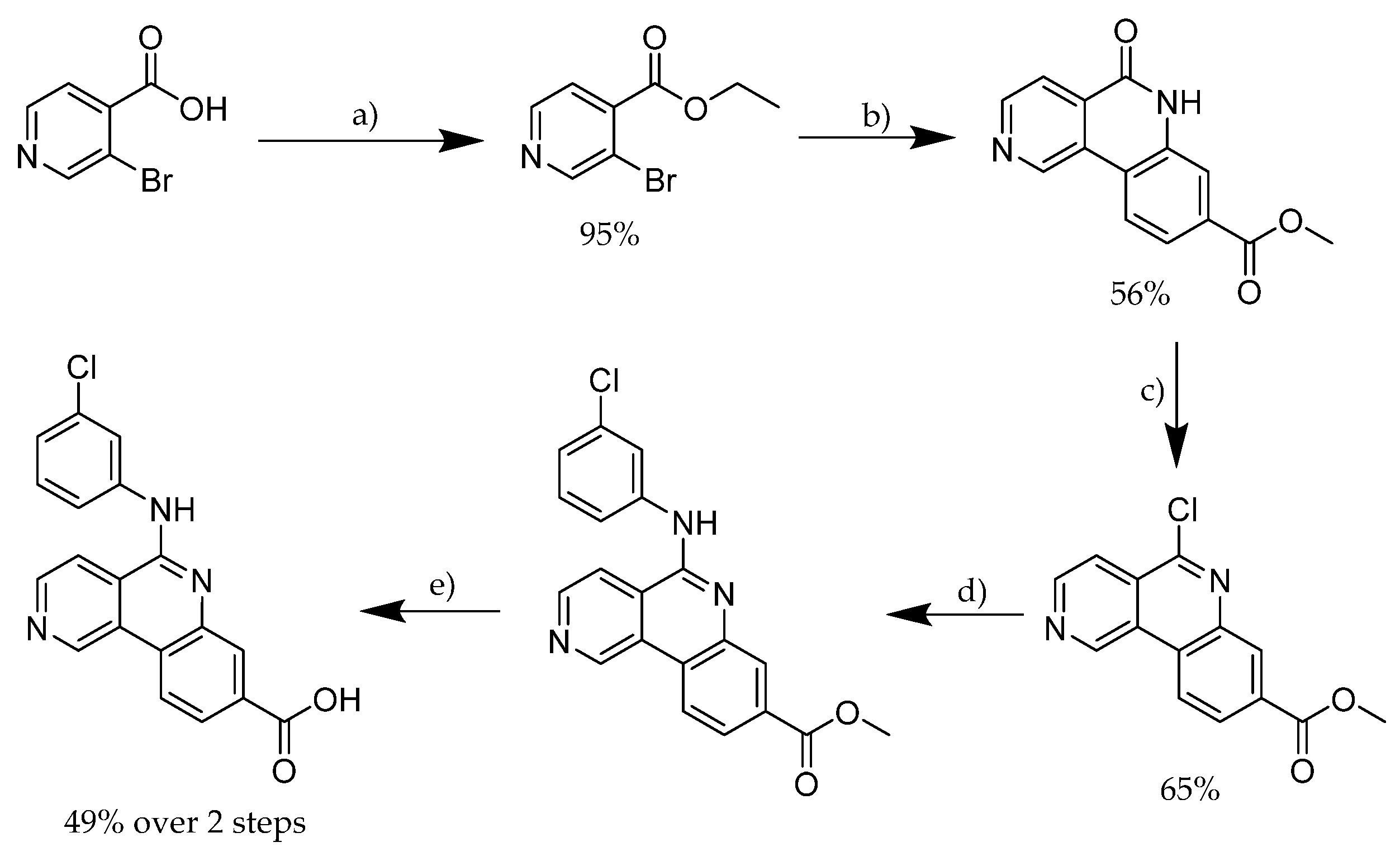
2.6. CX-4945
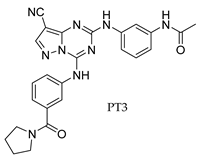
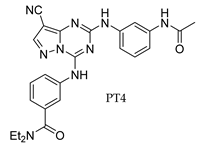
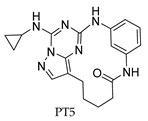
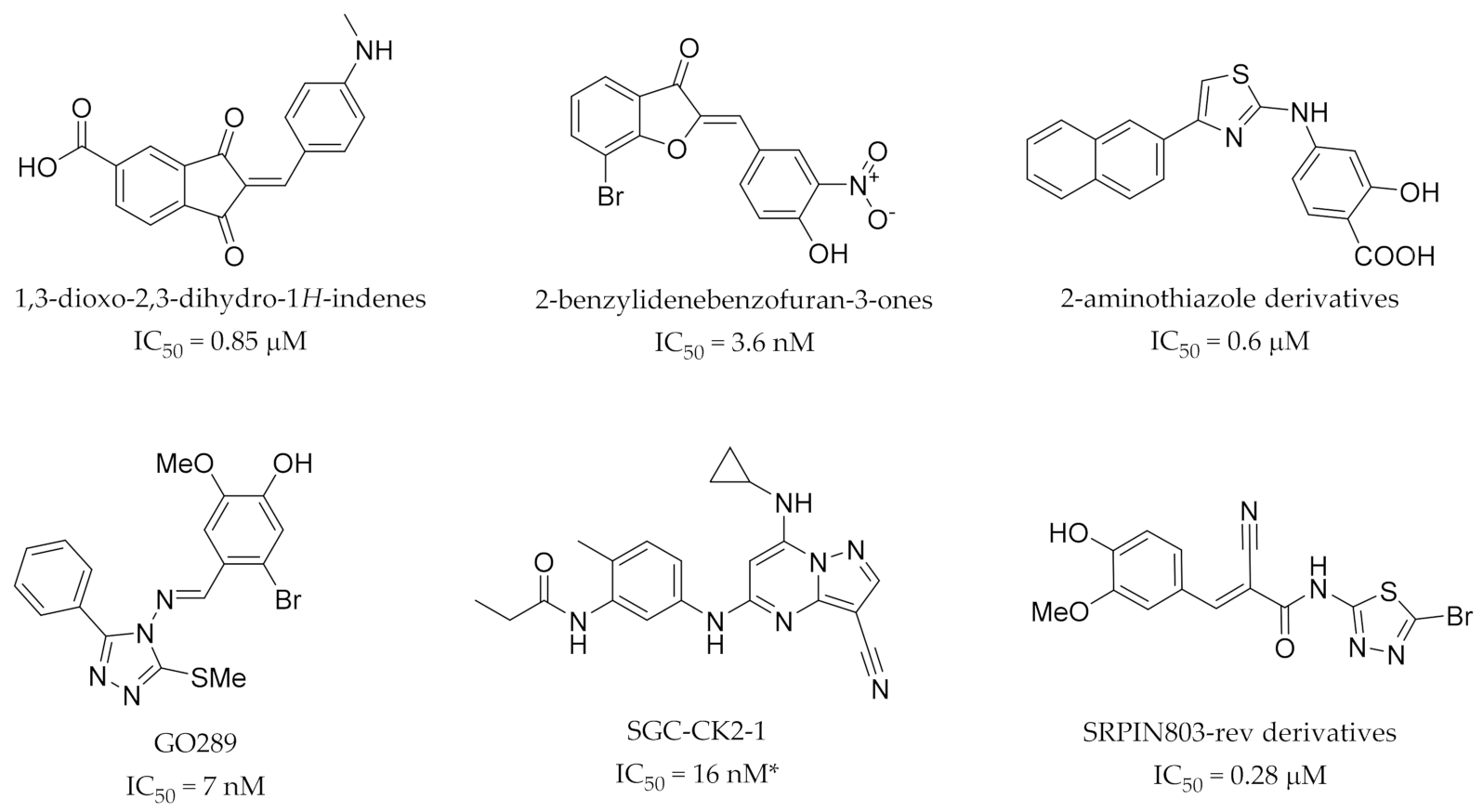
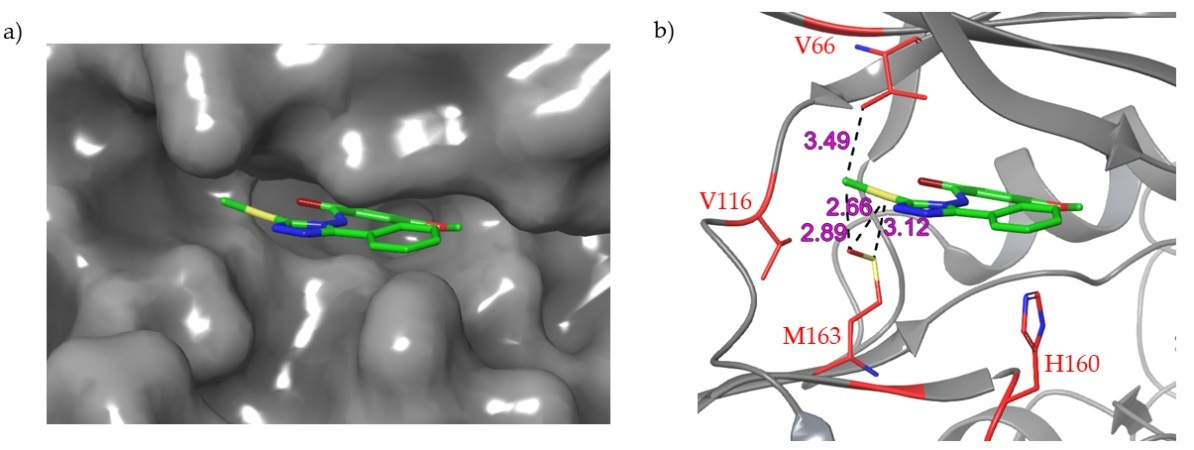
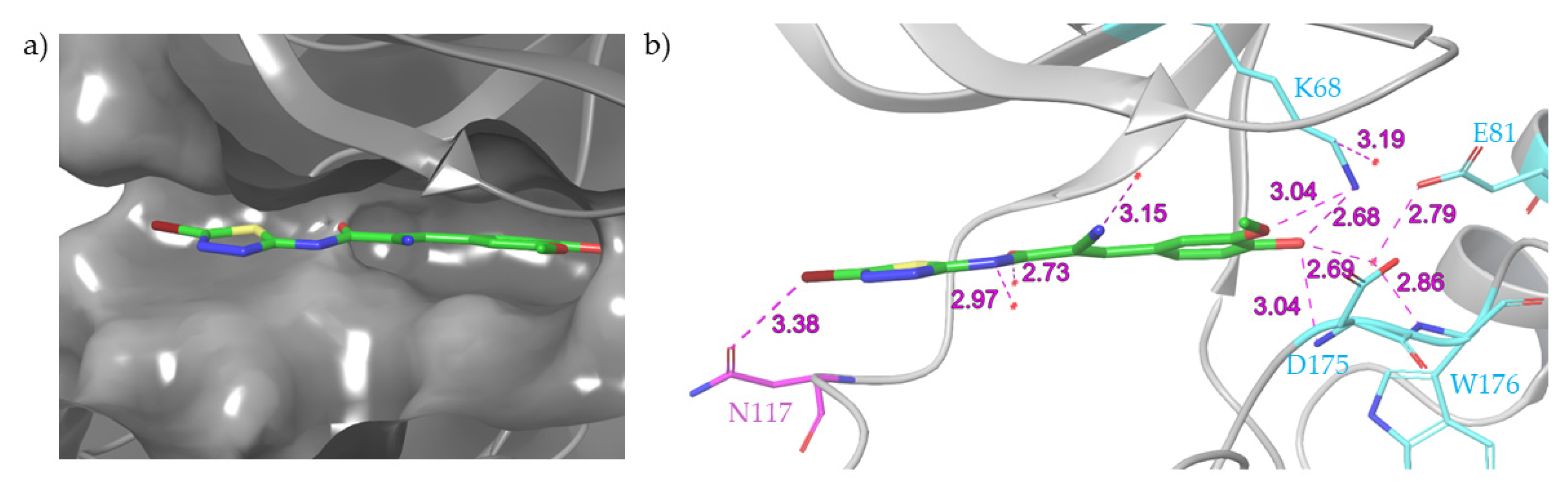
3. Recent Developments
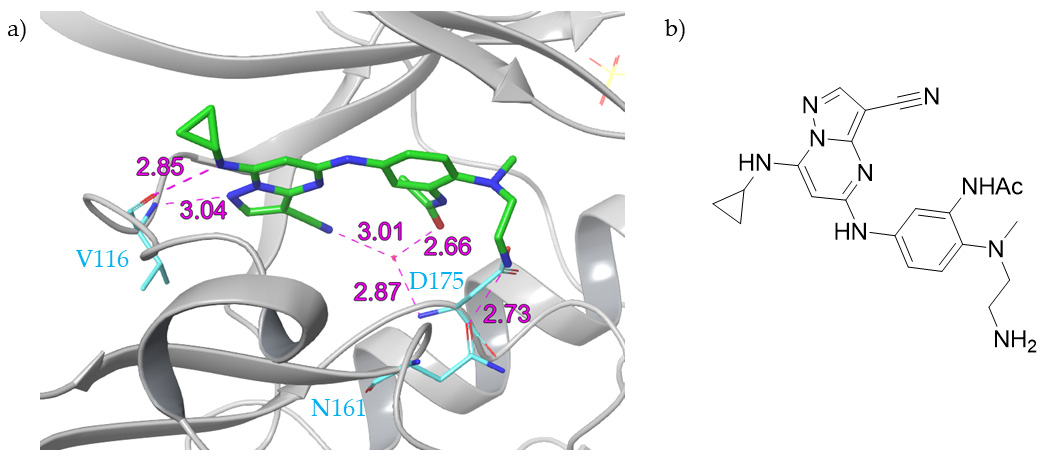
4. Dual Inhibitors
5. Bi-Substrate Inhibitors
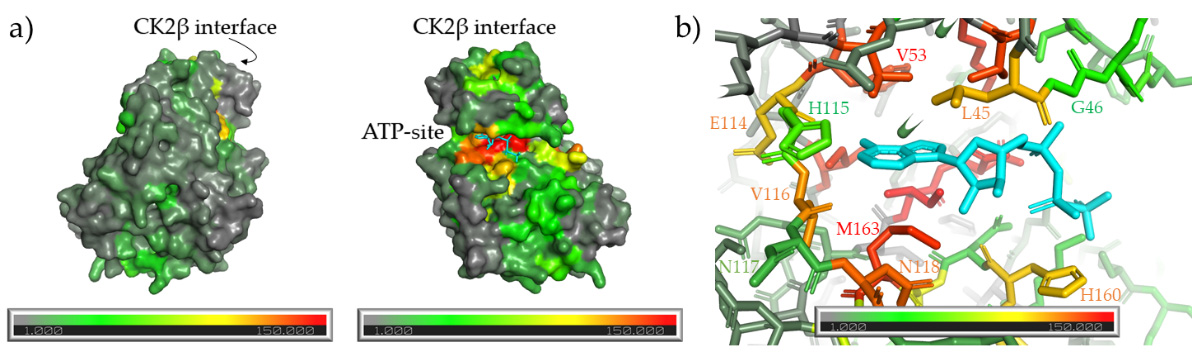
6. Inhibitors Extending Outside of the ATP-Site
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Kannaiyan, R.; Mahadevan, D. A comprehensive review of protein kinase inhibitors for cancer therapy. Expert Rev. Anticancer Ther. 2018, 18, 1249–1270. [Google Scholar] [CrossRef] [PubMed]
- Cohen, P. The origins of protein phosphorylation. Nat. Cell Biol. 2002, 4, E127–E130. [Google Scholar] [CrossRef] [PubMed]
- Endicott, J.A.; Noble, M.E.M.; Johnson, L.N. The Structural Basis for Control of Eukaryotic Protein Kinases. Annu. Rev. Biochem. 2012, 81, 587–613. [Google Scholar] [CrossRef] [PubMed]
- Johnson, L.N. Protein kinase inhibitors: Contributions from structure to clinical compounds. Q. Rev. Biophys. 2009, 42, 1–40. [Google Scholar] [CrossRef]
- Zhang, J.; Yang, P.L.; Gray, N.S. Targeting cancer with small molecule kinase inhibitors. Nat. Rev. Cancer 2009, 9, 28–39. [Google Scholar] [CrossRef]
- Carles, F.; Bourg, S.; Meyer, C.; Bonnet, P. PKIDB: A curated, annotated and updated database of protein kinase inhibitors in clinical trials. Molecules 2018, 23, 908. [Google Scholar] [CrossRef]
- Bournez, C.; Carles, F.; Peyrat, G.; Aci-Sèche, S.; Bourg, S.; Meyer, C.; Bonnet, P. Comparative assessment of protein kinase inhibitors in public databases and in PKIDB. Molecules 2020, 25, 3226. [Google Scholar] [CrossRef]
- Yang, C.-Y.; Chang, C.-H.; Yu, Y.-L.; Lin, T.-C.E.; Lee, S.-A.; Yen, C.-C.; Yang, J.-M.; Lai, J.-M.; Hong, Y.-R.; Tseng, T.-L.; et al. PhosphoPOINT: A comprehensive human kinase interactome and phospho-protein database. Bioinformatics 2008, 24, i14–i20. [Google Scholar] [CrossRef]
- Johnson, L.N.; Lewis, R.J. Structural basis for control by phosphorylation. Chem. Rev. 2001, 101, 2209–2242. [Google Scholar] [CrossRef]
- Lander, E.S.; Linton, L.M.; Birren, B.; Nusbaum, C.; Zody, M.C.; Baldwin, J.; Devon, K.; Dewar, K.; Doyle, M.; FitzHugh, W.; et al. Initial sequencing and analysis of the human genome. Nature 2001, 409, 860–921. [Google Scholar] [CrossRef]
- Manning, G.; Whyte, D.B.; Martinez, R.; Hunter, T.; Sudarsanam, S. The protein kinase complement of the human genome. Science 2002, 298, 1912–1934. [Google Scholar] [CrossRef]
- Subramani, S.; Jayapalan, S.; Kalpana, R.; Natarajan, J. HomoKinase: A Curated Database of Human Protein Kinases. ISRN Comput. Biol. 2013, 2013, 1–5. [Google Scholar] [CrossRef]
- Schwartz, P.A.; Murray, B.W. Protein kinase biochemistry and drug discovery. Bioorg. Chem. 2011, 39, 192–210. [Google Scholar] [CrossRef]
