Approaches toward the Separation, Modification, Identification and Scale up Purification of Tetracyclic Diterpene Glycosides from Stevia rebaudiana (Bertoni) Bertoni
Abstract
1. Introduction
2. Results
2.1. Chemical and Enzymatic Reactions of Diterpene Glycosides
2.1.1. Alkaline Hydrolysis
2.1.2. Acid Hydrolysis
2.1.3. Hydrogenation of the Exocyclic Double Bond
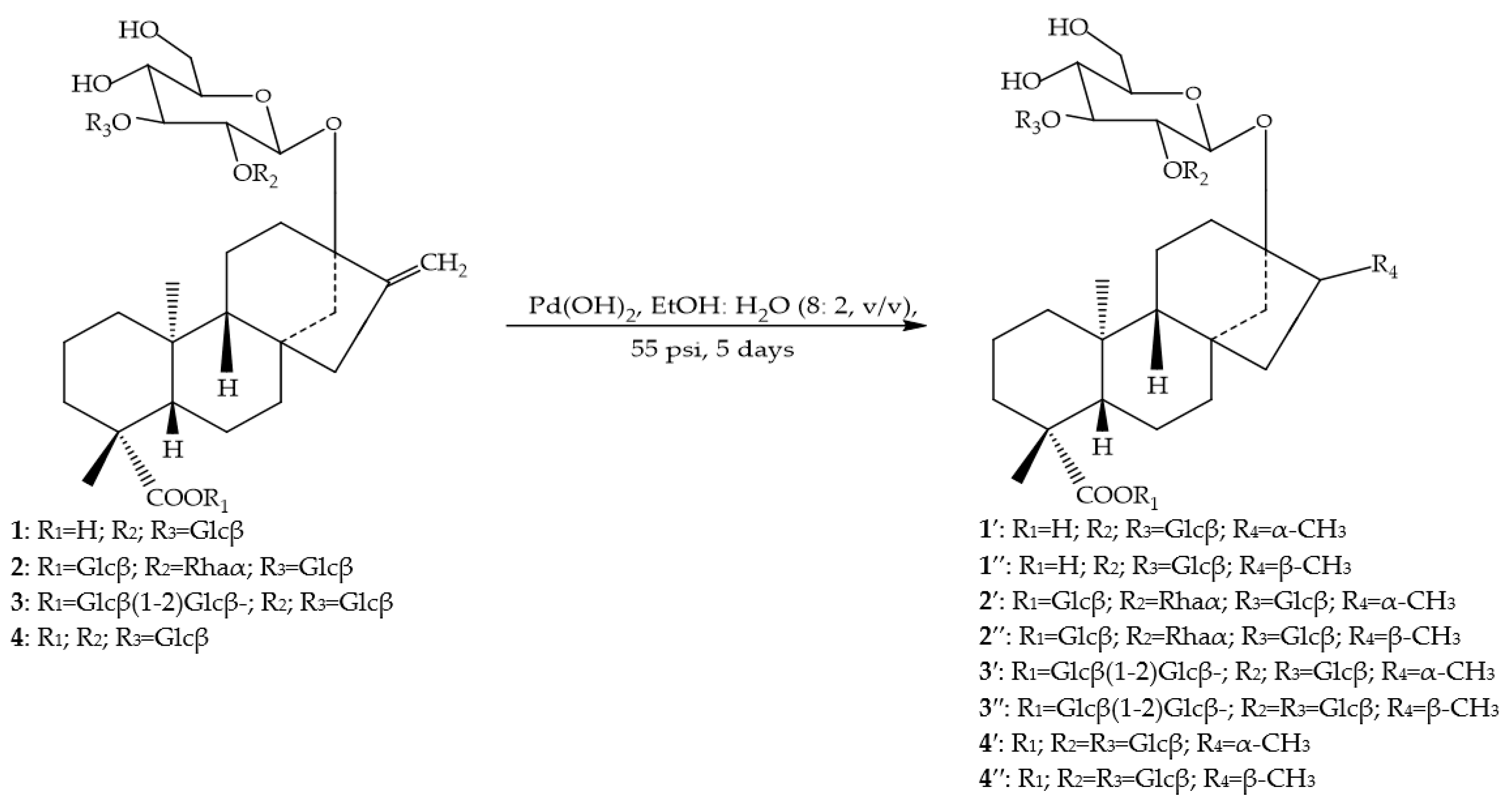
2.1.4. Enzymatic Reactions
3. Methods of Separation for Diterpene Glycosides
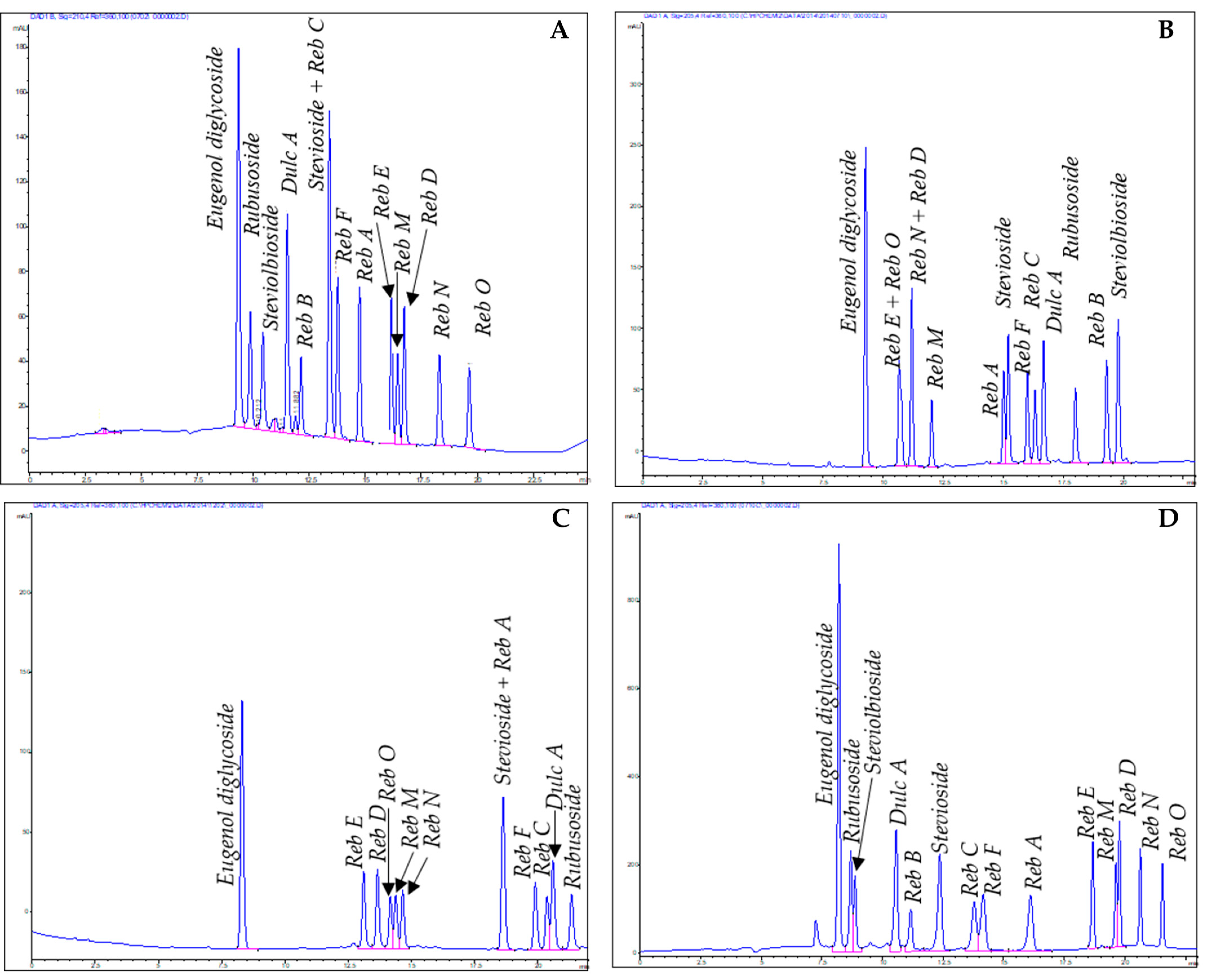
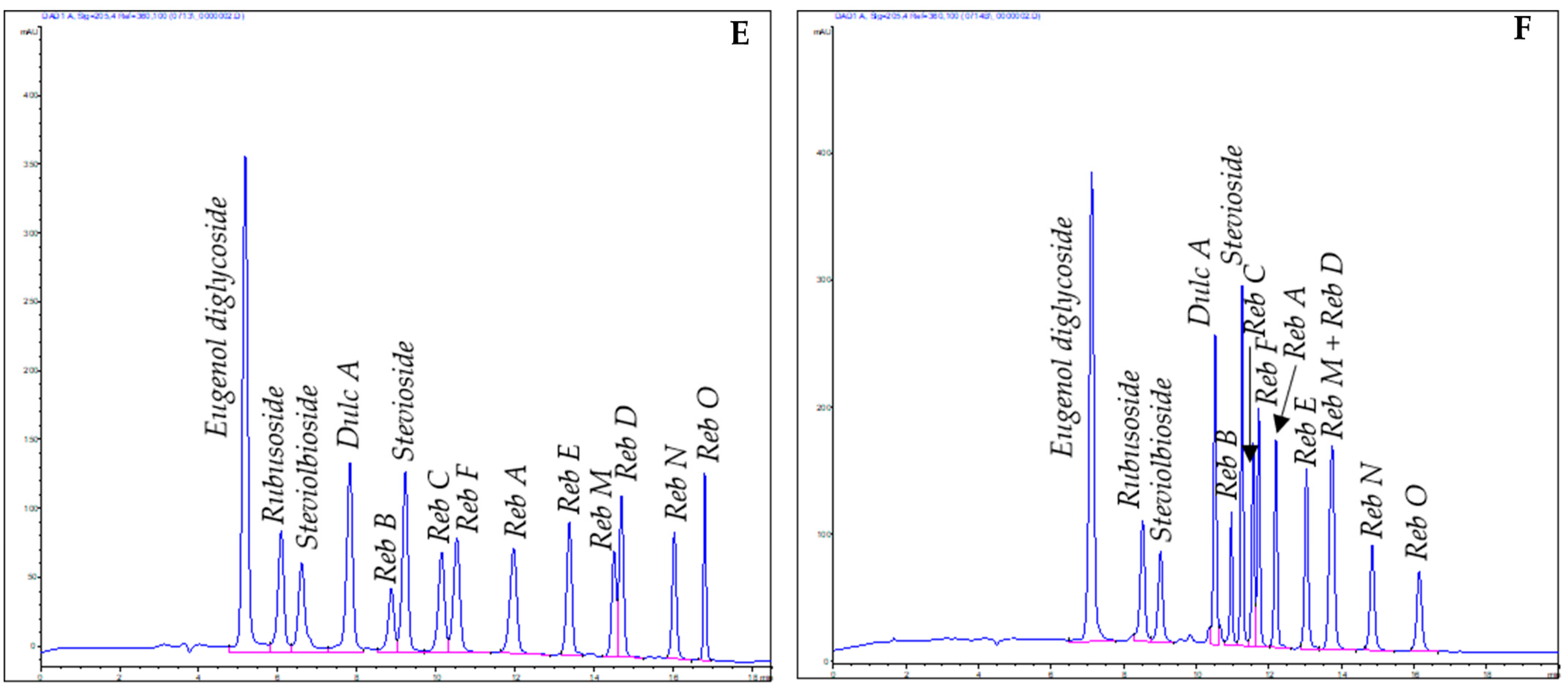
3.1. Analytical Methods
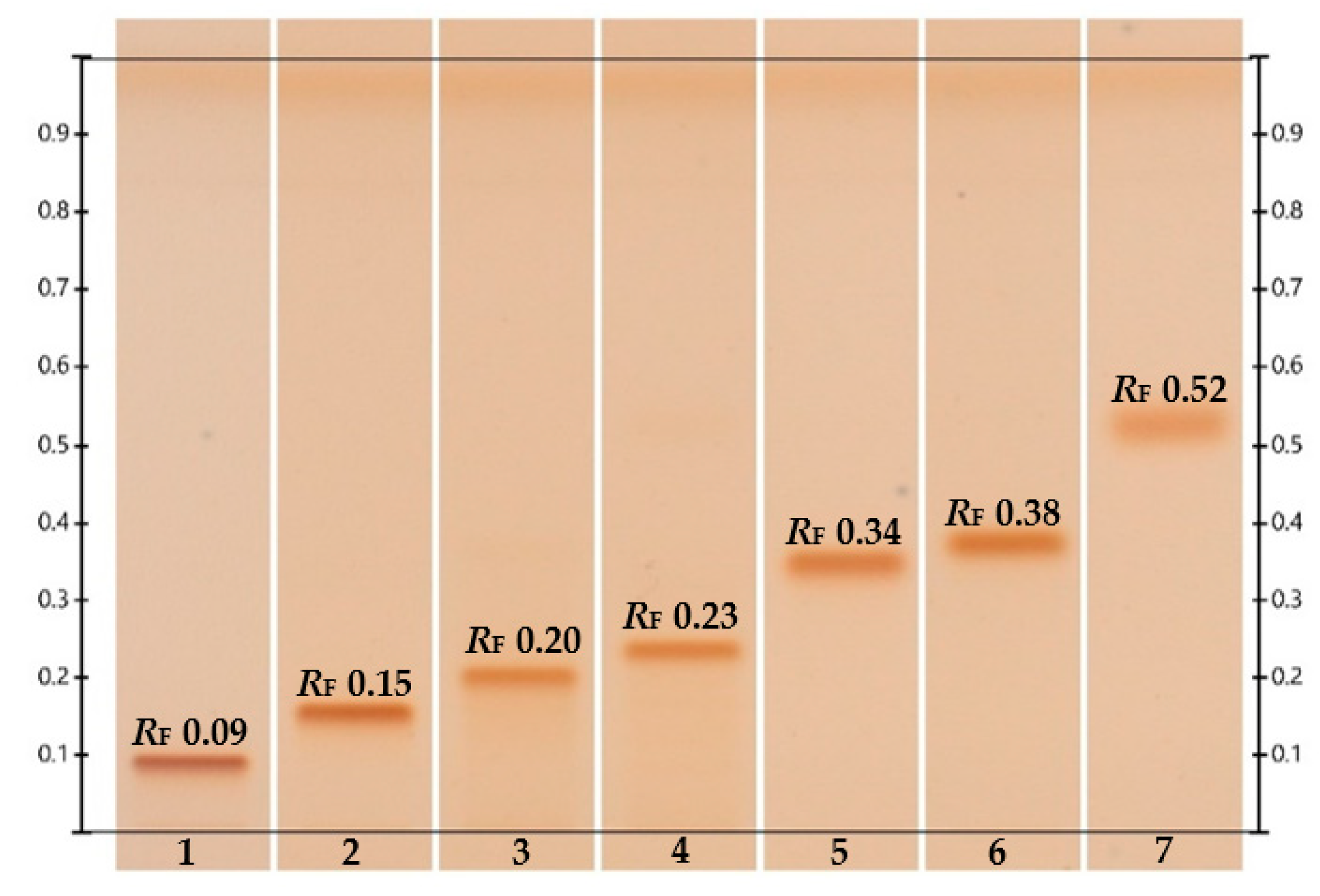
3.1.1. High-Performance Thin-Layer Chromatography
3.1.2. High-Performance Liquid Chromatography
3.2. Preparative Methods
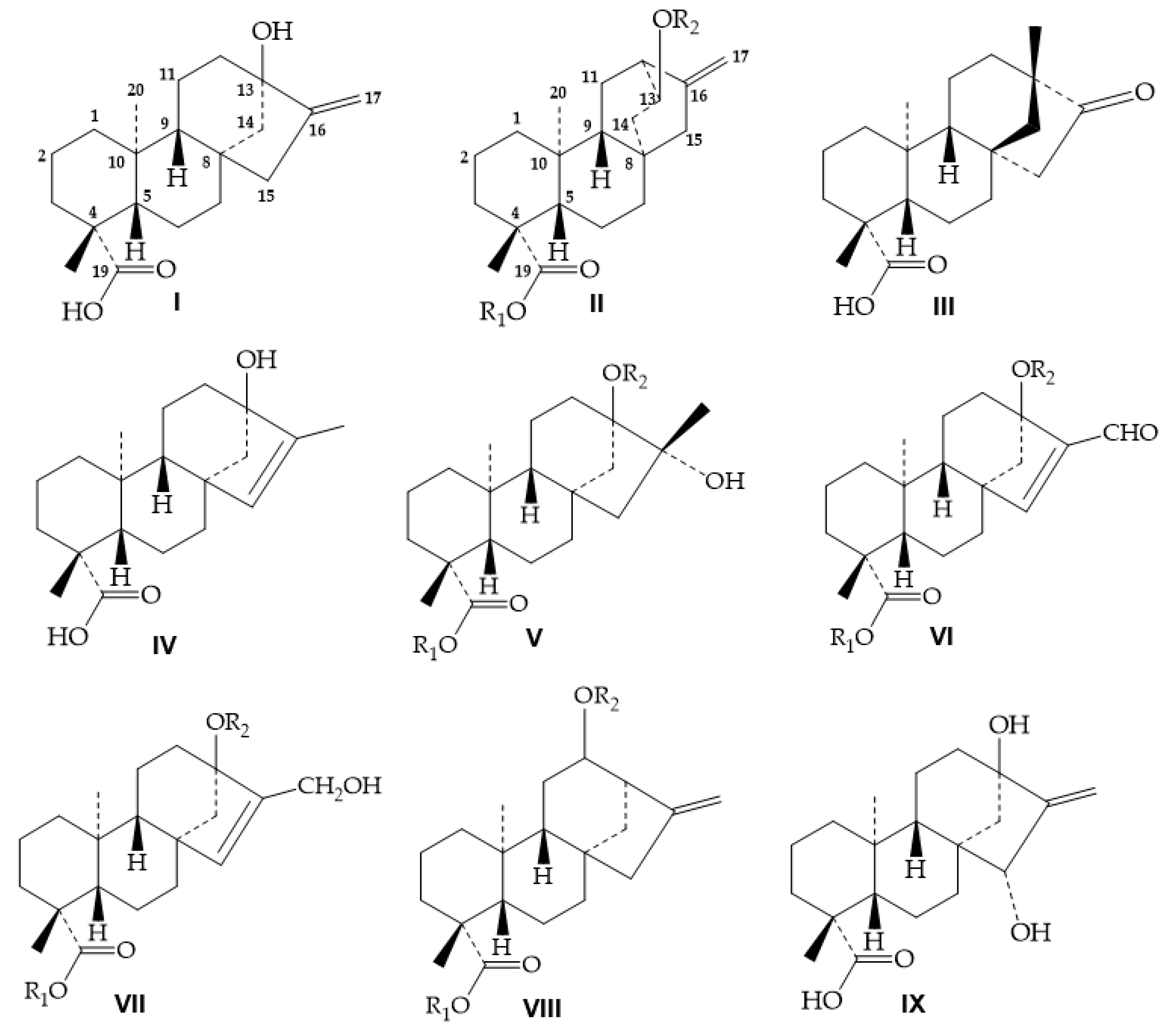
4. Structure Elucidation for Diterpene Glycosides
4.1. Reversed-Phase High-Performance Liquid Chromatography
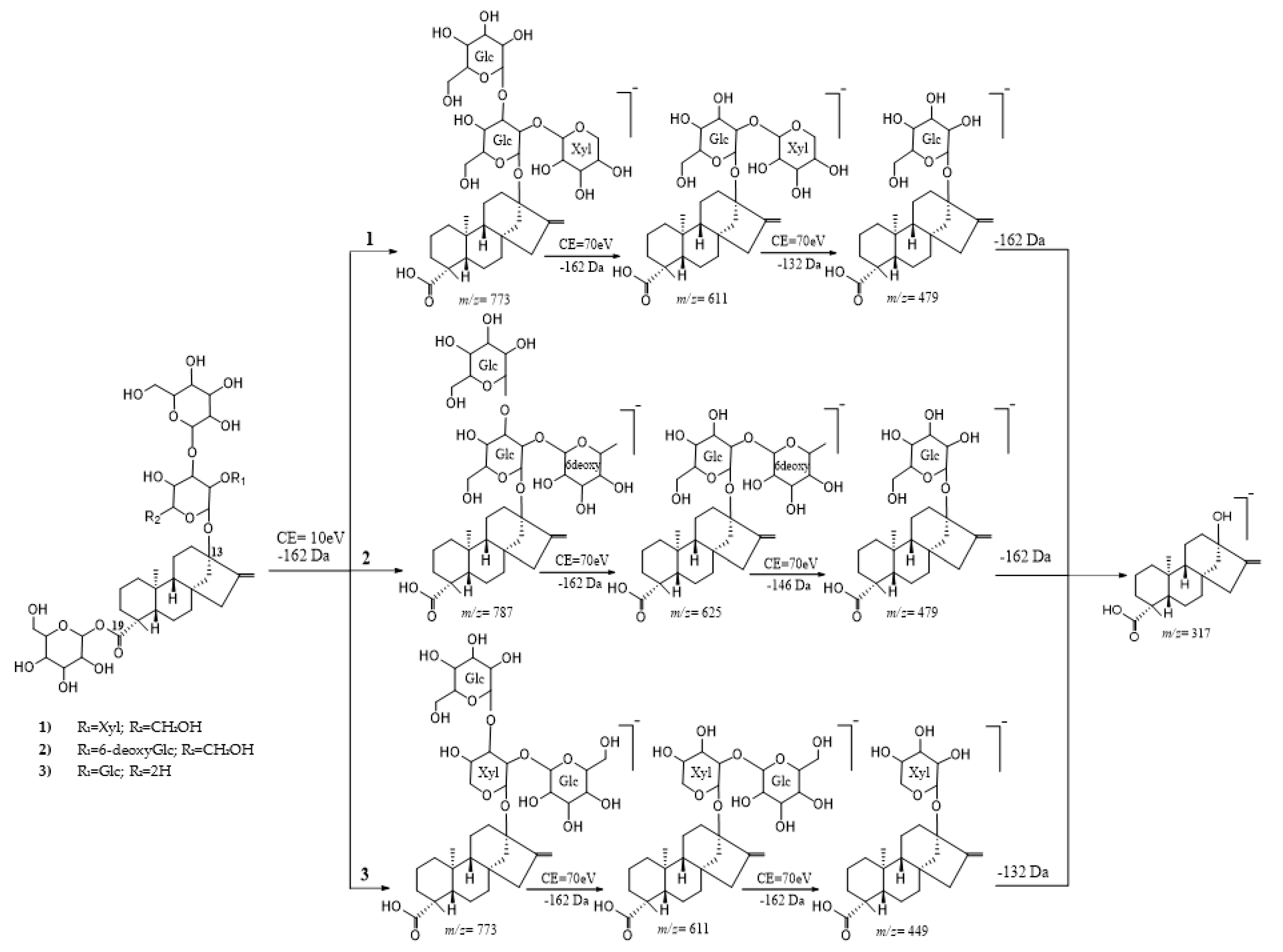
4.2. High-Resolution Electrospray Ionization Tandem Mass Spectrometry
4.2.1. Cleavage of the C-19 Moiety
4.2.2. Cleavage of the C-13 Moiety
4.3. Determination of the Absolute Configuration of the Monosaccharides.
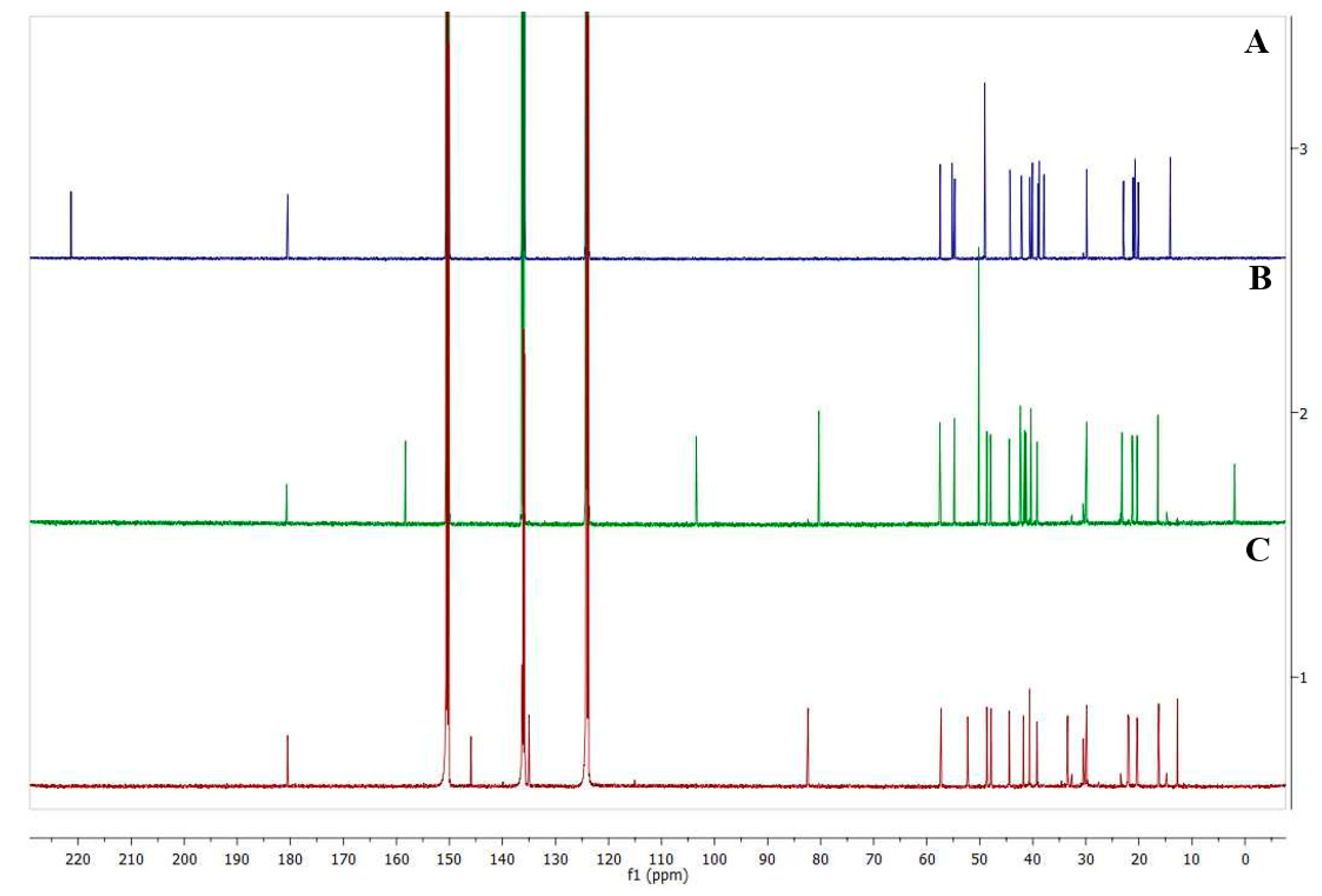
5. NMR Experiments
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
Sample Availability
References
- Kinghorn, A.D. Stevia: The Genus Stevia; Medicinal and Aromatic Plants—Industrial Profiles; Kinghorn, A.D., Ed.; Taylor & Francis: London, UK, 2002; Volume 19, pp. 1–17. [Google Scholar]
