Synergistic Effect by Combining a gp120-Binding Protein and a gp41-Binding Antibody to Inactivate HIV-1 Virions and Inhibit HIV-1 Infection
Abstract
1. Introduction
2. Results
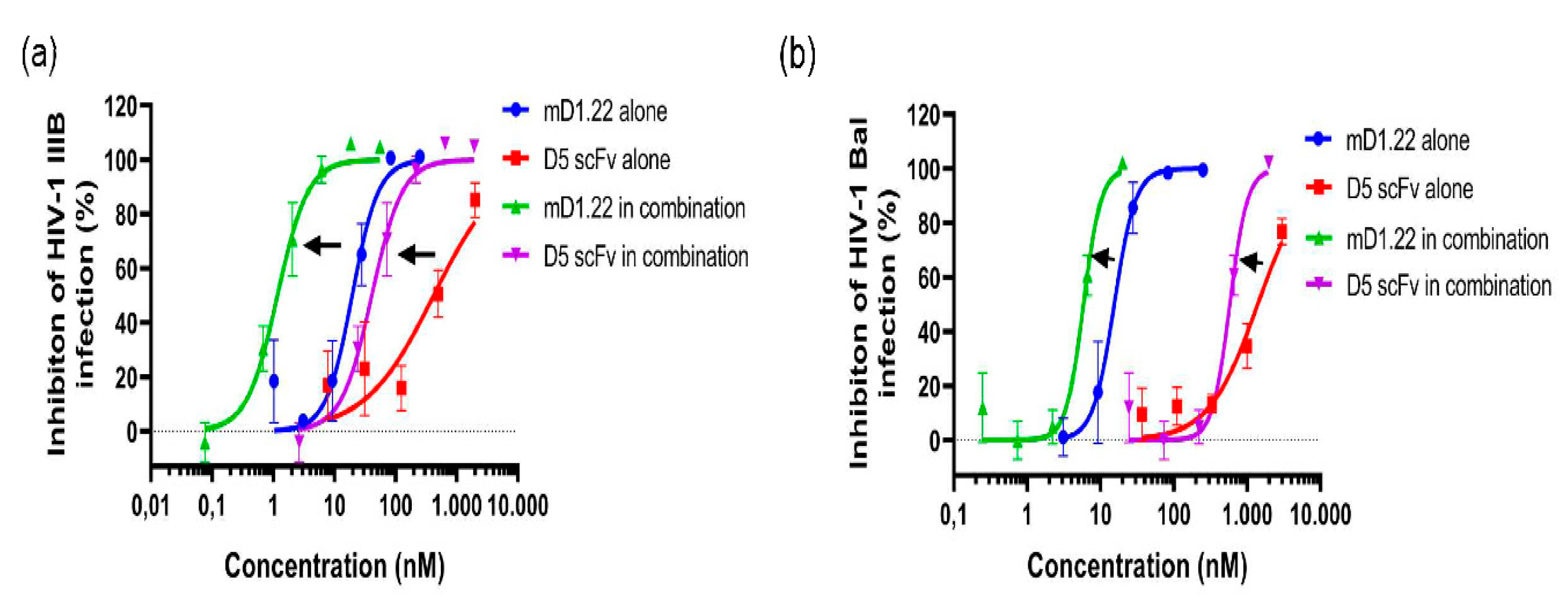
2.1. Combinatorial Use of mD1.22 and D5 scFv Exhibited Strong Synergism in Inhibiting Infection of HIV-1 Laboratory Strains
2.2. Combinatorial Use of mD1.22 and D5 scFv Exhibited Synergism in Inhibiting Infection by Divergent HIV-1 Strains
2.3. D5 scFv Enhances mD1.22-Mediated Inactivating Effect on HIV-1 Laboratory Strains and Primary HIV-1 Isolates
2.4. D5 scFv Enhances mD1.22-Mediated Inactivation Against T20-Resistant HIV-1 Strains and AZT-Resistant HIV-1 Strains
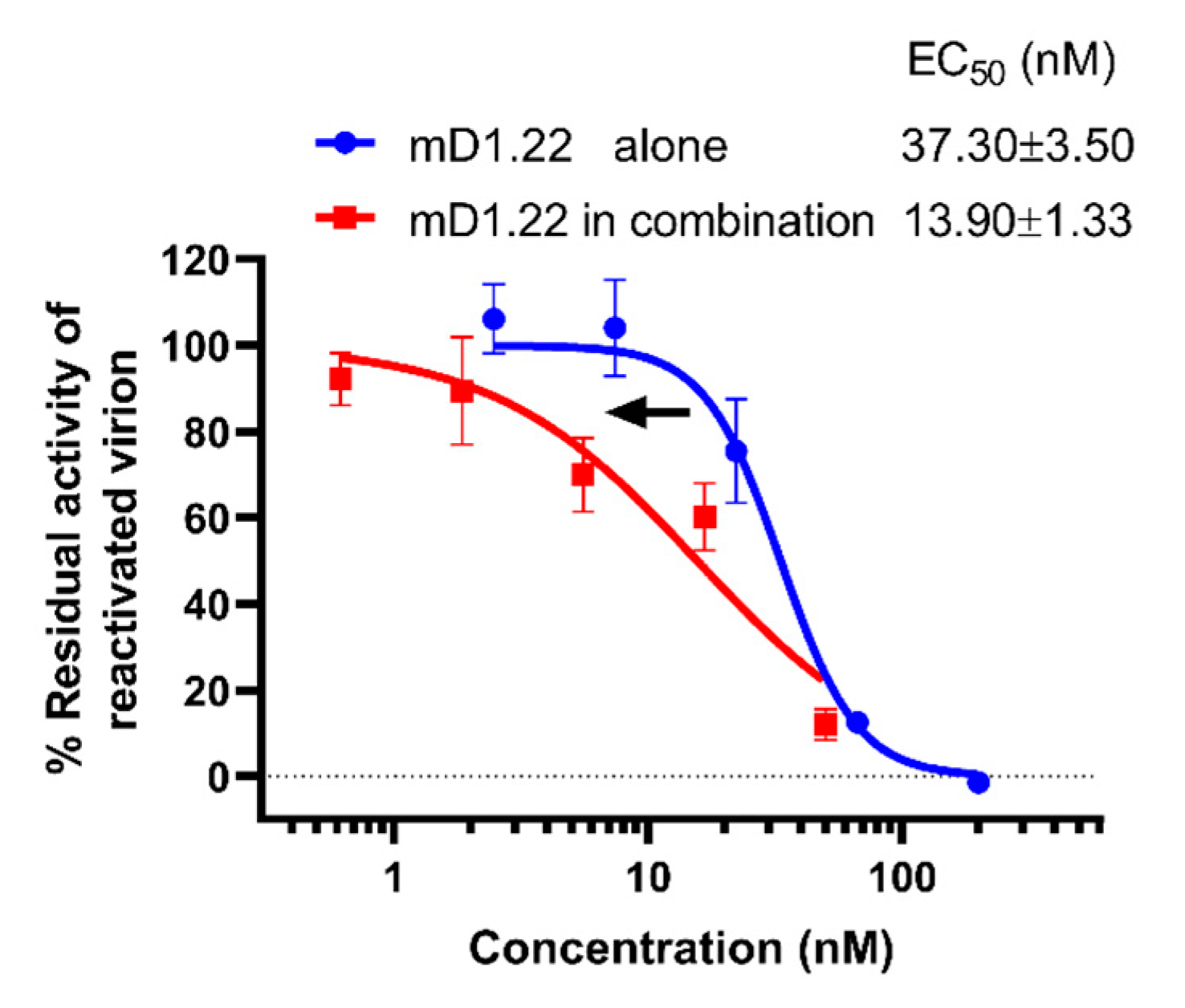
2.5. D5 scFv Enhances the Inactivation Activity of mD1.22 against the LRA-Reactivated HIV-1 Virions
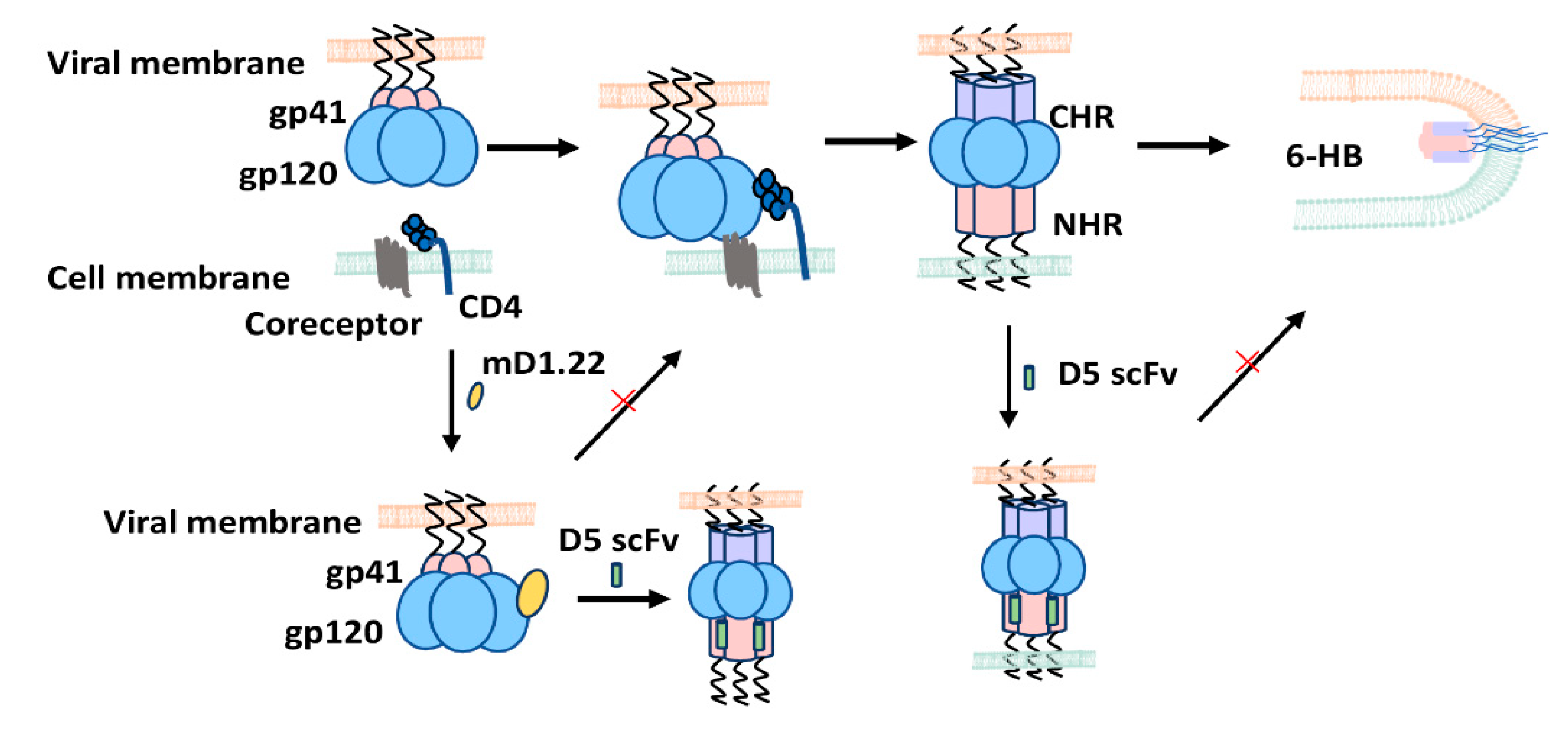
3. Discussion
4. Materials and Methods
4.1. Cells, Virus and Proteins
4.2. Inactivation of HIV-1 Virions
4.3. Inhibition of HIV-1 Infection
4.4. Detecting the Ability of mD1.22 to Inactivate LRA-Reactivated HIV-1 Virions Released from ACH-2 Cells
4.5. Combination Index (CI)
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Deeks, S.G.; Overbaugh, J.; Phillips, A.; Buchbinder, S. HIV infection. Nat. Rev. Dis. Primers 2015, 1, 1–22. [Google Scholar] [CrossRef] [PubMed]
- Vandamme, A.M.; Van Laethem, K.; De Clercq, E. Managing resistance to anti-HIV drugs: An important consideration for effective disease management. Drugs 1999, 57, 337–361. [Google Scholar] [CrossRef]
- Lu, L.; Tong, P.; Yu, X.; Pan, C.; Zou, P.; Chen, Y.H.; Jiang, S. HIV-1 variants with a single-point mutation in the gp41 pocket region exhibiting different susceptibility to HIV fusion inhibitors with pocket- or membrane-binding domain. Biochim. Biophys. Acta 2012, 1818, 2950–2957. [Google Scholar] [CrossRef]