- Liao, J.J. Lou Molecular recognition of protein kinase binding pockets for design of potent and selective kinase inhibitors. J. Med. Chem. 2007, 50, 409–424. [Google Scholar] [CrossRef]
- Huse, M.; Kuriyan, J. The conformational plasticity of protein kinases. Cell 2002, 109, 275–282. [Google Scholar] [CrossRef]
- Bossemeyer, D. Protein kinases—Structure and function. FEBS Lett. 1995, 369, 57–61. [Google Scholar] [CrossRef]
- Kornev, A.P.; Taylor, S.S. Defining the conserved internal architecture of a protein kinase. Biochim. Biophys. Acta Proteins Proteom. 2010, 1804, 440–444. [Google Scholar] [CrossRef]
- Karaman, M.W.; Herrgard, S.; Treiber, D.K.; Gallant, P.; Atteridge, C.E.; Campbell, B.T.; Chan, K.W.; Ciceri, P.; Davis, M.I.; Edeen, P.T.; et al. A quantitative analysis of kinase inhibitor selectivity. Nat. Biotechnol. 2008, 26, 127–132. [Google Scholar] [CrossRef]
- Huang, D.; Zhou, T.; Lafleur, K.; Nevado, C.; Caflisch, A. Kinase selectivity potential for inhibitors targeting the ATP binding site: A network analysis. Bioinformatics 2010, 26, 198–204. [Google Scholar] [CrossRef]
- Urich, R.; Wishart, G.; Kiczun, M.; Richters, A.; Tidten-Luksch, N.; Rauh, D.; Sherborne, B.; Wyatt, P.G.; Brenk, R. De Novo design of protein kinase inhibitors by in silico identification of hinge region-binding fragments. ACS Chem. Biol. 2013, 8, 1044–1052. [Google Scholar] [CrossRef]
- Tamaoki, T.; Nomoto, H.; Takahashi, I.; Kato, Y.; Morimoto, M.; Tomita, F. Staurosporine, a potent inhibitor of phospholipid Ca++dependent protein kinase. Biochem. Biophys. Res. Commun. 1986, 135, 397–402. [Google Scholar] [CrossRef]
- Davies, S.P.; Reddy, H.; Caivano, M.; Cohen, P. Specificity and mechanism of action of some commonly used protein kinase inhibitors. Biochem. J. 2000, 351, 95–105. [Google Scholar] [CrossRef] [PubMed]
- Bishop, A.C.; Ubersax, J.A.; Pøtsch, D.T.; Matheos, D.P.; Gray, N.S.; Blethrow, J.; Shimizu, E.; Tsien, J.Z.; Schultz, P.G.; Rose, M.D.; et al. A chemical switch for inhibitor-sensitive alleles of any protein kinase. Nature 2000, 407, 395–401. [Google Scholar] [CrossRef] [PubMed]
- Garske, A.L.; Peters, U.; Cortesi, A.T.; Perez, J.L.; Shokat, K.M. Chemical genetic strategy for targeting protein kinases based on covalent complementarity. Proc. Natl. Acad. Sci. USA 2011, 108, 15046–15052. [Google Scholar] [CrossRef]
- Roskoski, R. Properties of FDA-approved small molecule protein kinase inhibitors: A 2020 update. Pharmacol. Res. 2020, 152, 104609. [Google Scholar] [CrossRef]
- Blair, H.A. Fedratinib: First Approval. Drugs 2019, 79, 1719–1725. [Google Scholar] [CrossRef]
- Lamb, Y.N. Pexidartinib: First Approval. Drugs 2019, 79, 1805–1812. [Google Scholar] [CrossRef]
- Montazeri, K.; Bellmunt, J. Erdafitinib for the treatment of metastatic bladder cancer. Expert Rev. Clin. Pharmacol. 2020, 13, 1–6. [Google Scholar] [CrossRef]
- Entrectinib OK’d for Cancers with NTRK Fusions, NSCLC. Cancer Discov. 2019, 9, OF2. [CrossRef]
- Loriot, Y.; Necchi, A.; Park, S.H.; Garcia-Donas, J.; Huddart, R.; Burgess, E.; Fleming, M.; Rezazadeh, A.; Mellado, B.; Varlamov, S.; et al. Erdafitinib in Locally Advanced or Metastatic Urothelial Carcinoma. N. Engl. J. Med. 2019, 381, 338–348. [Google Scholar] [CrossRef]