- Cioni, P.L.; Morelli, I.; Andolfi, L.; Macchia, M.; Ceccarini, L. Qualitative and quantitative analysis of essential oils of five lines stevia rebaudiana bert. genotypes cultivated in pisa (italy). J. Essent. Oil. Res. 2006, 18, 76–79. [Google Scholar] [CrossRef]
- McGarvey, B.D.; Attygalle, A.B.; Starratt, A.N.; Xiang, B.; Schroeder, F.C.; Brandle, J.E.; Meinwald, J. New Non-Glycosidic Diterpenes from the Leaves of Stevia r Ebaudiana. J. Nat. Prod. 2003, 66, 1395–1398. [Google Scholar] [CrossRef]
- Rajbhandari, A.; Roberts, M.F. The Flavonoids of Stevia Rebaudiana. J. Nat. Prod. 1983, 46, 194–195. [Google Scholar] [CrossRef]
- Karaköse, H.; Jaiswal, R.; Kuhnert, N. Characterization and quantification of hydroxycinnamate derivatives in stevia rebaudiana leaves by LC-MSn. J. Agric. Food Chem. 2011, 59, 18. [Google Scholar] [CrossRef] [PubMed]
- Bridel, M.; Lavielle, R. Sur Le Principe Sucre Des Feuilles de Kaa-He-e (Stevia Rebaundiana B). Acad. Sci. Paris Comptes Rendus 1931, 192, 1123–1125. [Google Scholar]
- Kohda, H.; Kasai, R.; Yamasaki, K.; Murakami, K.; Tanaka, O. New sweet diterpene glucosides from stevia rebaudiana. Phytochemistry 1976, 15, 981–983. [Google Scholar] [CrossRef]
- Kobayashi, M.; Horikawa, S.; Degrandi, I.H.; Ueno, J.; Mitsuhashi, H. Dulcosides A and B, New diterpene glycosides from stevia rebaudiana. Phytochemistry 1977, 16, 1405–1408. [Google Scholar] [CrossRef]