- Zheng, B.; Wang, K.; Lu, L.; Yu, F.; Cheng, M.; Jiang, S.; Liu, K.; Cai, L. Hydrophobic mutations in buried polar residues enhance HIV-1 gp41 N-terminal heptad repeat-C-terminal heptad repeat interactions and C-peptides’ anti-HIV activity. AIDS 2014, 28, 1251–1260. [Google Scholar] [CrossRef]
- Su, S.; Rasquinha, G.; Du, L.; Wang, Q.; Xu, W.; Li, W.; Lu, L.; Jiang, S. A Peptide-Based HIV-1 Fusion Inhibitor with Two Tail-Anchors and Palmitic Acid Exhibits Substantially Improved In Vitro and Ex Vivo Anti-HIV-1 Activity and Prolonged In Vivo Half-Life. Molecules 2019, 24, 1134. [Google Scholar] [CrossRef]
- Lu, M.; Blacklow, S.C.; Kim, P.S. A Trimeric Structural Domain of the Hiv-1 Transmembrane Glycoprotein. Nat. Struct. Biol. 1995, 2, 1075–1082. [Google Scholar] [CrossRef]
- Lu, M.; Kim, P.S. A trimeric structural subdomain of the HIV-1 transmembrane glycoprotein. J. Biomol. Struct. Dyn. 1997, 15, 465–471. [Google Scholar] [CrossRef]
- Gardner, M.R.; Farzan, M. Engineering antibody-like inhibitors to prevent and treat HIV-1 infection. Curr. Opin. Hiv. Aids 2017, 12, 294–301. [Google Scholar] [CrossRef]
- Deen, K.C.; McDougal, J.S.; Inacker, R.; Folena-Wasserman, G.; Arthos, J.; Rosenberg, J.; Maddon, P.J.; Axel, R.; Sweet, R.W. A soluble form of CD4 (T4) protein inhibits AIDS virus infection. Nature 1988, 331, 82–84. [Google Scholar] [CrossRef]
- Gardner, M.R.; Kattenhorn, L.M.; Kondur, H.R.; von Schaewen, M.; Dorfman, T.; Chiang, J.J.; Haworth, K.G.; Decker, J.M.; Alpert, M.D.; Bailey, C.C.; et al. AAV-expressed eCD4-Ig provides durable protection from multiple SHIV challenges. Nature 2015, 519, 87–91. [Google Scholar] [CrossRef]