- Menichincheri, M.; Ardini, E.; Magnaghi, P.; Avanzi, N.; Banfi, P.; Bossi, R.; Buffa, L.; Canevari, G.; Ceriani, L.; Colombo, M.; et al. Discovery of Entrectinib: A New 3-Aminoindazole As a Potent Anaplastic Lymphoma Kinase (ALK), c-ros Oncogene 1 Kinase (ROS1), and Pan-Tropomyosin Receptor Kinases (Pan-TRKs) inhibitor. J. Med. Chem. 2016, 59, 3392–3408. [Google Scholar] [CrossRef]
- Kobe, B.; Kampmann, T.; Forwood, J.K.; Listwan, P.; Brinkworth, R.I. Substrate specificity of protein kinases and computational prediction of substrates. Biochim. Biophys. Acta Proteins Proteom. 2005, 1754, 200–209. [Google Scholar] [CrossRef]
- Hanks, S.K.; Hunter, T. The eukaryotic protein kinase superfamily: Kinase (catalytic) domain structure and classification 1. FASEB J. 1995, 9, 576–596. [Google Scholar] [CrossRef]
- Niefind, K.; Yde, C.W.; Ermakova, I.; Issinger, O.-G. Evolved to Be Active: Sulfate Ions Define Substrate Recognition Sites of CK2α and Emphasise its Exceptional Role within the CMGC Family of Eukaryotic Protein Kinases. J. Mol. Biol. 2007, 370, 427–438. [Google Scholar] [CrossRef]
- Cozza, G.; Bortolato, A.; Moro, S. How Druggable Is Protein Kinase CK2? Med. Res. Rev. 2010, 30, 419–462. [Google Scholar] [CrossRef]
- Filhol, O.; Cochet, C.; Niefind, K.; Raaf, J.; Issinger, O.-G.; Filhol, O.; Cochet, C.; Trembley, J.H.; Wang, G.; Unger, G.; et al. Protein Kinase CK2 in Health and Disease. Cell. Mol. Life Sci. 2009, 66, 1830–1839. [Google Scholar] [CrossRef]
- Ahmad, K.A.; Wang, G.; Unger, G.; Slaton, J.; Ahmed, K. Protein kinase CK2—A key suppressor of apoptosis. Adv. Enzyme Regul. 2008, 48, 179–187. [Google Scholar] [CrossRef]
- Sarno, S.; Ghisellini, P.; Pinna, L.A. Unique Activation Mechanism of Protein Kinase CK2. J. Biol. Chem. 2002, 277, 22509–22514. [Google Scholar] [CrossRef]
- Daya-Makin, M.; Sanghera, J.S.; Mogentale, T.L.; Lipp, M.; Parchomchuk, J.; Hogg, J.C.; Pelech, S.L. Activation of a Tumor-associated Protein Kinase (p40TAK) and Casein Kinase 2 in Human Squamous Cell Carcinomas and Adenocarcinomas of the Lung. Cancer Res. 1994, 54, 2262–2268. [Google Scholar]
- Pistorius, K.; Seitz, G.; Remberger, K.; Issinger, O.G. Differential CKII Activities in Human Colorectal Mucosa, Adenomas and Carcinomas. Oncol. Res. Treat. 1991, 14, 256–260. [Google Scholar] [CrossRef]
- Jin, S.K.; Ju, I.E.; Cheong, J.W.; Ae, J.C.; Jin, K.L.; Woo, I.Y.; Yoo, H.M. Protein kinase CK2α as an unfavorable prognostic marker and novel therapeutic target in acute myeloid leukemia. Clin. Cancer Res. 2007, 13, 1019–1028. [Google Scholar] [CrossRef]
- Trembley, J.H.; Wang, G.; Unger, G.; Slaton, J.; Ahmed, K. CK2: A key player in cancer biology. Cell. Mol. Life Sci. 2009, 66, 1858–1867. [Google Scholar] [CrossRef]
- Brear, P.; North, A.; Iegre, J.; Hadje Georgiou, K.; Lubin, A.; Carro, L.; Green, W.; Sore, H.F.; Hyvönen, M.; Spring, D.R. Novel non-ATP competitive small molecules targeting the CK2 α/β interface. Bioorg. Med. Chem. 2018, 26, 3016–3020. [Google Scholar] [CrossRef]