- Sakamoto, I.; Yamasaki, K.; Tanaka, O. Application of 13C NMR spectroscopy to chemistry of plant glycosides: Rebaudiosides-d and-e, new sweet diterpene-glucosides of stevia rebaudiana bertoni. Chem. Pharm. Bull. 1977, 25, 3437–3439. [Google Scholar] [CrossRef]
- Sakamoto, I.; Yamasaki, K.; Tanaka, O. Application of 13C NMR spectroscopy to chemistry of natural glycosides: Rebaudioside-c, a new sweet diterpene glycoside of stevia rebaudiana. Chem. Pharm. Bull. 1977, 25, 844–846. [Google Scholar] [CrossRef]
- Starratt, A.N.; Kirby, C.W.; Pocs, R.; Brandle, J.E. Rebaudioside F, A diterpene glycoside from stevia rebaudiana. Phytochemistry 2002, 59, 367–370. [Google Scholar] [CrossRef]
- Andress, S. Agency Response Letter GRAS Notice No. GRN 000252, CFSAN/Office of Food Additive Safety; Whole Earth Sweeten Company LLC: Chicago, IL, USA, 2008. [Google Scholar]
- Application A540—Steviol Glycosides as Intense Sweeteners. Available online: https://www.foodstandards.gov.au/code/applications/pages/applicationa540stevi3096.aspx (accessed on 21 March 2021).
- The European Commission. Commission Regulation (EU) No. 1131/2011: Amending Annex II to Regulation (EC) No. 1333/2008 of the European Parliament and of the Council with Regard to Steviol Glycosides. Off. J. Eur. Union 2011, 205–295. Available online: https://eur-lex.europa.eu/LexUriServ/LexUriServ.do?uri=OJ:L:2011:295:0205:0211:EN:PDF (accessed on 21 March 2021).