- Huang, J.; Kang, B.; Ishida, E.; Zhou, T.; Griesman, T.; Sheng, Z.; Wu, F.; Doria-Rose, N.A.; Zhang, B.; McKee, K.; et al. Identification of a CD4-Binding-Site Antibody to HIV that Evolved Near-Pan Neutralization Breadth. Immunity 2016, 45, 1108–1121. [Google Scholar] [CrossRef]
- Chen, W.; Zhu, Z.; Feng, Y.; Dimitrov, D.S. Human domain antibodies to conserved sterically restricted regions on gp120 as exceptionally potent cross-reactive HIV-1 neutralizers. Proc. Natl. Acad. Sci. USA 2008, 105, 17121–17126. [Google Scholar] [CrossRef]
- Jiang, S.; Lin, K.; Strick, N.; Neurath, A.R. HIV-1 inhibition by a peptide. Nature 1993, 365, 113. [Google Scholar] [CrossRef] [PubMed]
- Lalezari, J.P.; Henry, K.; O’Hearn, M.; Montaner, J.S.; Piliero, P.J.; Trottier, B.; Walmsley, S.; Cohen, C.; Kuritzkes, D.R.; Eron, J.J., Jr.; et al. Enfuvirtide, an HIV-1 fusion inhibitor, for drug-resistant HIV infection in North and South America. N. Engl. J. Med. 2003, 348, 2175–2185. [Google Scholar] [CrossRef]
- Dwyer, J.J.; Wilson, K.L.; Davison, D.K.; Freel, S.A.; Seedorff, J.E.; Wring, S.A.; Tvermoes, N.A.; Matthews, T.J.; Greenberg, M.L.; Delmedico, M.K. Design of helical, oligomeric HIV-1 fusion inhibitor peptides with potent activity against enfuvirtide-resistant virus. Proc. Natl. Acad. Sci. USA 2007, 104, 12772–12777. [Google Scholar] [CrossRef]
- Chen, W.; Feng, Y.; Prabakaran, P.; Ying, T.; Wang, Y.; Sun, J.; Macedo, C.D.; Zhu, Z.; He, Y.; Polonis, V.R.; et al. Exceptionally potent and broadly cross-reactive, bispecific multivalent HIV-1 inhibitors based on single human CD4 and antibody domains. J. Virol. 2014, 88, 1125–1139. [Google Scholar] [CrossRef]
- Lu, L.; Pan, C.; Li, Y.; Lu, H.; He, W.; Jiang, S. A bivalent recombinant protein inactivates HIV-1 by targeting the gp41 prehairpin fusion intermediate induced by CD4 D1D2 domains. Retrovirology 2012, 9, 104. [Google Scholar] [CrossRef] [PubMed]