- Ahmad, K.A.; Wang, G.; Ahmed, K. Molecular Cancer Research. Mol. Cancer Res. 2006, 2, 712–721. [Google Scholar] [CrossRef]
- Marschke, R.F.; Borad, M.J.; McFarland, R.W.; Alvarez, R.H.; Lim, J.K.; Padgett, C.S.; Von Hoff, D.D.; O’Brien, S.E.; Northfelt, D.W. Findings from the phase I clinical trials of CX-4945, an orally available inhibitor of CK2. J. Clin. Oncol. 2011, 29, 3087. [Google Scholar] [CrossRef]
- Bouhaddou, M.; Memon, D.; Meyer, B.; White, K.M.; Rezelj, V.V.; Correa Marrero, M.; Polacco, B.J.; Melnyk, J.E.; Ulferts, S.; Kaake, R.M.; et al. The Global Phosphorylation Landscape of SARS-CoV-2 Infection. Cell 2020, 182, 685–712.e19. [Google Scholar] [CrossRef]
- Gordon, D.E.; Jang, G.M.; Bouhaddou, M.; Xu, J.; Obernier, K.; White, K.M.; O’Meara, M.J.; Rezelj, V.V.; Guo, J.Z.; Swaney, D.L.; et al. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature 2020, 583, 459–468. [Google Scholar] [CrossRef]
- Seetoh, W.G.; Abell, C. Disrupting the Constitutive, Homodimeric Protein-Protein Interface in CK2β Using a Biophysical Fragment-Based Approach. J. Am. Chem. Soc. 2016, 138, 14303–14311. [Google Scholar] [CrossRef]
- Pinna, L.A. Protein kinase CK2: A challenge to canons. J. Cell Sci. 2002, 115, 3873–3878. [Google Scholar] [CrossRef]
- Siddiqui-Jain, A.; Drygin, D.; Streiner, N.; Chua, P.; Pierre, F.; O’brien, S.E.; Bliesath, J.; Omori, M.; Huser, N.; Ho, C.; et al. CX-4945, an orally bioavailable selective inhibitor of protein kinase CK2, inhibits prosurvival and angiogenic signaling and exhibits antitumor efficacy. Cancer Res. 2010, 70, 10288–10298. [Google Scholar] [CrossRef]
- Niefind, K.; Guerra, B.; Pinna, L.A.; Issinger, O.G.; Schomburg, D. Crystal structure of the catalytic subunit of protein kinase CK2 from Zea mays at 2.1 Å resolution. EMBO J. 1998, 17, 2451–2462. [Google Scholar] [CrossRef] [PubMed]
- Sarno, S.; Pinna, L.A. Protein kinase CK2 as a druggable target. Mol. Biosyst. 2008, 4, 889–894. [Google Scholar] [CrossRef] [PubMed]
- Sarno, S.; Salvi, M.; Battistutta, R.; Zanotti, G.; Pinna, L.A. Features and potentials of ATP-site directed CK2 inhibitors. Biochim. Biophys. Acta Proteins Proteom. 2005, 1754, 263–270. [Google Scholar] [CrossRef] [PubMed]
- Ruzzene, M.; Pinna, L.A. Addiction to protein kinase CK2: A common denominator of diverse cancer cells? Biochim. Biophys. Acta Proteins Proteom. 2010, 1804, 499–504. [Google Scholar] [CrossRef]
- Batool, M.; Ahmad, B.; Choi, S. A structure-based drug discovery paradigm. Int. J. Mol. Sci. 2019, 20, 2783. [Google Scholar] [CrossRef]
- Swinney, D.C.; Anthony, J. How were new medicines discovered? Nat. Rev. Drug Discov. 2011, 10, 507–519. [Google Scholar] [CrossRef]
- Study of CX-4945 in Combination with Gemcitabine and Cisplatin for Frontline Treatment of Cholangiocarcinoma—Full Text View—ClinicalTrials.gov. Available online: https://clinicaltrials.gov/ct2/show/NCT02128282 (accessed on 30 October 2020).