- McQuate, R.S. Agency Response Letter GRAS Notice No. GRN000304, CFSAN/Office of Food Additive Safety; GRAS Associates LLC: Bend, OR, USA, 2010. [Google Scholar]
- Ohta, M.; Morita, K.; Inoue, A.; Tamai, T.; Fujita, I.; Ohta, M.; Sasa, S. Characterization of novel steviol glycosides from leaves of stevia rebaudiana morita. J. Appl. Glycosci. 2010, 57, 199–209. [Google Scholar] [CrossRef]
- Philippaert, K.; Pironet, A.; Mesuere, M.; Sones, W.; Vermeiren, L.; Kerselaers, S.; Pinto, S.; Segal, A.; Antoine, N.; Gysemans, C.; et al. Steviol glycosides enhance pancreatic Beta-Cell function and taste sensation by potentiation of TRPM5 channel activity. Nat. Commun. 2017, 8, 14733. [Google Scholar] [CrossRef]
- Gu, W.; Rebsdorf, A.; Anker, C.; Gregersen, S.; Hermansen, K.; Geuns, J.M.C.; Jeppesen, P.B. Steviol glucuronide, a metabolite of steviol glycosides, potently stimulates insulin secretion from isolated mouse islets: Studies in vitro. Endocrinol. Diabetes Metab. 2019, 2, e00093. [Google Scholar] [CrossRef] [PubMed]
- Geuns, J.M.C.; Buyse, J.; Vankeirsbilck, A.; Temme, E.H.M.; Compernolle, F.; Toppet, S. Identification of Steviol Glucuronide in Human Urine. J. Agric. Food Chem. 2006, 54, 2794–2798. [Google Scholar] [CrossRef]
- Chen, T.-H.; Chen, S.-C.; Chan, P.; Chu, Y.-L.; Yang, H.-Y.; Cheng, J.-T. Mechanism of the hypoglycemic effect of stevioside, a glycoside of stevia rebaudiana. Planta Med. 2005, 71, 108–113. [Google Scholar] [CrossRef]
- Jeppesen, P.B.; Gregersen, S.; Rolfsen, S.E.D.; Jepsen, M.; Colombo, M.; Agger, A.; Xiao, J.; Kruhøffer, M.; Orntoft, T.; Hermansen, K. Antihyperglycemic and blood pressure-reducing effects of stevioside in the diabetic goto-kakizaki rat. Metabolism 2003, 52, 372–378. [Google Scholar] [CrossRef] [PubMed]
- Ibrahim, M.A.; Rodenburg, D.L.; Alves, K.; Fronczek, F.R.; McChesney, J.D.; Wu, C.; Nettles, B.J.; Venkataraman, S.K.; Jaksch, F. Minor diterpene glycosides from the leaves of stevia rebaudiana. J. Nat. Prod. 2014, 77, 1231–1235. [Google Scholar] [CrossRef] [PubMed]
- Ibrahim, M.A.; Rodenburg, D.L.; Alves, K.; Perera, W.H.; Fronczek, F.R.; Bowling, J.; McChesney, J.D. Rebaudiosides R and S, Minor diterpene glycosides from the leaves of stevia rebaudiana. J. Nat. Prod. 2016, 79, 1468–1472. [Google Scholar] [CrossRef]
- Perera, W.H.; Ghiviriga, I.; Rodenburg, D.L.; Alves, K.; Bowling, J.J.; Avula, B.; Khan, I.A.; McChesney, J.D. Rebaudiosides T and U, Minor C-19 xylopyranosyl and arabinopyranosyl steviol glycoside derivatives from stevia rebaudiana (bertoni) bertoni. Phytochemistry 2017, 135, 106–114. [Google Scholar] [CrossRef]
- Purkayastha, S.; Clos, J.F.; Prakash, I.; Yin, C.S.; Markosyan, A. Additional minor diterpene glycosides from stevia rebaudiana. IOSR J. Appl. Chem. 2019, 12, 48–55. [Google Scholar]
- Perera, W.H.; Ghiviriga, I.; Rodenburg, D.L.; Carvalho, R.; Alves, K.; McChesney, J.D. Development of a high-performance liquid chromatography procedure to identify known and detect novel c-13 oligosaccharide moieties in diterpene glycosides from stevia rebaudiana (bertoni) bertoni (asteraceae): Structure elucidation of rebaudiosides V and W. J. Sep. Sci. 2017, 40, 3771–3781. [Google Scholar] [CrossRef] [PubMed]
- Prakash, I.; Ma, G.; Bunders, C.; Charan, R.D.; Ramirez, C.; Devkota, K.P.; Snyder, T.M. A novel diterpene glycoside with nine glucose units from stevia rebaudiana bertoni. Biomolecules 2017, 7, 10. [Google Scholar] [CrossRef] [PubMed]
- Prakash, I.; Hong, S.; Ma, G.; Bunders, C.; Devkota, K.P.; Charan, R.D.; Ramirez, C.; Snyder, T.M. Complete structure elucidation of new steviol glycosides possessing 9 glucose units isolated from stevia rebaudiana. Nat. Prod. Commun. 2017, 12. [Google Scholar] [CrossRef]
- Ma, G.; Bechman, A.; Bunders, C.; Devkota, K.P.; Charan, R.D.; Ramirez, C.; Snyder, T.M.; Priedemann, C.; Prakash, I. New diterpene glycosides from stevia rebaudiana bertoni: Rebaudioside VIII and Rebaudioside IXd. Nat. Prod. Commun. 2018, 13. [Google Scholar] [CrossRef]
- Prakash, I.; Ma, G.; Bunders, C.; Devkota, K.P.; Charan, R.D.; Ramirez, C.; Snyder, T.M.; Priedemann, C. A new diterpene glycoside: 15α-hydroxy-rebaudioside m isolated from stevia rebaudiana–Indra prakash. Nat. Prod. Commun. 2015, 10, 1177. [Google Scholar] [CrossRef]
- Perera, W.H.; Ghiviriga, I.; Rodenburg, D.L.; Alves, K.; Wiggers, F.T.; Hufford, C.D.; Fronczek, F.R.; Ibrahim, M.A.; Muhammad, I.; Avula, B.; et al. Tetra-Glucopyranosyl diterpene Ent-Kaur-16-En-19-Oic acid and Ent-13(S)-Hydroxyatisenoic acid derivatives from a commercial extract of stevia rebaudiana (bertoni) bertoni. Molecules 2018, 23, 3328. [Google Scholar] [CrossRef]
- Devkota, K.P.; Charan, R.D.; Priedemann, C.; Donovan, R.; Snyder, T.M.; Ramirez, C.; Harrigan, G.; Ma, G.; Prakash, I. Five new ent-atisene glycosides from stevia rebaudiana. Nat. Prod. Commun. 2019, 14. [Google Scholar] [CrossRef]
- Lee, T. Steviol Glycoside Isomers 2009. U.S. Patent No. 7964232 B2, 19 March 2009. [Google Scholar]
- Avent, A.G.; Hanson, J.; Oliveira, B.H. Hydrolysis of the diterpenoid glycoside, stevioside. Phytochemistry 1990, 29, 2712–2715. [Google Scholar] [CrossRef]
- Prakash, I.; Chaturvedula, V.S.P.; Markosyan, A. Structural characterization of the degradation products of a minor natural sweet diterpene glycoside rebaudioside m under acidic conditions. Int. J. Mol. Sci. 2014, 15, 1014–1025. [Google Scholar] [CrossRef] [PubMed]
- Prakash, I.; Clos, J.F.; Chaturvedula, V.S.P. Stability of rebaudioside a under acidic conditions and its degradation products. Food Res. Int. 2012, 48, 65–75. [Google Scholar] [CrossRef]
- Chaturvedula, V.S.P.; Clos, J.F.; Rhea, J.; Milanowski, D.; Mocek, U.; DuBois, G.E.; Prakash, I. Minor diterpenoid glycosides from the leaves of stevia rebaudiana. Phytochem. Lett. 2011, 4, 209–212. [Google Scholar] [CrossRef]
- Perera, W.H.; Docampo, M.L.; Wiggers, F.T.; Hufford, C.D.; Fronczek, F.R.; Avula, B.; Khan, I.A.; McChesney, J.D. Endocyclic double bond isomers and by-products from rebaudioside a and stevioside formed under acid conditions. Phytochem. Lett. 2018, 25, 163–170. [Google Scholar] [CrossRef]
- Prakash, I.; Bunders, C.; Devkota, K.P.; Charan, R.D.; Hartz, R.M.; Sears, T.L.; Snyder, T.M.; Ramirez, C. Degradation products of rubusoside under acidic conditions. Nat. Prod. Commun. 2015, 10. [Google Scholar] [CrossRef]
- Rodenburg, D.L.; Alves, K.; Perera, W.H.; Ramsaroop, T.; Carvalho, R.; McChesney, J.D. Development of hplc analytical techniques for diterpene glycosides from stevia rebaudiana (bertoni) bertoni: Strategies to scale-up. J. Braz. Chem. Soc. 2016, 27, 1406–1412. [Google Scholar]
- Chaturvedula, V.S.P.; Prakash, I. Utilization of RP-HPLC fingerprinting analysis for the identification of diterpene glycosides from stevia rebaudiana. Int. J. Res. Phytochem. Pharmacol. 2011, 1, 88–92. [Google Scholar]
- Prakash Chaturvedula, V.S.; Upreti, M.; Prakash, I. Diterpene glycosides from stevia rebaudiana. Molecules 2011, 16, 3552–3562. [Google Scholar] [CrossRef] [PubMed]
- Chaturvedula, V.S.P.; Prakash, I. Hydrogenation of the exocyclic olefinic bond at C-16/C-17 position of Ent-Kaurane diterpene glycosides of stevia rebaudiana using various catalysts. Int. J. Mol. Sci. 2013, 14, 15669–15680. [Google Scholar] [CrossRef]