- Kagiampakis, I.; Gharibi, A.; Mankowski, M.K.; Snyder, B.A.; Ptak, R.G.; Alatas, K.; LiWang, P.J. Potent strategy to inhibit HIV-1 by binding both gp120 and gp41. Antimicrob. Agents Chemother. 2011, 55, 264–275. [Google Scholar] [CrossRef]
- Haim, H.; Si, Z.; Madani, N.; Wang, L.; Courter, J.R.; Princiotto, A.; Kassa, A.; DeGrace, M.; McGee-Estrada, K.; Mefford, M.; et al. Soluble CD4 and CD4-mimetic compounds inhibit HIV-1 infection by induction of a short-lived activated state. PLoS Pathog. 2009, 5, e1000360. [Google Scholar] [CrossRef] [PubMed]
- Qi, Q.; Wang, Q.; Chen, W.; Du, L.; Dimitrov, D.S.; Lu, L.; Jiang, S. HIV-1 gp41-targeting fusion inhibitory peptides enhance the gp120-targeting protein-mediated inactivation of HIV-1 virions. Emerg. Microbes Infect. 2017, 6, e59. [Google Scholar] [CrossRef] [PubMed]
- Su, X.; Wang, Q.; Wen, Y.; Jiang, S.; Lu, L. Protein- and Peptide-Based Virus Inactivators: Inactivating Viruses Before Their Entry into Cells. Front. Microbiol. 2020, 11, 1063. [Google Scholar] [CrossRef]
- Blair, H.A. Ibalizumab: A Review in Multidrug-Resistant HIV-1 Infection. Drugs 2020, 80, 189–196. [Google Scholar] [CrossRef]
- Tiller, K.E.; Tessier, P.M. Advances in Antibody Design. Annu. Rev. Biomed. Eng. 2015, 17, 191–216. [Google Scholar] [CrossRef]
- Huang, J.; Ofek, G.; Laub, L.; Louder, M.K.; Doria-Rose, N.A.; Longo, N.S.; Imamichi, H.; Bailer, R.T.; Chakrabarti, B.; Sharma, S.K.; et al. Broad and potent neutralization of HIV-1 by a gp41-specific human antibody. Nature 2012, 491, 406–412. [Google Scholar] [CrossRef]