- Cozza, G.; Pinna, L.; Moro, S. Kinase CK2 Inhibition: An Update. Curr. Med. Chem. 2013, 20, 671–693. [Google Scholar] [CrossRef]
- Zandomenig, R.; Carrera Zandomenil, M.; Shugars, D.; Weinmannst, R. Casein Kinase Type II Is Involved in the Inhibition by 5,6-Dichloro-1-,8-D-ribofuranosylbenzimidazole of Specific RNA Polymerase I1 Transcription. J. Biol. Chem. 1986, 261, 3414–3419. [Google Scholar] [CrossRef]
- Sarno, S.; De Moliner, E.; Ruzzene, M.; Pagano, M.A.; Battistutta, R.; Bain, J.; Fabbro, D.; Schoepfer, J.; Elliott, M.; Furet, P.; et al. Biochemical and three-dimensional-structural study of the specific inhibition of protein kinase CK2 by [5-oxo-5,6-dihydroindolo-(1,2-a)quinazolin-7-yl]acetic acid (IQA). Biochem. J. 2003, 374, 639–646. [Google Scholar] [CrossRef]
- Yim, H.; Lee, C.H.; Lee, Y.H.; Lee, S.K. Emodin, an Anthraquinone Derivative Isolated from the Rhizomes of Rheum palmatum, Selectively Inhibits the Activity of Casein Kinase II as a Competitive Inhibitor. Planta Med. 1999, 65, 9–13. [Google Scholar] [CrossRef]
- Meggio, F.; Pagano, M.A.; Moro, S.; Zagotto, G.; Ruzzene, M.; Sarno, S.; Cozza, G.; Bain, J.; Elliott, M.; Deana, A.D.; et al. Inhibition of protein kinase CK2 by condensed polyphenolic derivatives. An in vitro and in vivo study. Biochemistry 2004, 43, 12931–12936. [Google Scholar] [CrossRef]
- Nie, Z.; Perretta, C.; Erickson, P.; Margosiak, S.; Almassy, R.; Lu, J.; Averill, A.; Yager, K.M.; Chu, S. Structure-based design, synthesis, and study of pyrazolo[1,5-a][1,3,5]triazine derivatives as potent inhibitors of protein kinase CK2. Bioorg. Med. Chem. Lett. 2007, 17, 4191–4195. [Google Scholar] [CrossRef]
- Dowling, J.E.; Chuaqui, C.; Pontz, T.W.; Lyne, P.D.; Larsen, N.A.; Block, M.H.; Chen, H.; Su, N.; Wu, A.; Russell, D.; et al. Potent and selective inhibitors of CK2 kinase identified through structure-guided hybridization. ACS Med. Chem. Lett. 2012, 3, 278–283. [Google Scholar] [CrossRef]
- Vangrevelinghe, E.; Zimmermann, K.; Schoepfer, J.; Portmann, R.; Fabbro, D.; Furet, P. Discovery of a potent and selective protein kinase CK2 inhibitor by high-throughput docking. J. Med. Chem. 2003, 46, 2656–2662. [Google Scholar] [CrossRef]
- Tamm, I.; Folkers, K.; Shunk, C.H.; Horsfall, F.L. Inhibition of influenza virus multiplication by N-glycosides of benzimidazoles-N. J. Exp. Med. 1954, 99, 227–250. [Google Scholar] [CrossRef]
- Bookser, B.C.; Raffaele, N.B. High-throughput five minute microwave accelerated glycosylation approach to the synthesis of nucleoside libraries. J. Org. Chem. 2007, 72, 173–179. [Google Scholar] [CrossRef]
- Meggio, F.; Shugar, D.; Pinna, L.A. Ribofuranosyl-benzimidazole derivatives as inhibitors of casein kinase-2 and casein kinase-1. Eur. J. Biochem. 1990, 187, 89–94. [Google Scholar] [CrossRef]
- Raaf, J.; Brunstein, E.; Issinger, O.-G.; Niefind, K. The CK2α/CK2β Interface of Human Protein Kinase CK2 Harbors a Binding Pocket for Small Molecules. Chem. Biol. 2008, 15, 111–117. [Google Scholar] [CrossRef]
- Szyszka, R.; Grankowski, N.; Felczak, K.; Shugar, D. Halogenated Benzimidazoles and Benzotriazoles as Selective Inhibitors of Protein Kinases CK-I and CK-II from Saccharomyces Cerevisiae and Other Sources. Biochem. Biophys. Res. Commun. 1995, 208, 418–424. [Google Scholar] [CrossRef]
- Wiley, R.H.; Hussung, K.F. Halogenated Benzotriazoles. J. Am. Chem. Soc. 1957, 79, 4395–4400. [Google Scholar] [CrossRef]
- Sarno, S.; Reddy, H.; Meggio, F.; Ruzzene, M.; Davies, S.P.; Donella-Deana, A.; Shugar, D.; Pinna, L.A. Selectivity of 4,5,6,7-tetrabromobenzotriazole, an ATP site-directed inhibitor of protein kinase CK2 (‘casein kinase-2′). FEBS Lett. 2001, 496, 44–48. [Google Scholar] [CrossRef]
- Bain, J.; Plater, L.; Elliott, M.; Shpiro, N.; Hastie, C.J.; Mclauchlan, H.; Klevernic, I.; Arthur, J.S.C.; Alessi, D.R.; Cohen, P. The selectivity of protein kinase inhibitors: A further update. Biochem. J. 2007, 408, 297–315. [Google Scholar] [CrossRef]
- Pagano, M.A.; Bain, J.; Kazimierczuk, Z.; Sarno, S.; Ruzzene, M.; Di Maira, G.; Elliott, M.; Orzeszko, A.; Cozza, G.; Meggio, F.; et al. The selectivity of inhibitors of protein kinase CK2: An update. Biochem. J. 2008, 415, 353–365. [Google Scholar] [CrossRef]
- DePierre, J.W. Mammalian Toxicity of Organic Compounds of Bromine and Iodine. In Organic Bromine and Iodine Compounds. The Handbook of Environmental Chemistry; Springer: Berlin/Heidelberg, Germany, 2003; Volume 3R, pp. 205–251. [Google Scholar] [CrossRef]
- Vilela, B.; Nájar, E.; Lumbreras, V.; Leung, J.; Pagès, M. Casein kinase 2 negatively regulates abscisic acid-activated SnRK2s in the core abscisic acid-signaling module. Mol. Plant 2015, 8, 709–721. [Google Scholar] [CrossRef]
- Treharne, K.J.; Xu, Z.; Chen, J.H.; Best, O.G.; Cassidy, D.M.; Gruenert, D.C.; Hegyi, P.; Gray, M.A.; Sheppard, D.N.; Kunzelmann, K.; et al. Inhibition of protein kinase CK2 closes the CFTR Cl channel, but has no effect on the cystic fibrosis mutant Δf508-CFTR. Cell. Physiol. Biochem. 2009, 24, 347–360. [Google Scholar] [CrossRef] [PubMed]
- Chapman, J.R.; Jackson, S.P. Phospho-dependent interactions between NBS1 and MDC1 mediate chromatin retention of the MRN complex at sites of DNA damage. EMBO Rep. 2008, 9, 795–801. [Google Scholar] [CrossRef]
- Zień, P.; Bretner, M.; Zastapiło, K.; Szyszka, R.; Shugar, D. Selectivity of 4,5,6,7-tetrabromobenzimidazole as an ATP-competitive potent inhibitor of protein kinase CK2 from various sources. Biochem. Biophys. Res. Commun. 2003, 306, 129–133. [Google Scholar] [CrossRef]
- Andrzejewska, M.; Pagano, M.A.; Meggio, F.; Brunati, A.M.; Kazimierczuk, Z. Polyhalogenobenzimidazoles: Synthesis and their inhibitory activity against casein kinases. Bioorganic Med. Chem. 2003, 11, 3997–4002. [Google Scholar] [CrossRef]
- Pagano, M.A.; Meggio, F.; Ruzzene, M.; Andrzejewska, M.; Kazimierczuk, Z.; Pinna, L.A. 2-Dimethylamino-4,5,6,7-tetrabromo-1H-benzimidazole: A novel powerful and selective inhibitor of protein kinase CK2. Biochem. Biophys. Res. Commun. 2004, 321, 1040–1044. [Google Scholar] [CrossRef]
- Pagano, M.A.; Andrzejewska, M.; Ruzzene, M.; Sarno, S.; Cesaro, L.; Bain, J.; Elliott, M.; Meggio, F.; Kazimierczuk, Z.; Pinna, L.A. Optimization of protein kinase CK2 inhibitors derived from 4,5,6,7-tetrabromobenzimidazole. J. Med. Chem. 2004, 47, 6239–6247. [Google Scholar] [CrossRef]