- Prakash, I.; Campbell, M.; Chaturvedula, V.S.P. Catalytic hydrogenation of the sweet principles of stevia rebaudiana, rebaudioside b, rebaudioside c, and rebaudioside d and sensory evaluation of their reduced derivatives. Int. J. Mol. Sci. 2012, 13, 15126–15136. [Google Scholar] [CrossRef]
- Brandle, J.E.; Telmer, P.G. Steviol glycoside biosynthesis. Phytochemistry 2007, 68, 1855–1863. [Google Scholar] [CrossRef]
- Ceunen, S.; Geuns, J.M. Steviol glycosides: Chemical Diversity, metabolism, and function. J. Nat. Prod. 2013, 76, 1201–1228. [Google Scholar] [CrossRef]
- Gerwig, G.J.; Te Poele, E.M.; Dijkhuizen, L.; Kamerling, J.P. Stevia glycosides: Chemical and enzymatic modifications of their carbohydrate moieties to improve the sweet-tasting quality. Adv. Carbohydr. Chem. Biochem. 2016, 73, 1–72. [Google Scholar] [CrossRef] [PubMed]
- Ruddat, M.; Heftmann, E.; Lang, A. biosynthesis of steviol. Arch. Biochem. Biophys. 1965, 110, 496–499. [Google Scholar] [CrossRef]
- Mizukami, H.; Shiiba, K.; Ohashi, H. Enzymatic determination of stevioside in stevia rebaudiana. Phytochemistry 1982, 21, 1927–1930. [Google Scholar] [CrossRef]
- Reich, E.; Schibli, A. High.-Performance Thin-Layer Chromatography for the Analysis of Medicinal Plants; Thieme Medical Publisher Inc.: New York, NY, USA, 2007; ISBN 978-1-58890-409-6. [Google Scholar]
- Jarne, C.; Savirón, M.; Lapieza, M.P.; Membrado, L.; Orduna, J.; Galbán, J.; Garriga, R.; Morlock, G.E.; Cebolla, V.L. High-Performance thin-layer chromatography coupled with electrospray ionization tandem mass spectrometry for identifying neutral lipids and sphingolipids in complex samples. J. AOAC Int. 2018, 101, 1993–2000. [Google Scholar] [CrossRef]
- Jarne, C.; Cebolla, V.L.; Membrado, L.; Galbán, J.; Savirón, M.; Orduna, J.; Garriga, R. Separation and profiling of monoglycerides in biodiesel using a hyphenated technique based on high-performance thin-layer chromatography. Fuel 2016, 177, 244–250. [Google Scholar] [CrossRef]
- Barret, L.-A.; Polidori, A.; Bonneté, F.; Bernard-Savary, P.; Jungas, C. A new high-performance thin layer chromatography-based assay of detergents and surfactants commonly used in membrane protein studies. J. Chromatogr. A 2013, 1281, 135–141. [Google Scholar] [CrossRef] [PubMed]
- Wald, J.P.; Morlock, G.E. Quantification of steviol glycosides in food products, stevia leaves and formulations by planar chromatography, including proof of absence for steviol and isosteviol. J. Chromatogr. A 2017, 1506, 109–119. [Google Scholar] [CrossRef]
- Morlock, G.E.; Meyer, S.; Zimmermann, B.F.; Roussel, J.-M. High-Performance thin-layer chromatography analysis of steviol glycosides in stevia formulations and sugar-free food products, and benchmarking with (ultra) high-performance liquid chromatography. J. Chromatogr. A 2014, 1350, 102–111. [Google Scholar] [CrossRef]
- Wölwer-Rieck, U. The leaves of stevia rebaudiana (bertoni), their constituents and the analyses thereof: A review. J. Agric. Food Chem. 2012, 60, 886–895. [Google Scholar] [CrossRef]
- Kolb, N.; Herrera, J.L.; Ferreyra, D.J.; Uliana, R.F. Analysis of sweet diterpene glycosides from stevia rebaudiana: Improved HPLC method. J. Agric. Food Chem. 2001, 49, 4538–4541. [Google Scholar] [CrossRef] [PubMed]
- Perera, W.H.; Ramsaroop, T.; Carvalho, R.; Rodenburg, D.L.; McChesney, J.D. A silica gel orthogonal high-performance liquid chromatography method for the analyses of steviol glycosides: Novel tetra-glucopyranosyl steviol. Nat. Prod. Res. 2019, 33, 1876–1884. [Google Scholar] [CrossRef] [PubMed]
- McChesney, J.D.; Rodenburg, D.L. Preparative chromatography and natural products discovery. Curr. Opin. Biotechnol. 2014, 25, 111–113. [Google Scholar] [CrossRef] [PubMed]
- McChesney, J.D.; Rodenburg, D.L. Chromatography Methods 2014. U.S Patent 8801924 B2, 12 August 2014. [Google Scholar]
- Minne, V.J.; Compernolle, F.; Toppet, S.; Geuns, J.M. Steviol quantification at the picomole level by high-performance liquid chromatography. J. Agric. Food Chem. 2004, 52, 2445–2449. [Google Scholar] [CrossRef] [PubMed]
- Chaturvedula, V.S.P.; Kinger, K.P.; Campbell, M.R.; Prakash, I. NMR spectral assignments of steviol and steviol monoacetate. J. Chem. Pharm. Res. 2012, 4, 2666–2670. [Google Scholar]
- Gardana, C.; Scaglianti, M.; Simonetti, P. Evaluation of steviol and its glycosides in stevia rebaudiana leaves and commercial sweetener by ultra-high-performance liquid chromatography-mass spectrometry. J. Chromatogr. A 2010, 1217, 1463–1470. [Google Scholar] [CrossRef]
- Chaturvedula, V.S.P.; Prakash, I. Structure elucidation of three new diterpene glycosides from stevia rebaudiana. Int. J. Phys. Sci. 2011, 6, 6698–6705. [Google Scholar]
- Chaturvedula, V.S.P.; Rhea, J.; Milanowski, D.; Mocek, U.; Prakash, I. Two minor diterpene glycosides from the leaves of stevia rebaudiana. Nat. Prod. Commun. 2011, 6. [Google Scholar] [CrossRef]
- Chaturvedula, V.S.P.; Meneni, S.R. A new penta β-D-Glucopyranosyl diterpene from stevia rebaudiana. Int. J. Org. Chem. 2017, 7, 91–98. [Google Scholar] [CrossRef]
- Chaturvedula, V.S.P.; Prakash, I. Additional minor diterpene glycosides from stevia rebaudiana. Nat. Prod. Commun. 2011, 6. [Google Scholar] [CrossRef]
- Prakash, I.; Chaturvedula, V.S.P. Additional minor diterpene glycosides from stevia rebaudiana bertoni. Molecules 2013, 18, 13510–13519. [Google Scholar] [CrossRef]
- Chaturvedula, V.S.P. Indra prakash diterpene glycosides from stevia rebaudiana. J. Med. Plants Res. 2011, 5, 4838–4842. [Google Scholar]
- Chaturvedula, V.S.P.; Upreti, M.; Prakash, I. Structures of the novel α-glucosyl linked diterpene glycosides from stevia rebaudiana. Carbohydr. Res. 2011, 346, 2034–2038. [Google Scholar] [CrossRef]
- Chaturvedula, V.S.P.; Prakash, I. Structures of the novel diterpene glycosides from stevia rebaudiana. Carbohydr. Res. 2011, 346, 1057–1060. [Google Scholar] [CrossRef]
- Chaturvedula, V.; Rhea, J.; Milanowski, D.; Mocek, U.; Prakash, I. Isolation and structure elucidation of two new minor diterpene glycosdies from stevia rebaudiana. Org. Chem. Curr. Res. 2011, 1, 2161-0401. [Google Scholar]
- Zimmermann, B.F. Tandem mass spectrometric fragmentation patterns of known and new steviol glycosides with structure proposals—zimmermann—2011—Rapid communications in mass spectrometry. Rapid Commun. Mass. Spectrom. 2011, 25, 1575–1582. [Google Scholar] [CrossRef] [PubMed]
- Perera, W.H.; Avula, B.; Khan, I.A.; McChesney, J.D. Assignment of sugar arrangement in branched steviol glycosides using electrospray ionization quadrupole time-of-flight tandem mass spectrometry. Rapid Commun. Mass. Spectrom. 2017, 31, 315–324. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, T.; Nakashima, T.; Ueda, T.; Tomii, K.; Kouno, I. Facile discrimination of aldose enantiomers by reversed-phase HPLC. Chem. Pharm. Bull. 2007, 55, 899–901. [Google Scholar] [CrossRef]
- Wang, Y.-H.; Avula, B.; Fu, X.; Wang, M.; Khan, I.A. Simultaneous determination of the absolute configuration of twelve monosaccharide enantiomers from natural products in a single injection by a UPLC-UV/MS method. Planta Med. 2012, 78, 834–837. [Google Scholar] [CrossRef] [PubMed]