- Miller, M.D.; Geleziunas, R.; Bianchi, E.; Lennard, S.; Hrin, R.; Zhang, H.; Lu, M.; An, Z.; Ingallinella, P.; Finotto, M.; et al. A human monoclonal antibody neutralizes diverse HIV-1 isolates by binding a critical gp41 epitope. Proc. Natl. Acad. Sci. USA 2005, 102, 14759–14764. [Google Scholar] [CrossRef]
- Eckert, D.M.; Shi, Y.; Kim, S.; Welch, B.D.; Kang, E.; Poff, E.S.; Kay, M.S. Characterization of the steric defense of the HIV-1 gp41 N-trimer region. Protein Sci. 2008, 17, 2091–2100. [Google Scholar] [CrossRef]
- Wei, D.G.; Chiang, V.; Fyne, E.; Balakrishnan, M.; Barnes, T.; Graupe, M.; Hesselgesser, J.; Irrinki, A.; Murry, J.P.; Stepan, G.; et al. Histone deacetylase inhibitor romidepsin induces HIV expression in CD4 T cells from patients on suppressive antiretroviral therapy at concentrations achieved by clinical dosing. PLoS Pathog. 2014, 10, e1004071. [Google Scholar] [CrossRef]
- Barresinoussi, F.; Chermann, J.C.; Rey, F.; Nugeyre, M.T.; Chamaret, S.; Gruest, J.; Dauguet, C.; Axlerblin, C.; Vezinetbrun, F.; Rouzioux, C.; et al. Isolation of a T-Lymphotropic Retrovirus from a Patient at Risk for Acquired Immune-Deficiency Syndrome (Aids). Science 1983, 220, 868–871. [Google Scholar] [CrossRef]
- Gu, S.X.; Zhu, Y.Y.; Wang, C.; Wang, H.F.; Liu, G.Y.; Cao, S.; Huang, L. Recent discoveries in HIV-1 reverse transcriptase inhibitors. Curr. Opin. Pharmacol. 2020, 54, 166–172. [Google Scholar] [CrossRef]
- Finzi, D.; Hermankova, M.; Pierson, T.; Carruth, L.M.; Buck, C.; Chaisson, R.E.; Quinn, T.C.; Chadwick, K.; Margolick, J.; Brookmeyer, R.; et al. Identification of a reservoir for HIV-1 in patients on highly active antiretroviral therapy. Science 1997, 278, 1295–1300. [Google Scholar] [CrossRef] [PubMed]
- Zhan, P.; Pannecouque, C.; De Clercq, E.; Liu, X. Anti-HIV Drug Discovery and Development: Current Innovations and Future Trends. J. Med. Chem. 2016, 59, 2849–2878. [Google Scholar] [CrossRef] [PubMed]