- Mishra, S.; Pertz, V.; Zhang, B.; Kaur, P.; Shimada, H.; Groffen, J.; Kazimierczuk, Z.; Pinna, L.A.; Heisterkamp, N. Treatment of P190 Bcr/Abl lymphoblastic leukemia cells with inhibitors of the serine/threonine kinase CK2. Leukemia 2007, 21, 178–180. [Google Scholar] [CrossRef] [PubMed]
- Siddiqui, Y.H.; Kershaw, R.M.; Humphreys, E.H.; Assis Junior, E.M.; Chaudhri, S.; Jayaraman, P.S.; Gaston, K. CK2 abrogates the inhibitory effects of PRH/HHEX on prostate cancer cell migration and invasion and acts through PRH to control cell proliferation. Oncogenesis 2017, 6, e293. [Google Scholar] [CrossRef] [PubMed]
- Jung, D.H.; Kim, K.H.; Byeon, H.E.; Park, H.J.; Park, B.; Rhee, D.K.; Um, S.H.; Pyo, S. Involvement of ATF3 in the negative regulation of iNOS expression and NO production in activated macrophages. Immunol. Res. 2015, 62, 35–45. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.; Hao, W.; Xiao, C.; Wang, R.; Xu, X.; Lu, H.; Chen, W.; Deng, C.X. SIRT6 Is Essential for Adipocyte Differentiation by Regulating Mitotic Clonal Expansion. Cell Rep. 2017, 18, 3155–3166. [Google Scholar] [CrossRef]
- Ulgesa, A.; Witsch, E.J.; Pramanik, G.; Klein, M.; Birkner, K.; Bühler, U.; Wasser, B.; Luessi, F.; Stergiou, N.; Dietzen, S.; et al. Protein kinase CK2 governs the molecular decision between encephalitogenic TH 17 cell and Treg cell development. Proc. Natl. Acad. Sci. USA 2016, 113, 10145–10150. [Google Scholar] [CrossRef]
- Jayasuriya, H.; Koonchanok, N.M.; Geahlen, R.L.; Mclaughlin, J.L.; Chang, C.J. Emodin, a protein tyrosine kinase inhibitor from polygonum cuspidatum. J. Nat. Prod. 1992, 55, 696–698. [Google Scholar] [CrossRef]
- Zhang, L.; Lau, Y.K.; Xi, L.; Hong, R.L.; Kim, D.S.H.L.; Chen, C.F.; Hortobagyi, G.N.; Chang, C.J.; Hung, M.C. Tyrosine kinase inhibitors, emodin and its derivative repress HER-2/neu-induced cellular transformation and metastasis-associated properties. Oncogene 1998, 16, 2855–2863. [Google Scholar] [CrossRef]
- Baell, J.B. Feeling Nature’s PAINS: Natural Products, Natural Product Drugs, and Pan Assay Interference Compounds (PAINS). J. Nat. Prod. 2016, 79, 616–628. [Google Scholar] [CrossRef]
- Baell, J.B.; Holloway, G.A. New Substructure Filters for Removal of Pan Assay Interference Compounds (PAINS) from Screening Libraries and for Their Exclusion in Bioassays. J. Med. Chem. 2010, 53, 2719–2740. [Google Scholar] [CrossRef]
- Paudel, P.; Shrestha, S.; Park, S.E.; Seong, S.H.; Fauzi, F.M.; Jung, H.A.; Choi, J.S. Emodin Derivatives as Multi-Target-Directed Ligands Inhibiting Monoamine Oxidase and Antagonizing Vasopressin V1A Receptors. ACS Omega 2020, 5, 26720–26731. [Google Scholar] [CrossRef]
- Cozza, G.; Mazzorana, M.; Papinutto, E.; Bain, J.; Elliott, M.; Di Maira, G.; Gianoncelli, A.; Pagano, M.A.; Sarno, S.; Ruzzene, M.; et al. Quinalizarin as a potent, selective and cell-permeable inhibitor of protein kinase CK2. Biochem. J. 2009, 421, 387–395. [Google Scholar] [CrossRef]
- Nie, Z.; Perretta, C.; Erickson, P.; Margosiak, S.; Lu, J.; Averill, A.; Almassy, R.; Chu, S. Structure-based design and synthesis of novel macrocyclic pyrazolo[1,5-a] [1,3,5]triazine compounds as potent inhibitors of protein kinase CK2 and their anticancer activities. Bioorganic Med. Chem. Lett. 2008, 18, 619–623. [Google Scholar] [CrossRef]
- Dowling, J.E.; Alimzhanov, M.; Bao, L.; Chuaqui, C.; Denz, C.R.; Jenkins, E.; Larsen, N.A.; Lyne, P.D.; Pontz, T.; Ye, Q.; et al. Potent and Selective CK2 Kinase Inhibitors with Effects on Wnt Pathway Signaling in Vivo. ACS Med. Chem. Lett. 2016, 7, 300–305. [Google Scholar] [CrossRef]
- Chua, P.C.; Pierre, F.; Whitten, J.P. Serine-Threonine Protein Kinase and PARP Modulators. WO2007US77464 31 August 2008. [Google Scholar]
- Pierre, F.; Chua, P.C.; Obrien, S.E.; Siddiqui-Jain, A.; Bourbon, P.; Haddach, M.; Michaux, J.; Nagasawa, J.; Schwaebe, M.K.; Stefan, E.; et al. Discovery and SAR of 5-(3-Chlorophenylamino)benzo[ c ][2,6]naphthyridine-8- carboxylic Acid (CX-4945), the first clinical stage inhibitor of protein kinase CK2 for the Treatment of Cancer. J. Med. Chem. 2011, 54, 635–654. [Google Scholar] [CrossRef]
- Kim, H.; Choi, K.; Kang, H.; Lee, S.-Y.; Chi, S.-W.; Lee, M.-S.; Song, J.; Im, D.; Choi, Y.; Cho, S. Identification of a Novel Function of CX-4945 as a Splicing Regulator. PLoS ONE 2014, 9, e94978. [Google Scholar] [CrossRef]
- Kim, H.; Lee, K.S.; Kim, A.K.; Choi, M.; Choi, K.; Kang, M.; Chi, S.W.; Lee, M.S.; Lee, J.S.; Lee, S.Y.; et al. A chemical with proven clinical safety rescues down-syndromerelated phenotypes in through DYRK1A inhibition. DMM Dis. Models Mech. 2016, 9, 839–848. [Google Scholar] [CrossRef]
- Pierre, F.; Stefan, E.; Nédellec, A.S.; Chevrel, M.C.; Regan, C.F.; Siddiqui-Jain, A.; MacAlino, D.; Streiner, N.; Drygin, D.; Haddach, M.; et al. 7-(4H-1,2,4-Triazol-3-yl)benzo[c][2,6]naphthyridines: A novel class of Pim kinase inhibitors with potent cell antiproliferative activity. Bioorganic Med. Chem. Lett. 2011, 21, 6687–6692. [Google Scholar] [CrossRef]
- Mojzych, M.; Šubertová, V.; Bielawska, A.; Bielawski, K.; Bazgier, V.; Berka, K.; Gucký, T.; Fornal, E.; Kryštof, V. Synthesis and kinase inhibitory activity of new sulfonamide derivatives of pyrazolo[4,3-e][1,2,4]triazines. Eur. J. Med. Chem. 2014, 78, 217–224. [Google Scholar] [CrossRef]
- Brear, P.; De Fusco, C.; Hadje Georgiou, K.; Francis-Newton, N.J.; Stubbs, C.J.; Sore, H.F.; Venkitaraman, A.R.; Abell, C.; Spring, D.R.; Hyvönen, M. Specific inhibition of CK2α from an anchor outside the active site. Chem. Sci. 2016, 7, 6839–6845. [Google Scholar] [CrossRef]
- Zecchin, D.; Moore, C.; Michailidis, F.; Horswell, S.; Rana, S.; Howell, M.; Downward, J. Combined targeting of G protein-coupled receptor and EGF receptor signaling overcomes resistance to PI 3K pathway inhibitors in PTEN -null triple negative breast cancer. EMBO Mol. Med. 2020, 12, e11987. [Google Scholar] [CrossRef]
- Von Morgen, P.V.; Burdova, K.; Flower, T.G.; O’Reilly, N.J.; Boulton, S.J.; Smerdon, S.J.; MacUrek, L.; Hoøejší, Z. MRE11 stability is regulated by CK2-dependent interaction with R2TP complex. Oncogene 2017, 36, 4943–4950. [Google Scholar] [CrossRef]
- Borgo, C.; D’Amore, C.; Cesaro, L.; Itami, K.; Hirota, T.; Salvi, M.; Pinna, L.A. A N-terminally deleted form of the CK2α’ catalytic subunit is sufficient to support cell viability. Biochem. Biophys. Res. Commun. 2020, 531, 409–415. [Google Scholar] [CrossRef]