- Perera, W.H.; Shivanagoudra, S.R.; Pérez, J.L.; Kim, D.M.; Sun, Y.K.; Jayaprakasha, G.S.; Patil, B. Anti-Inflammatory, antidiabetic properties and in silico modeling of cucurbitane-type triterpene glycosides from fruits of an indian cultivar of Momordica charantia L. Molecules 2021, 26, 1038. [Google Scholar] [CrossRef]
- Tangpaisarnkul, N.; Tuchinda, P.; Wilairat, P.; Siripinyanond, A.; Shiowattana, J.; Nobsathian, S. Development of pure certified reference material of stevioside. Food Chem. 2018, 255, 75–80. [Google Scholar] [CrossRef] [PubMed]
- Steinmetz, W.E.; Lin, A. NMR studies of the conformation of the natural sweetener rebaudioside A. Carbohydr. Res. 2009, 344, 2533–2538. [Google Scholar] [CrossRef] [PubMed]
| DGs from Alkaline Hydrolysis | |||||||
|---|---|---|---|---|---|---|---|
| Common Name | Oligosaccharide Moieties | AS | Chemical Formula | Accurate MW | Starting Material | Ref | |
| C-13 | C-19 | ||||||
| - | Xylβ(1-2)Glcβ1- | - | I | C31H48O12 | 612.3146 | a | [26] |
| Dulcoside A1 | Rhaα(1-2)Glcβ1- | - | I | C32H50O12 | 626.3303 | Dulcoside A | [26] |
| Rebaudioside G1 | Glcβ(1-3)Glcβ1- | - | I | C32H50O13 | 642.3252 | Rebaudioside G | [26] |
| Rebaudioside F1 | Xylβ(1-2)[Glcβ(1-3)]Glcβ1- | - | I | C37H58O17 | 774.3675 | Rebaudioside F | [11,26] |
| Rebaudioside R1 | Glc(1-2)[Glcβ(1-3)]Xylβ1- | - | I | C37H58O17 | 774.3675 | Rebaudioside R | [26] |
| Rebaudioside Z1 | Glcβ(1-6)[Glcβ(1-2)]Glcβ1- | - | I | C38H60O18 | 804.3781 | Rebaudioside Z | [31] |
| - | Glcβ(1-2)[Glcβ(1-3)]Glcβ1- | II | C38H60O18 | 804.3781 | - | [31] | |
| - | 6-deoxyGlcβ(1-2)[Glcβ(1-3)]Glcβ1- | - | I | C38H60O17 | 788.3831 | b | [26] |
| Rebaudioside H1 | Glcβ(1-6)Glcβ(1-3)[Glcβ(1-3)]Glcβ1- | - | I | C44H70O23 | 966.4309 | Rebaudioside H | [26] |
| Rebaudioside L1 | Glcβ(1-3)Rhaα(1-2)[Glcβ(1-3)]Glcβ1- | - | I | C44H70O23 | 966.4309 | Rebaudioside L | [26] |
| DGs from acid hydrolysis | |||||||
| Isosteviol | - | - | III | C20H30O3 | 318.2195 | Rebaudioside A/Stevioside | [34,38] |
| Endo-steviol | - | - | IV | C20H30O3 | 318.2195 | Rebaudioside A/Stevioside | [34,38] |
| Endo-steviolmonoside | Glcβ1- | - | IV | C26H40O8 | 480.2724 | Rebaudioside A/Rubusoside | [38,39] |
| Endo-rebaudioside G1 | Glcβ(1-3)Glcβ1- | - | IV | C32H50O13 | 642.3252 | Rebaudioside A | [38] |
| Endo-steviolbioside | Glcβ(1-2)Glcβ1- | - | IV | C32H50O13 | 642.3252 | Rebaudioside A | [38] |
| Endo-rubusoside | Glcβ1- | Glcβ1- | IV | C32H50O13 | 642.3252 | Rebaudioside A/Rubusoside | [38,39] |
| Iso-stevioside/Endo-stevioside | Glcβ(1-2)Glcβ1- | Glcβ1- | IV | C38H60O18 | 804.3781 | Rebaudioside A | [38,41] |
| Iso-rebaudioside B/Endo-rebaudioside B | Glcβ(1-2)[Glcβ(1-3)]-Glcβ1- | - | IV | C38H60O18 | 804.3781 | Rebaudioside A | [38,42] |
| - | - | Glcβ(1-2)[Glcβ(1-3)]Glcβ1- | III | C38H60O18 | 804.3781 | Rebaudioside M | [35] |
| Iso-rebaudioside AEndo-rebaudioside A | Glcβ(1-2)[Glcβ(1-3)]Glcβ1- | Glcβ1- | IV | C44H70O23 | 966.4309 | Rebaudioside A | [38] |
| - | Glcβ(1-2)[Glcβ(1-3)]Glcβ1- | Glcβ(1-2)[Glcβ(1-3)]Glcβ1- | IV | C56H90O33 | 1290.5366 | Rebaudioside M | [35] |
| - | Glcβ(1-2)[Glcβ(1-3)]Glcβ1- | Glcβ(1-2)[Glcβ(1-3)]Glcβ1- | V | C56H92O34 | 1308.5472 | Rebaudioside M | [35] |
| Common Name | Oligosaccharide Moieties | AS | Chemical Formula | MW | Ref | |
|---|---|---|---|---|---|---|
| C-13 | C-19 | |||||
| Steviol | H | H | I | C20H30O3 | 318.2195 | [61] |
| Steviol monoacetate | H | COCH3 | I | C22H32O4 | 360.2192 | [62] |
| Steviolmonoside | Glcβ1- | H | I | C26H40O8 | 480.2724 | [16] |
| Steviol-19-O-β-D glucoside | H | Glcβ1- | I | C26H40O8 | 480.2724 | [63] |
| Rubusoside | Glcβ1- | Glcβ1- | I | C32H50O13 | 642.3252 | [16] |
| Oxidized core family | ||||||
| Isosteviol-19-O-β-glucoside | - | Glcβ1- | III | C26H40O8 | 480.2724 | [42] |
| - | Glcβ(1-2)Glcβ1- | Glcβ1- | VI | C38H58O19 | 818.3573 | [37] |
| - | Glcβ(1-2)Glcβ1- | Glcβ1- | VII | C38H60O19 | 820.3728 | [37] |
| - | Glcβ(1-2)[Glcβ(1-3)]Glcβ1- | Glcβ1- | VII | C44H70O24 | 982.4258 | [64] |
| - | Glcβ(1-2)[Glcβ(1-3)]Glcβ1- | Glcβ1- | IV | C44H72O24 | 984.4415 | [41] |
| Rebaudioside A family | ||||||
| Steviolbioside | Glcβ(1-2)Glcβ1- | H | I | C32H50O13 | 642.3252 | [7] |
| - | Glcβ1- | Glcβ(1-2)Glcβ1- | I | C38H60O18 | 804.3781 | [22] |
| Rebaudioside KA | Glcβ(1-2)Glcβ1- | Glcβ1- | VIII | C38H60O18 | 804.3781 | [22] |
| Stevioside | Glcβ(1-2)Glcβ1- | Glcβ1- | I | C38H60O18 | 804.3781 | [6] |
| Rebaudioside B | Glcβ(1-2)[Glcβ(1-3)]Glcβ1- | H | I | C38H60O18 | 804.3781 | [7] |
| Rebaudioside E | Glcβ(1-2)Glcβ1- | Glcβ(1-2)Glcβ1- | I | C44H70O23 | 966.4309 | [9] |
| Rebaudioside A | Glcβ(1-2)[Glcβ(1-3)]Glcβ1- | Glcβ1- | I | C44H70O23 | 966.4309 | [7] |
| - | Glcβ(1-6)Glcβ(1-2)Glcβ1- | Glcβ1- | I | C44H70O23 | 966.4309 | [65] |
| Rebaudioside Y | Glcβ(1-2)Glcβ1- | Glcβ(1-6)Glcβ1- | I | C44H70O23 | 966.4309 | [58] |
| Rebaudioside Z | Glcβ(1-6)[Glcβ(1-2)]Glcβ1- | Glcβ1- | I | C44H70O23 | 966.4309 | [31] |
| Rebaudioside D | Glcβ(1-2)[Glcβ(1-3)]Glcβ1- | Glcβ(1-2)Glcβ1- | I | C50H80O28 | 1128.4838 | [9] |
| Rebaudioside I | Glcβ(1-2)[Glcβ(1-3)]Glcβ1- | Glcβ(1-3)Glcβ1- | I | C50H80O28 | 1128.4838 | [16] |
| Rebaudioside M | Glcβ(1-2)[Glcβ(1-3)]Glcβ1- | Glcβ(1-2)[Glcβ(1-3)]Glcβ1- | I | C56H90O33 | 1128.4838 | [16] |
| Rebaudioside L | Glcβ(1-6)Glcβ(1-2)[Glcβ(1-3)]Glcβ1- | Glcβ1- | I | C50H80O28 | 1128.4838 | [16] |
| - | Glcβ(1-6)[Glcβ(1-3)]Glcβ1- | Glcβ(1-2)Glcβ1- | I | C50H80O28 | 1128.4838 | [41] |
| - | Glcβ(1-2)Glcβ1- | Glcβ(1-6)[Glcβ(1-2)Glcβ1- | I | C50H80O28 | 1128.4838 | [66] |
| 15α-hydroxy-Reb M | Glcβ(1-2)[Glcβ(1-3)]Glcβ1- | Glcβ(1-2)[Glcβ(1-3)]Glcβ1- | IX | C56H90O34 | 1306.5313 | [30] |
| Rebaudioside VIII and IX families | ||||||
| Rebaudioside VIII | Glcβ(1-2)[Glcβ(1-3)]-Glcα(1-6)-Glcβ(1-2)[Glcβ(1-3)]Glcβ1- | Glcβ(1-2)Glcβ1- | I | C68H110O43 | 1614.6420 | [29] |
| Rebaudioside IX | Glcβ(1-2)[Glcβ(1-3)]-Glcα(1-6)-Glcβ(1-2)[Glcβ(1-3)]Glcβ1- | Glcβ(1-2)[Glcβ(1-3)]Glcβ1- | I | C74H119O48 | 1776.6949 | [27] |
| Rebaudioside IXa | Glcβ(1-2)[Glcβ(1-3)]Glcβ1- | Glcβ(1-2)[Glcβ(1-3)]Glcβ(1-2)[Glcβ(1-3)]-Glcα(1-6)-Glcβ1- | I | C74H119O48 | 1776.6949 | [28] |
| Rebaudioside IXb | Glcβ(1-2)[Glcβ(1-3)]Glcβ1- | Glcβ(1-2)[Glcβ(1-3)]-Glcα(1-6)-Glcβ(1-2)[Glcβ(1-3)]Glcβ1- | I | C74H119O48 | 1776.6949 | [28] |
| Rebaudioside IXc | Glcβ(1-2)[Glcβ(1-3)]Glcβ1- | Glcβ(1-2)[Glcβ(1-2)[Glcβ(1-3)]-Glcα(1-3)-Glcβ(1-3)]Glcβ1- | I | C74H119O48 | 1776.6949 | [28] |
| Rebaudioside IXd | Glcβ(1-2)[Glcβ(1-3)]Glcβ(1-2)[Glcβ(1-3)]-Glcα(1-6)-Glcβ1- | Glcβ(1-2)[Glcβ(1-3)]Glcβ1- | I | C74H119O48 | 1776.6949 | [29] |
| Rebaudioside C family | ||||||
| Dulcoside A | Rhaα(1-2)Glcβ1- | Glcβ1- | I | C38H60O17 | 788.3831 | [8] |
| Dulcoside B | Rhaα(1-2)[Glcβ(1-3)]Glcβ1- | H | I | C38H60O17 | 788.3831 | [16] |
| Rebaudioside C/Dulcoside B | Rhaα(1-2)[Glcβ(1-3)]Glcβ1- | Glcβ1- | I | C44H70O22 | 950.4356 | [8,10] |
| Rebaudioside S | Glcα(1-2)Glcβ1- | Rhaα(1-2)Glcβ1- | I | C44H70O22 | 950.4356 | [23] |
| - | Glcβ(1-2)Glcβ1- | Rhaα(1-2)Glcβ1- | I | C44H70O22 | 950.4358 | [25] |
| Rebaudioside H | Glcβ(1-3)Rhaα(1-2)[Glcβ(1-3)]Glcβ1- | Glcβ1- | I | C50H80O27 | 1112.4888 | |
| Rebaudioside K | Rhaα(1-2)[Glcβ(1-3)]Glcβ1- | Glcβ(1-2)Glcβ1- | I | C50H80O27 | 1112.4888 | [16] |
| Rebaudioside J | Glcβ(1-2)[Glcβ(1-3)]Glcβ1- | Rhaα(1-2)Glcβ1- | I | C50H80O27 | 1112.4888 | [16] |
| Rebaudioside C family | ||||||