- Bandera, A.; Gori, A.; Clerici, M.; Sironi, M. Phylogenies in ART: HIV reservoirs, HIV latency and drug resistance. Curr. Opin. Pharmacol. 2019, 48, 24–32. [Google Scholar] [CrossRef] [PubMed]
- Allaway, G.P.; Davis-Bruno, K.L.; Beaudry, G.A.; Garcia, E.B.; Wong, E.L.; Ryder, A.M.; Hasel, K.W.; Gauduin, M.C.; Koup, R.A.; McDougal, J.S.; et al. Expression and characterization of CD4-IgG2, a novel heterotetramer that neutralizes primary HIV type 1 isolates. AIDS Res. Hum. Retrovir. 1995, 11, 533–539. [Google Scholar] [CrossRef]
- Trkola, A.; Ketas, T.; Kewalramani, V.N.; Endorf, F.; Binley, J.M.; Katinger, H.; Robinson, J.; Littman, D.R.; Moore, J.P. Neutralization sensitivity of human immunodeficiency virus type 1 primary isolates to antibodies and CD4-based reagents is independent of coreceptor usage. J. Virol. 1998, 72, 1876–1885. [Google Scholar] [CrossRef]
- Gauduin, M.C.; Allaway, G.P.; Olson, W.C.; Weir, R.; Maddon, P.J.; Koup, R.A. CD4-immunoglobulin G2 protects Hu-PBL-SCID mice against challenge by primary human immunodeficiency virus type 1 isolates. J. Virol. 1998, 72, 3475–3478. [Google Scholar] [CrossRef] [PubMed]
- Jacobson, J.M.; Israel, R.J.; Lowy, I.; Ostrow, N.A.; Vassilatos, L.S.; Barish, M.; Tran, D.N.H.; Sullivan, B.M.; Ketas, T.J.; O’Neill, T.J.; et al. Treatment of advanced human immunodeficiency virus type 1 disease with the viral entry inhibitor PRO 542. Antimicrob. Agents Chemother. 2004, 48, 423–429. [Google Scholar] [CrossRef][Green Version]
- Su, S.; Ma, Z.; Hua, C.; Li, W.; Lu, L.; Jiang, S. Adding an Artificial Tail-Anchor to a Peptide-Based HIV-1 Fusion Inhibitor for Improvement of Its Potency and Resistance Profile. Molecules 2017, 22, 1996. [Google Scholar] [CrossRef]