- Kendall, J.J.; Chaney, K.E.; Patel, A.V.; Rizvi, T.A.; Largaespada, D.A.; Ratner, N. CK2 blockade causes MPNST cell apoptosis and promotes degradation of β-catenin. Oncotarget 2016, 7, 53191–53203. [Google Scholar] [CrossRef]
- Chen, F.; Pei, S.; Wang, X.; Zhu, Q.; Gou, S. Emerging JWA-targeted Pt(IV) prodrugs conjugated with CX-4945 to overcome chemo-immune-resistance. Biochem. Biophys. Res. Commun. 2020, 521, 753–761. [Google Scholar] [CrossRef]
- Pierre, F.; O’Brien, S.E.; Haddach, M.; Bourbon, P.; Schwaebe, M.K.; Stefan, E.; Darjania, L.; Stansfield, R.; Ho, C.; Siddiqui-Jain, A.; et al. Novel potent pyrimido[4,5-c]quinoline inhibitors of protein kinase CK2: SAR and preliminary assessment of their analgesic and anti-viral properties. Bioorganic Med. Chem. Lett. 2011, 21, 1687–1691. [Google Scholar] [CrossRef]
- Battistutta, R.; Cozza, G.; Pierre, F.; Papinutto, E.; Lolli, G.; Sarno, S.; Obrien, S.E.; Siddiqui-Jain, A.; Haddach, M.; Anderes, K.; et al. Unprecedented selectivity and structural determinants of a new class of protein kinase CK2 inhibitors in clinical trials for the treatment of cancer. Biochemistry 2011, 50, 8478–8488. [Google Scholar] [CrossRef]
- Liu, Z.L.; Zhang, R.M.; Meng, Q.G.; Zhang, X.C.; Sun, Y. Discovery of new protein kinase CK2 inhibitors with 1,3-dioxo-2,3-dihydro-1: H -indene core. MedChemComm 2016, 7, 1352–1355. [Google Scholar] [CrossRef]
- Protopopov, M.V.; Vdovin, V.S.; Starosyla, S.A.; Borysenko, I.P.; Prykhod’ko, A.O.; Lukashov, S.S.; Bilokin, Y.V.; Bdzhola, V.G.; Yarmoluk, S.M. Flavone inspired discovery of benzylidenebenzofuran-3(2H)-ones (aurones) as potent inhibitors of human protein kinase CK2. Bioorg. Chem. 2020, 102, 104062. [Google Scholar] [CrossRef]
- Bestgen, B.; Krimm, I.; Kufareva, I.; Kamal, A.A.M.; Seetoh, W.G.; Abell, C.; Hartmann, R.W.; Abagyan, R.; Cochet, C.; Le Borgne, M.; et al. 2-Aminothiazole Derivatives as Selective Allosteric Modulators of the Protein Kinase CK2. 1. Identification of an Allosteric Binding Site. J. Med. Chem. 2019, 62, 1803–1816. [Google Scholar] [CrossRef]
- Bestgen, B.; Kufareva, I.; Seetoh, W.; Abell, C.; Hartmann, R.W.; Abagyan, R.; Le Borgne, M.; Filhol, O.; Cochet, C.; Lomberget, T.; et al. 2-Aminothiazole Derivatives as Selective Allosteric Modulators of the Protein Kinase CK2. 2. Structure-Based Optimization and Investigation of Effects Specific to the Allosteric Mode of Action. J. Med. Chem. 2019, 62, 1817–1836. [Google Scholar] [CrossRef]
- Brear, P.; Ball, D.; Stott, K.; D’Arcy, S.; Hyvönen, M. Proposed Allosteric Inhibitors Bind to the ATP Site of CK2α. J. Med. Chem. 2020, 63, 12786–12798. [Google Scholar] [CrossRef] [PubMed]
- Oshima, T.; Niwa, Y.; Kuwata, K.; Srivastava, A.; Hyoda, T.; Tsuchiya, Y.; Kumagai, M.; Tsuyuguchi, M.; Tamaru, T.; Sugiyama, A.; et al. Cell-based screen identifies a new potent and highly selective CK2 inhibitor for modulation of circadian rhythms and cancer cell growth. Sci. Adv. 2019, 5, 9060–9083. [Google Scholar] [CrossRef] [PubMed]
- Dalle Vedove, A.; Zonta, F.; Zanforlin, E.; Demitri, N.; Ribaudo, G.; Cazzanelli, G.; Ongaro, A.; Sarno, S.; Zagotto, G.; Battistutta, R.; et al. A novel class of selective CK2 inhibitors targeting its open hinge conformation. Eur. J. Med. Chem. 2020, 195, 112267. [Google Scholar] [CrossRef] [PubMed]
- Wells, C.; Drewry, D.; Picket, J.E.; Axtman, A.D. SGC-CK2-1: The First Selective Chemical Probe for the Pleiotropic Kinase CK2. ChemRxiv 2020. [Google Scholar] [CrossRef]
- Xing, L.; Klug-Mcleod, J.; Rai, B.; Lunney, E.A. Kinase hinge binding scaffolds and their hydrogen bond patterns. Bioorganic Med. Chem. 2015, 23, 6520–6527. [Google Scholar] [CrossRef]
- Morooka, S.; Hoshina, M.; Kii, I.; Okabe, T.; Kojima, H.; Inoue, N.; Okuno, Y.; Denawa, M.; Yoshida, S.; Fukuhara, J.; et al. Identification of a dual inhibitor of SRPK1 and CK2 that attenuates pathological angiogenesis of macular degeneration in mice. Mol. Pharmacol. 2015, 88, 316–325. [Google Scholar] [CrossRef]
- Battistutta, R.; Lolli, G. Structural and functional determinants of protein kinase CK2α: Facts and open questions. Mol. Cell. Biochem. 2011, 356, 67–73. [Google Scholar] [CrossRef]
- Cozza, G.; Sarno, S.; Ruzzene, M.; Girardi, C.; Orzeszko, A.; Kazimierczuk, Z.; Zagotto, G.; Bonaiuto, E.; Di Paolo, M.L.; Pinna, L.A. Exploiting the repertoire of CK2 inhibitors to target DYRK and PIM kinases. Biochim. Biophys. Acta Proteins Proteom. 2013, 1834, 1402–1409. [Google Scholar] [CrossRef]
- Cozza, G.; Girardi, C.; Ranchio, A.; Lolli, G.; Sarno, S.; Orzeszko, A.; Kazimierczuk, Z.; Battistutta, R.; Ruzzene, M.; Pinna, L.A. Cell-permeable dual inhibitors of protein kinases CK2 and PIM-1: Structural features and pharmacological potential. Cell. Mol. Life Sci. 2014, 71, 3173–3185. [Google Scholar] [CrossRef]
- Chojnacki, K.; Wińska, P.; Wielechowska, M.; Łukowska-Chojnacka, E.; Tölzer, C.; Niefind, K.; Bretner, M. Biological properties and structural study of new aminoalkyl derivatives of benzimidazole and benzotriazole, dual inhibitors of CK2 and PIM1 kinases. Bioorg. Chem. 2018, 80, 266–275. [Google Scholar] [CrossRef]
- Purwin, M.; Hernández-Toribio, J.; Coderch, C.; Panchuk, R.; Skorokhyd, N.; Filipiak, K.; De Pascual-Teresa, B.; Ramos, A. Design and synthesis of novel dual-target agents for HDAC1 and CK2 inhibition. RSC Adv. 2016, 6, 66595–66608. [Google Scholar] [CrossRef]
- Martínez, R.; Di Geronimo, B.; Pastor, M.; Zapico, J.M.; Coderch, C.; Panchuk, R.; Skorokhyd, N.; Maslyk, M.; Ramos, A.; de Pascual-Teresa, B.; et al. Multitarget Anticancer Agents Based on Histone Deacetylase and Protein Kinase CK2 Inhibitors Regina. Molecules 2020, 25, 1497. [Google Scholar] [CrossRef]
- Rangasamy, L.; Ortín, I.; Zapico, J.M.; Coderch, C.; Ramos, A.; De Pascual-Teresa, B. New Dual CK2/HDAC1 Inhibitors with Nanomolar Inhibitory Activity against Both Enzymes. ACS Med. Chem. Lett. 2020, 11, 713–719. [Google Scholar] [CrossRef]
- Lavogina, D.; Enkvist, E.; Uri, A. Bisubstrate Inhibitors of Protein Kinases: From Principle to Practical Applications. ChemMedChem 2010, 5, 23–34. [Google Scholar] [CrossRef]
- Iegre, J.; Atkinson, E.L.; Brear, P.; Cooper, B.M.; Hyvönen, M.; Spring, D.R. Chemical probes targeting the kinase CK2: A journey outside the catalytic box. Org. Biomol. Chem.