| - | Rhaα(1-2)[Glcβ(1-3)]Glcβ1- | Glcβ(1-6)Glcβ1- | I | C50H80O27 | 1112.4887 | [25] |
| Rebaudioside N | Glcβ(1-2)[Glcβ(1-3)]Glcβ1- | Rhaα(1-2)[Glcβ(1-3)]Glcβ1- | I | C56H90O32 | 1274.5417 | [16] |
| Rebaudioside O | Glcβ(1-2)[Glcβ(1-3)]Glcβ1- | Glcβ(1-3)Rhaα(1-2)[Glcβ(1-3)]Glcβ1- | I | C62H100O37 | 1436.5945 | [16] |
| Glcβ(1-3)Glcβ1- family | ||||||
| Rebaudioside G | Glcβ(1-3)Glcβ1- | Glcβ1- | I | C38H60O18 | 788.3831 | [16] |
| Rebaudioside F family | ||||||
| - | Xylβ(1-2)Glcβ1- | Glcβ1- | I | C37H58O17 | 774.3675 | [67] |
| Rebaudioside F | Xylβ(1-2)[Glcβ(1-3)]Glcβ1- | Glcβ1- | I | C43H68O22 | 936.4203 | [11] |
| - | Glcβ(1-2)[Xylβ(1-3)]Glcβ1- | Glcβ1- | I | C43H68O22 | 936.4203 | [67] |
| - | Glcβ(1-2)Glcβ1- | Xylβ(1-6)Glcβ1- | I | C43H68O22 | 936.4203 | [37] |
| Rebaudioside R | Glcβ(1-2)[Glcβ(1-3)]Xylβ1- | Glcβ1- | I | C43H68O22 | 936.4203 | [23] |
| Rebaudioside V | Xylβ(1-2)[Glcβ(1-3)]Glcβ1- | Glcβ(1-2)Glcβ1- | I | C49H78O27 | 1098.4730 | [26] |
| - | Glcβ(1-2)[Glcβ(1-3)]Glcβ1- | Xylβ(1-2)Glcβ1- | I | C49H78O27 | 1098.4730 | [25] |
| - | Glcβ(1-2)Glcβ1- | Xylβ(1-2)[Glcβ(1-4)]Glcβ1- | I | C49H78O27 | 1098.4730 | [25] |
| Rebaudioside T | Glcβ(1-2)[Glcβ(1-3)]Glcβ1- | Xylβ(1-2)[Glcβ(1-3)]Glcβ1- | I | C55H88O32 | 1260.5260 | [24,68] |
| - | Glcβ(1-2)[Xylβ (1-3)]Glcβ1- | Glcβ(1-2)[Glcβ(1-3)]Glcβ1- | I | C55H88O32 | 1260.5260 | [25] |
| Fruβ family | ||||||
| - | Glcβ(1-2)[Fruβ(1-3)]Glcβ1- | Glcβ1- | I | C44H70O23 | 966.4309 | [65] |
| Glcα family | ||||||
| - | Glcβ(1-2)Glcβ1- | Glcα(1-2)[Glcα(1-4)]-Glcβ1- | I | C50H80O28 | 1128.4838 | [69] |
| - | Glcα(1-3)Glcβ(1-2)[Glcβ(1-3)]Glcβ1- | Glcβ1- | I | C50H80O28 | 1128.4838 | [70] |
| - | Glcα(1-4)Glcβ(1-3)[Glcβ(1-2)]Glcβ1- | Glcβ1- | I | C50H80O28 | 1128.4838 | [70] |
| 6-deoxyGlcβ family | ||||||
| - | 6-deoxyGlcβ(1-2)Glcβ1- | Glcβ1- | I | C38H60O17 | 788.3831 | [71] |
| - | 6-deoxyGlcβ(1-2)[Glcβ(1-3)]Glcβ1- | Glcβ1- | I | C44H70O22 | 950.4356 | [71] |
| - | Glcβ(1-2)[Glcβ(1-3)]Glcβ1- | 6-deoxyGlcβ1- | I | C44H70O22 | 950.4356 | [72] |
| Rebaudioside W family (three different sugar moieties) | ||||||
| Rebaudioside W | Xylβ(1-2)[Glcβ(1-3)]-Glcβ1- | Rhaα(1-2)[Glcβ(1-3)]Glcβ1- | I | C55H88O31 | 1244.5311 | [26] |
| Rebaudioside U Araα family | ||||||
| Rebaudioside U/6 | Glcβ(1-2)[Glcβ(1-3)]Glcβ1- | Araα(1-6)Glcβ1- | I | C49H78O27 | 1098.4730 | [24,25] |
| - | Glcβ(1-2)[Glcβ(1-3)]Glcβ1- | Araα(1-6)[Glcβ(1-2)]Glcβ1- | I | C55H88O32 | 1260.5258 | [25] |
| Ent-atisene-core family | ||||||
| - | Glcβ(1-2)[Glcβ(1-3)]Glcβ1- | Glcβ1- | II | C44H70O23 | 966.4309 | [31] |
| Stevatisene J | Glcβ(1-2)[Glcβ(1-3)]Glcβ1- | Rhaα(1-2)Glcβ1- | II | C50H80O27 | 1112.4888 | [32] |
| Stevatisene K | Rhaα(1-2)[Glcβ(1-3)]Glcβ1- | Glcβ(1-2)Glcβ1- | II | C50H80O27 | 1112.4888 | [32] |
| Stevatisene T | Glcβ(1-2)[Glcβ(1-3)]Glcβ1- | Xylβ(1-2)[Glcβ(1-3)]Glcβ1- | II | C55H88O32 | 1260.5260 | [32] |
| Stevatisene N | Glcβ(1-2)[Glcβ(1-3)]Glcβ1- | Rhaα(1-2)[Glcβ(1-3)]Glcβ1- | II | C56H90O32 | 1274.5417 | [32] |
| Stevatisene O | Glcβ(1-2)[Glcβ(1-3)]Glcβ1- | Glcβ(1-3)Rhaα(1-2)[Glcβ(1-3)]Glcβ1- | II | C62H100O37 | 1436.5945 | [32] |
| DG | RT (min) | ΔRT (min) | Oligosaccharide Positions | |
|---|---|---|---|---|
| C-13 | C-19 | |||
| Reb O | 2.92 | 0.16 | Glcβ(1-2)[Glcβ(1-3)]Glcβ1- | Glcβ(1-3) Rhaα(1-2)[Glcβ(1-3)]-Glcβ1- |
| Reb N | 3.08 | - | Glcβ(1-2)[Glcβ(1-3)]Glcβ1- | Rhaα(1-2) [Glcβ(1-3)]-Glcβ1- |
| Reb E | 3.08 | 0.18 | Glcβ1(1-2)Glcβ1- | Glcβ1(1-2)Glcβ1 - |
| Reb D | 3.26 | 0.23 | Glcβ(1-2)[Glcβ(1-3)]Glcβ1- | Glcβ1(1-2)Glcβ1 - |
| Reb J | 3.49 | 0.07 | Glcβ(1-2)[Glcβ(1-3)]Glcβ1- | Rhaα (1-2)Glcβ1- |
| Reb W | 3.56 | 0.20 | Xylβ(1-2)[Glcβ(1-3)]Glcβ1- | Rhaα(1-2) [Glcβ(1-3)]-Glcβ1- |
| Reb M | 3.76 | 0.04 | Glcβ(1-2)[Glcβ(1-3)]Glcβ1- | Glcβ(1-2)[Glcβ(1-3)]-Glcβ1- |
| Reb Y | 3.80 | 0.09 | Glcβ(1-2)Glcβ1- | Glcβ1(1-6)Glcβ1- |
| Reb V | 3.89 | 0.05 | Xylβ(1-2)[Glcβ(1-3)]Glcβ1- | Glcβ1(1-2)Glcβ1 - |
| Reb Z | 3.94 | 0.24 | Glcβ(1-2)[Glcβ(1-6)]-Glcβ1- | Glcβ1- |
| Reb U | 4.18 | 0.17 | Glcβ(1-2)[Glcβ(1-3)]Glcβ1- | Araα(1-6)-Glcβ1- |
| a | 4.35 | 0.50 | Glcβ(1-2)[Glcβ(1-3)]Glcβ1- | Glcβ1- |
| Reb T | 4.85 | 0.61 | Glcβ(1-2)[Glcβ(1-3)]Glcβ1- | Xylβ(1-2)[Glcβ(1-3)]-Glcβ1- |
| Reb H | 5.46 | 0.18 | Glcβ(1-3)Rhaα(1-2)[Glcβ(1-3)]Glcβ1- | Glcβ1- |
| Reb L | 5.64 | 0.71 | Glcβ(1-6)Glcβ(1-3)[Glcβ(1-3)]Glcβ1- | Glcβ1- |
| Reb I | 6.35 | 0.76 | Glcβ(1-2)[Glcβ(1-3)]Glcβ1- | Glcβ1(1-3)Glcβ1- |
| Reb A | 7.11 | 0.42 | Glcβ(1-2)[Glcβ(1-3)]Glcβ1- | Glcβ1- |
| Stev | 7.53 | 0.38 | Glcβ(1-2)Glcβ1- | Glcβ1- |
| Iso-Reb A | 7.91 | 0.65 | Glcβ(1-2)[Glcβ(1-3)]Glcβ1- | Glcβ1- |
| Iso-Stev | 8.56 | 0.52 | Glcβ(1-2)Glcβ1- | Glcβ1- |
| Reb F | 9.08 | 0.24 | Xylβ(1-2)[Glcβ(1-3)]Glcβ1- | Glcβ1- |
| Reb Z1 | 9.32 | 0.47 | Glcβ(1-2)[Glcβ(1-6)]Glcβ1- | H- |
| Reb C | 9.79 | 0.13 | Rhaα(1-2)[Glcβ(1-3)]Glcβ1- | Glcβ1- |
| Reb R | 9.92 | 0.34 | Glc(1-2)[Glcβ(1-3)]Xylβ1- | Glcβ1- |
| b | 10.26 | 0.16 | Xylβ(1-2)Glcβ1- | Glcβ1- |
| Dulc A | 10.42 | 0.08 | Rhaα(1-2)Glcβ1- | Glcβ1- |
| c | 10.50 | 0.69 | Glcβ(1-2)[Glcβ(1-3)]Glcβ1- | - |
| Reb G | 11.19 | 0.26 | Glcβ(1-3)Glcβ1- | Glcβ1- |
| d | 11.45 | 0.56 | 6-deoxyGlcβ(1-2)[Glcβ(1-3)]Glcβ1- | Glcβ1- |
| Reb L1 | 12.01 | 0.28 | Glcβ(1-3) Rhaα(1-2)[Glcβ(1-3)]Glcβ1- | H- |
| Reb H1 | 12.29 | 0.23 | Glcβ(1-6) Glcβ(1-3)[Glcβ(1-3)]Glcβ1- | H- |
| Rub | 12.52 | 1.80 | Glcβ1- | Glcβ1- |
| Reb B | 14.32 | 0.12 | Glcβ(1-2)[Glcβ(1-3)]Glcβ1- | H- |
| Iso-Reb B | 14.44 | 0.44 | Glcβ(1-2)[Glcβ(1-3)]Glcβ1- | H- |
| Stev-bio | 14.88 | 0.29 | Glcβ(1-2)Glcβ1- | H- |
| Iso-Stevbio | 15.17 | 0.15 | Glcβ(1-2)Glcβ1- | H- |
| Dulc B | 15.31 | 0.52 | Rhaα(1-2)[Glcβ(1-3)]Glcβ1- | H- |
| Reb F1 | 15.83 | 0.61 | Xylβ(1-2)[Glcβ(1-3)]Glcβ1- | H- |
| Dulc A1 | 16.45 | 0.12 | Rhaα(1-2)Glcβ1- | H- |
| Reb R1 | 16.57 | 0.70 | Glc(1-2)[Glcβ(1-3)]Xylβ1- | H- |
| e | 17.26 | 0.02 | Xylβ(1-2)Glcβ1- | H- |
| Reb G1 | 17.28 | 0.1 | Glcβ(1-3)Glcβ1- | H- |
| f | 17.36 | 2.19 | 6-deoxyGlcβ(1-2)[Glcβ(1-3)]Glcβ1- | H- |
| Stev-mono | 19.55 | Glcβ1- | H- | |
| Position | 1 (a) | 2 (b) | 3 (a) | 4 (a) | 5 (a) | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| δH | δC | δH | δC | δH | δC | δC | δH | δC | ||
| 1 | 0.87; 1.87 | 41.5 | 0.79 | 40.2 | 0.92; 1.92 | 41.8 | 0.78; 1.75 | 40.3 | 0.88; 1.67 | 40.6 |
| 2 | 2.28; 1.52 | 20.3 | 2.13; 1.38 | 19.7 | 2.32; 1.56 | 20.4 | 1.34; 2.23 | 19.3 | 2.23; 1.46 | 20.1 |
| 3 | 2.05; 1.87 | 41.3 | 2.36 | 39.1 | 2.50; 1.11 | 39.2 | 1.00; 2.32 | 38.4 | 1.61; 1.33 | 37.9 |
| 4 | - | 44.4 | - | 44.6 | - | 44.4 | - | 43.8 | - | 44.3 |
| 5 | 1.11 | 57.5 | 1.03 | 57.9 | 1.14 | 57.3 | 1.04 | 57.1 | 1.14 | 57.4 |
| 6 | 2.07; 2.21 | 23.2 | 2.41; 1.84 | 21.1 | 2.23; 2.07 | 22.1 | 2.11; 2.43 | 23.1 | 2.04; 2.04 | 22.9 |
| 7 | 1.55; 1.45 | 42.4 | 2.78 | 39.6 | 1.90; 1.79 | 33.4 | 1.37; 1.88 | 42.8 | 1.66; 1.66 | 42.1 |
| 8 | - | 42.3 | - | 34.9 | - | 48.7 | - | NA | - | 49.1 |
| 9 | 1.02 | 54.8 | 1.01 | 51.7 | 0.97 | 47.9 | 0.84 | 54.8 | 1.14 | 55.2 |
| 10 | - | 40.3 | - | 44.6 | - | 40.6 | - | NA | - | 38.8 |
| 11 | 1.79; 1.55 | 21.3 | 1.59 | 27.3 | 1.72; 1.72 | 21.9 | 1.52; 1.71 | 19.8 | 1.60; 1.20 | 21.1 |
| 12 | 2.48; 1.09 | 39.2 | 4.11 | 78.4 | 1.75; 1.60 | 40.6 | 1.85; 2.67 | 31.6 | 2.45; 1.09 | 39.0 |
| 13 | - | 80.3 | 2.94 | 42.0 | - | 82.4 | - | 87.6 | - | 40.1 |
| 14 | 2.35; 1.58 | 48.7 | 1.42; 1.05 | 39.9 | 2.47; 1.77 | 52.3 | 2.44; 2.58 | 40.3 | 1.49; 1.38 | 54.7 |