- Xu, W.; Wang, Q.; Yu, F.; Lu, L.; Jiang, S. Synergistic effect resulting from combinations of a bifunctional HIV-1 antagonist with antiretroviral drugs. J. Acquir. Immune Defic. Syndr. 2014, 67, 1–6. [Google Scholar] [CrossRef]
| Virus Strain | CI | mD1.22 | D5 scFv | ||||
|---|---|---|---|---|---|---|---|
| IC50 (nM) | Fold of Enhancement | IC50 (nM) | Fold of Enhancement | ||||
| Alone | In Combination | Alone | In Combination | ||||
| HIV-1 Laboratory Strains | |||||||
| IIIB (X4) | 0.16 | 16.57 | 1.23 | 12.47 | 518.03 | 42.97 | 11.06 |
| Bal (R5) | 0.57 | 13.49 | 3.59 | 2.76 | 1190.51 | 359.11 | 2.32 |
| HIV-1 Primary Isolates | |||||||
| 91US_4 | 0.59 | 26.88 | 7.87 | 2.42 | 674.25 | 196.81 | 2.43 |
| 92UG024 | 0.38 | 7.58 | 0.32 | 22.69 | 318.82 | 108.01 | 1.95 |
| NP1525 | 0.20 | 22.97 | 2.66 | 7.64 | 3070.19 | 265.68 | 10.56 |
| T20-Resistant HIV-1 Strains | |||||||
| HIV-1 NL4-3 D36G (WT) | 0.47 | 14.00 | 2.71 | 4.17 | 386.82 | 108.42 | 2.57 |
| (D36G) N42T, N43K | 0.79 | 14.58 | 5.72 | 1.55 | 286.19 | 114.47 | 1.50 |
| (D36G) V38A, N42T | 0.64 | 12.69 | 4.20 | 2.02 | 345.41 | 105.01 | 2.29 |
| AZT-Resistant HIV-1 Strains | |||||||
| 629 | 0.79 | 17.34 | 6.44 | 1.69 | 764.82 | 322.18 | 1.37 |
| 964 | 0.24 | 7.19 | 0.90 | 6.99 | 389.64 | 45.03 | 7.65 |
| Virus Strains | Concentration (nM) of mD1.22 | Fold of Enhancement | ||
|---|---|---|---|---|
| Alone | In Combination | |||
| HIV-1 Laboratory-Adapted Strains | ||||
| IIIB (X4) | EC50 | 2.09 | 0.64 | 2.27 |
| EC90 | 11.54 | 1.21 | 8.54 | |
| Bal (R5) | EC50 | 9.12 | 1.75 | 4.21 |
| EC90 | 17.46 | 4.68 | 2.73 | |
| Primary HIV-1 Isolates | ||||
| 91US_4 | EC50 | 4.28 | 1.99 | 1.15 |
| EC90 | 8.51 | 3.80 | 1.24 | |
| 97TH_NP1525 | EC50 | 5.92 | 2.51 | 1.36 |
| EC90 | 31.23 | 15.74 | 0.98 | |