- Enkvist, E.; Viht, K.; Bischoff, N.; Vahter, J.; Saaver, S.; Raidaru, G.; Issinger, O.G.; Niefind, K.; Uri, A. A subnanomolar fluorescent probe for protein kinase CK2 interaction studies. Org. Biomol. Chem. 2012, 10, 8645–8653. [Google Scholar] [CrossRef]
- Viht, K.; Saaver, S.; Vahter, J.; Enkvist, E.; Lavogina, D.; Sinijärv, H.; Raidaru, G.; Guerra, B.; Issinger, O.G.; Uri, A. Acetoxymethyl Ester of Tetrabromobenzimidazole-Peptoid Conjugate for Inhibition of Protein Kinase CK2 in Living Cells. Bioconjug. Chem. 2015, 26, 2324–2335. [Google Scholar] [CrossRef]
- Vahter, J.; Viht, K.; Uri, A.; Enkvist, E. Oligo-aspartic acid conjugates with benzo[c][2,6]naphthyridine-8-carboxylic acid scaffold as picomolar inhibitors of CK2. Bioorganic Med. Chem. 2017, 25, 2277–2284. [Google Scholar] [CrossRef]
- Pietsch, M.; Viht, K.; Schnitzler, A.; Ekambaram, R.; Steinkrüger, M.; Enkvist, E.; Nienberg, C.; Nickelsen, A.; Lauwers, M.; Jose, J.; et al. Unexpected CK2β-antagonistic functionality of bisubstrate inhibitors targeting protein kinase CK2. Bioorg. Chem. 2020, 96, 103608. [Google Scholar] [CrossRef]
- De Fusco, C.; Brear, P.; Iegre, J.; Georgiou, K.H.; Sore, H.F.; Hyvönen, M.; Spring, D.R. A fragment-based approach leading to the discovery of a novel binding site and the selective CK2 inhibitor CAM4066. Bioorganic Med. Chem. 2017, 25, 3471–3482. [Google Scholar] [CrossRef]


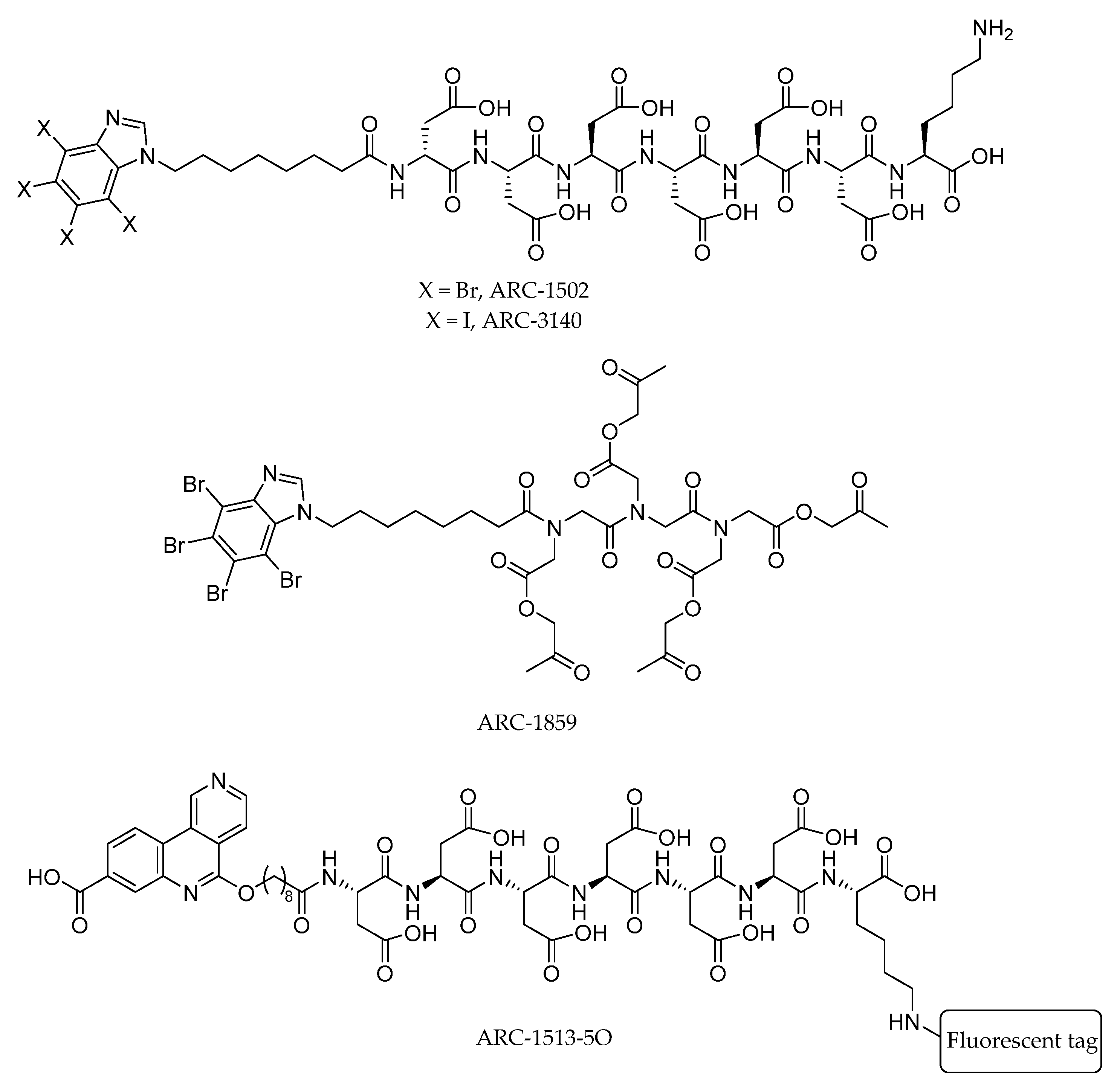


| Compound | Structure | CK2 Ki (μM) |
|---|---|---|
| DRB | 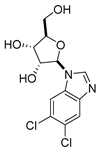 | 4.50 |
| TBB | 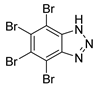 | 0.40 |
| TBI | 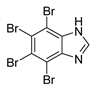 | 0.30 |
| DMAT | 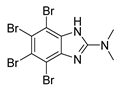 | 0.04 |
| Compound | IC50 (μM) | Ki (μM) | Number of other Proteins Inhibited > 40% (Concentration Tested) |
|---|---|---|---|
| Emodin | ~2 | 7.2 | 2 (10 μM) |
| MNX | 0.40 | 0.80 | 1 (10 μM) |
| NBC | 0.30 | 0.22 | 0 (10 μM) |
| DBC | 0.10 | 0.06 | 4 (10 μM) |
| Quinalizarin | 0.11 | 0.055 | 1 * (1 μM) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Atkinson, E.L.; Iegre, J.; Brear, P.D.; Zhabina, E.A.; Hyvönen, M.; Spring, D.R. Downfalls of Chemical Probes Acting at the Kinase ATP-Site: CK2 as a Case Study. Molecules 2021, 26, 1977. https://doi.org/10.3390/molecules26071977
Atkinson EL, Iegre J, Brear PD, Zhabina EA, Hyvönen M, Spring DR. Downfalls of Chemical Probes Acting at the Kinase ATP-Site: CK2 as a Case Study. Molecules. 2021; 26(7):1977. https://doi.org/10.3390/molecules26071977
Chicago/Turabian StyleAtkinson, Eleanor L., Jessica Iegre, Paul D. Brear, Elizabeth A. Zhabina, Marko Hyvönen, and David R. Spring. 2021. "Downfalls of Chemical Probes Acting at the Kinase ATP-Site: CK2 as a Case Study" Molecules 26, no. 7: 1977. https://doi.org/10.3390/molecules26071977
APA StyleAtkinson, E. L., Iegre, J., Brear, P. D., Zhabina, E. A., Hyvönen, M., & Spring, D. R. (2021). Downfalls of Chemical Probes Acting at the Kinase ATP-Site: CK2 as a Case Study. Molecules, 26(7), 1977. https://doi.org/10.3390/molecules26071977