| 92UG024 | EC50 | 13.43 | 2.20 | 5.10 |
| EC90 | 41.49 | 12.09 | 2.43 | |
| T20-Resistant HIV-1 Strains | ||||
| HIV-1 NL4-1 D36G (WT) | EC50 | 1.10 | 0.66 | 0.67 |
| EC90 | 2.12 | 1.16 | 0.83 | |
| (D36G) N42T, N43K | EC50 | 2.56 | 0.58 | 3.41 |
| EC90 | 3.82 | 1.24 | 2.08 | |
| (D36G) V38A, N42T | EC50 | 5.08 | 2.49 | 1.04 |
| EC90 | 10.03 | 6.28 | 0.60 | |
| AZT- Resistant HIV-1 Strains | ||||
| 629 | EC50 | 11.28 | 4.78 | 1.36 |
| EC90 | 20.08 | 8.84 | 1.27 | |
| 964 | EC50 | 3.19 | 1.56 | 1.04 |
| EC90 | 23.80 | 3.23 | 6.37 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, X.; Cao, M.; Wu, Y.; Xu, W.; Wang, Q.; Ying, T.; Lu, L.; Jiang, S. Synergistic Effect by Combining a gp120-Binding Protein and a gp41-Binding Antibody to Inactivate HIV-1 Virions and Inhibit HIV-1 Infection. Molecules 2021, 26, 1964. https://doi.org/10.3390/molecules26071964
Wang X, Cao M, Wu Y, Xu W, Wang Q, Ying T, Lu L, Jiang S. Synergistic Effect by Combining a gp120-Binding Protein and a gp41-Binding Antibody to Inactivate HIV-1 Virions and Inhibit HIV-1 Infection. Molecules. 2021; 26(7):1964. https://doi.org/10.3390/molecules26071964
Chicago/Turabian StyleWang, Xinling, Miao Cao, Yanling Wu, Wei Xu, Qian Wang, Tianlei Ying, Lu Lu, and Shibo Jiang. 2021. "Synergistic Effect by Combining a gp120-Binding Protein and a gp41-Binding Antibody to Inactivate HIV-1 Virions and Inhibit HIV-1 Infection" Molecules 26, no. 7: 1964. https://doi.org/10.3390/molecules26071964
APA StyleWang, X., Cao, M., Wu, Y., Xu, W., Wang, Q., Ying, T., Lu, L., & Jiang, S. (2021). Synergistic Effect by Combining a gp120-Binding Protein and a gp41-Binding Antibody to Inactivate HIV-1 Virions and Inhibit HIV-1 Infection. Molecules, 26(7), 1964. https://doi.org/10.3390/molecules26071964