| 15 | 2.25; 2.22 | 48.0 | 2.23; 1.91 | 49.0 | 5.24 | 135.0 | 1.41; 1.83 | 54.3 | 2.71; 2.66 | 49.1 |
| 16 | - | 158.3 | - | 148.4 | - | 145.9 | - | 77.1 | - | 221.3 |
| 17 | 5.48; 5.04 | 103.5 | 5.24; 4.90 | 109.5 | 1.89 | 12.7 | 1.32 | 22.2 | 1.04 | 20.7 |
| 18 | 1.28 | 29.9 | 1.26 | 29.1 | 1.38 | 29.8 | 1.28 | 27.7 | 1.40 | 29.9 |
| 19 | - | 180.7 | - | 177.4 | - | 180.6 | - | 176.9 | - | 180.7 |
| 20 | 1.20 | 16.4 | 1.04 | 13.3 | 1.24 | 16.3 | 1.31 | 16.0 | 0.98 | 14.1 |
| Position | 6 (b) | 7 (b) | 8 (b) | 9 (b) | ||||||
| δH | δH | δH | δC | δH | δC | δC | δH | |||
| 1 | 0.92; 1.59 | 39.5 | 0.89; 1.88 | 41.7 | 0.86; 1.86 | 41.8 | 0.86; 1.89 | 40.9 | ||
| 2 | 1.92; 1.40 | 18.4 | 1.42; 1.95 | 19.7 | 1.41; 1.96 | 19.8 | 1.45; 2.15 | 20.1 | ||
| 3 | 2.20; 1.09 | 37.7 | 1.07; 2.17 | 38.6 | 1.06; 2.14 | 38.8 | 1.09; 2.06 | 39.0 | ||
| 4 | 1.12 | 43.7 | - | 44.3 | - | 44.7 | - | 45.1 | ||
| 5 | 1.78; 2.05 | 57.1 | 1.18 | 57.6 | 1.12 | 58.0 | 1.07 | 58.2 | ||
| 6 | 1.57; 1.16 | 19.9 | 1.88; 2.05 | 21.1 | 1.83; 2.00 | 21.5 | 1.72; 2.08 | 23.6 | ||
| 7 | 1.09 | 38.8 | 1.66; 1.75 | 39.0 | 1.55; 1.65 | 40.2 | 1.33; 1.68 | 36.7 | ||
| 8 | 1.63; 1.45 | 33.9 | - | 50.6 | - | 49.5 | - | 46.4 | ||
| 9 | 2.51 | 51.0 | 1.00 | 46.1 | 0.90 | 47.7 | 0.94 | 54.3 | ||
| 10 | 4.04 | 38.1 | - | 40.5 | - | 40.1 | - | 40.9 | ||
| 11 | 1.15; 2.51 | 26.3 | 1.77; 1.82 | 20.6 | 1.63; 1.72 | 21.8 | 1.40; 1.76 | 20.0 | ||
| 12 | 1.89; 2.11 | 41.0 | 1.74; 1.87 | 31.4 | 1.69; 1.81 | 30.4 | 1.45; 2.16 | 38.7 | ||
| 13 | 4.70; 4.84 | 77.8 | - | 88.5 | - | 90.2 | - | 88.7 | ||
| 14 | 1.24 | 37.7 | 1.95; 2.32 | 47.7 | 1.73; 2.32 | 49.6 | 1.66; 2.16 | 40.8 | ||
| 15 | 0.89 | 47.4 | 6.57 | 158.0 | 5.36 | 136.6 | 3.70 | 80.8 | ||
| 16 | 0.92; 1.59 | 146.7 | - | 147.7 | - | 146.9 | - | 157.2 | ||
| 17 | 1.92; 1.40 | 108.0 | 9.61 | 191.4 | 4.11; 4.29 | 59.2 | 5.31; 5.54 | 110.2 | ||
| 18 | 2.20; 1.09 | 27.5 | - | 178.0 | - | 178.4 | 1.25 | 28.5 | ||
| 19 | 1.12 | 176.8 | 1.22 | 28.5 | 1.21 | 28.6 | - | 178.8 | ||
| 20 | 1.78; 2.05 | 11.9 | 1.02 | 16.0 | 1.00 | 15.8 | 0.97 | 17.3 | ||
| Moiety | Position | 10 (a) | 11 (b) | 12 (b) | 13 (a) | ||||
|---|---|---|---|---|---|---|---|---|---|
| δH | δC | δH | δC | δH | δC | δH | δC | ||
| Aglycone | 1 | 1.71; 0.73 | 40.9 | 0.76; 1.75 | 40.8 | 0.79 | 40.2 | NA | 40.7 |
| 2 | 2.21; 1.42 | 19.5 | 1.70; 2.17 | 20.2 | 1.38; 2.13 | 19.7 | NA | 19.2 | |
| 3 | 2.33; 1.02 | 38.5 | 1.82; 2.14 | 38.9 | 2.36 | 39.1 | NA | 38.1 | |
| 4 | -- | 44.1 | -- | 44.5 | -- | 44.6 | NA | 43.9 | |
| 5 | 1.05; 2.45 | 57.5 | 0.99 | 57.6 | 1.03 | 57.9 | NA | 57.3 | |
| 6 | 2.45; 1.90 | 22.3 | 1.91; 2.20 | 22.2 | 1.84; 2.41 | 21.1 | NA | 22.0 | |
| 7 | 1.26 | 41.8 | 1.31; 1.51 | 41.9 | 2.78 | 39.6 | NA | 41.5 | |
| 8 | -- | 42.7 | -- | 42.2 | -- | 34.9 | NA | 42.5 | |
| 9 | 0.86 | 54.0 | 0.93 | 54.2 | 1.01 | 51.7 | NA | 53.8 | |
| 10 | -- | 39.9 | -- | 39.8 | -- | 44.6 | NA | 39.7 | |
| 11 | 1.60 | 20.7 | 1.48 | 20.7 | 1.59 | 27.3 | NA | 20.6 | |
| 12 | 2.22; 1.92 | 36.8 | 2.75; 1.10 | 38.0 | 4.11 | 78.4 | NA | 36.6 | |
| 13 | -- | 86.2 | -- | 87.2 | 2.94 | 42.0 | NA | 85.9 | |
| 14 | 2.70; 1.77 | 44.6 | 1.94; 2.45 | 44.8 | 1.42; 1.05 | 39.9 | NA | 44.3 | |
| 15 | 2.09; 2.02 | 47.7 | 2.10 | 48.6 | 1.91; 2.23 | 49.0 | NA | 47.5 | |
| 16 | -- | 154.5 | -- | 153.8 | -- | 148.4 | NA | 154.3 | |
| 17 | 5.68; 5.04 | 104.7 | 5.10; 5.64 | 105.5 | 4.90; 5.40 | 109.5 | NA | 104.5 | |
| 18 | 1.22 | 28.4 | 1.42 | 29.4 | 1.26 | 29.1 | NA | 28.2 | |
| 19 | -- | 177.2 | -- | 176.1 | -- | 177.4 | NA | 177.0 | |
| 20 | 1.27 | 15.7 | 0.99 | 16.5 | 1.04 | 13.3 | NA | 15.4 | |
| Glcβ-C19 | 1′ | 6.08 | 95.9 | 6.23 | 93.8 | 6.18 | 96.4 | 6.16 | 95.6 |
| 2′ | 4.15 | 74.0 | NA | NA | NA | NA | NA | NA | |
| 3′ | 4.17 | 79.0 | NA | NA | NA | NA | NA | NA | |
| 4′ | 4.27 | 71.1 | NA | NA | NA | NA | NA | NA | |
| 5′ | 3.93 | 79.3 | NA | NA | NA | NA | NA | NA | |
| 6′ | 4.39; 4.30 | 62.2 | NA | NA | NA | NA | NA | NA | |
| Glcβ(1-X) | 1″ | -- | -- | 5.10 | 105.8 | -- | -- | -- | -- |
| 2″ | -- | -- | NA | NA | -- | -- | -- | -- | |
| 3″ | -- | -- | NA | NA | -- | -- | -- | -- | |
| 4″ | -- | -- | NA | NA | -- | -- | -- | -- | |
| 5″ | -- | -- | NA | NA | -- | -- | -- | -- | |
| 6″ | -- | -- | NA | NA | -- | -- | -- | -- | |
| Glcβ-C13 | 1‴ | 5.12 | 98.0 | 5.12 | 99.7 | 5.01 | 103.0 | 5.08 | 97.6 |
| 2‴ | 4.14 | 84.6 | -- | -- | NA | NA | NA | NA | |
| 3‴ | 4.25 | 78.24 | -- | -- | NA | NA | NA | NA | |
| 4‴ | 4.00 | 72.2 | -- | -- | NA | NA | NA | NA | |
| 5‴ | 3.88 | 77.9 | -- | -- | NA | NA | NA | NA | |
| 6‴ | 4.55; 4.19 | 62.9 | -- | -- | NA | NA | NA | NA | |
| Glcβ(1-Y) | 1′′′′ | 5.27 | 106.8 | -- | -- | 5.24 | 106.4 | 95.6 | 104.7 |
| 2′′′′ | 4.18 | 77.0 | -- | -- | NA | NA | NA | NA | |
| 3′′′′ | 4.23 | 78.16 | -- | -- | NA | NA | NA | NA | |
| 4′′′′ | 4.39 | 71.6 | -- | -- | NA | NA | NA | NA | |
| 5′′′′ | 3.94 | 78.6 | -- | -- | NA | NA | NA | NA | |
| 6′′′′ | 4.46; 4.41 | 62.7 | -- | -- | NA | NA | NA | NA | |
| Moiety | Position | 14 (a) | 15 (a) | 16 (a) | 17 (b) | 18 (b) | 19 (b) | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| δH | δC | δH | δC | δH | δC | δH | δC | δH | δC | δH | δC | ||
| Aglycone | 1 | 2.34; 0.98 | 39.1 | 0.78; 1.77 | 41.2 | 0.73; 1.73 | 41.2 | 0.92; 1.59 | 39.5 | 0.87; 1.87 | 41.4 | 0.86; 1.87 | 41.5 |
| 2 | 2.18; 1.14 | 20.1 | 2.22; 1.45 | 19.9 | 2.20; 1.43 | 19.9 | 1.92; 1.40 | 18.4 | 1.41; 1.92 | 19.8 | 1.41; 1.94 | 19.7 | |
| 3 | 2.21; 1.95 | 37.4 | 1.03; 2.35 | 37.3 | 2.28; 1.79 | 36.8 | 2.20; 1.09 | 37.7 | 1.06; 2.14 | 38.8 | 1.05; 2.17 | 38.8 | |
| 4 | -- | 43.4 | -- | 44.5 | -- | 44.5 | -- | 43.7 | -- | 43.8 | -- | 44.6 | |
| 5 | 1.01 | 57.9 | 1.05 | 57.8 | 1.03 | 57.8 | 1.12 | 57.1 | 1.12 | 58.1 | 1.12 | 58.2 | |
| 6 | 2.47; 1.90 | 22.9 | 2.46; 1.92 | 22.6 | 1.91; 2.50 | 22.7 | 1.78; 2.05 | 19.9 | 1.84; 1.97 | 22.6 | 1.84; 2.04 | 22.8 | |
| 7 | 1.25; 1.25 | 42.4 | 1.30 | 42.2 | 1.28; 1.28 | 42.2 | 1.57; 1.16 | 38.8 | 1.41; 1.54 | 42.2 | 1.42; 1.55 | 42.4 | |
| 8 | -- | 44.7 | -- | 43.1 | -- | 43.3 | -- | 33.9 | -- | 43.0 | -- | 43.4 | |
| 9 | 0.86 | 54.6 | 0.88 | 54.5 | 0.87 | 54.3 | 1.09 | 51.0 | 0.97 | 54.7 | 0.98 | 55.0 | |
| 10 | -- | 40.5 | -- | 40.3 | -- | 40.3 | -- | 38.1 | -- | 39.4 | -- | 40.5 | |
| 11 | 1.60; 1.60 | 21.4 | 1.68 | 21.1 | 1.68; 1.68 | 21.1 | 1.63; 1.45 | 26.3 | 1.64; 1.80 | 21.0 | 1.65; 1.81 | 21.1 | |
| 12 | 1.70; 0.73 | 41.4 | 2.25; 2.00 | 37.3 | 1.02; 2.35 | 38.9 | 2.51 | 41.0 | 1.52; 1.97 | 37.9 | 1.55; 1.98 | 37.8 | |
| 13 | -- | 86.8 | -- | 86.9 | -- | 86.4 | -- | 77.8 | -- | 87.8 | -- | 87.7 | |
| 14 | 2.72; 1.78 | 45.1 | 2.66; 1.81 | 45.0 | 1.97; 2.74 | 45.1 | 4.04 | 37.7 | 1.50; 2.27 | 45.0 | 1.51; 2.26 | 45.0 | |
| 15 | 2.04; 2.04 | 48.3 | 2.05 | 48.2 | 2.06; 2.06 | 48.0 | 1.15; 2.51 | 47.4 | 2.04; 2.12 | 48.2 | 2.04; 2.14 | 48.4 | |
| 16 | -- | 155.2 | -- | 154.7 | -- | 155.0 | -- | 146.7 | -- | 154.5 | -- | 154.4 | |
| 17 | 5.70; 5.07 | 105.4 | 5.01; 5.64 | 105.1 | 5.10; 5.73 | 105.4 | 1.89; 2.11 | 108.0 | 4.85; 5.18 | 105.3 | 4.84; 5.17 | 105.2 | |
| 18 | 1.25 | 28.9 | 1.25 | 28.8 | 1.23 | 28.8 | 4.70; 4.84 | 27.5 | 1.21 | 28.6 | 1.23 | 28.5 | |
| 19 | -- | 178.0 | -- | 177.5 | -- | 177.7 | 1.24 | 176.8 | -- | 178.4 | -- | 178.4 | |
| 20 | 1.28 | 16.3 | 1.32 | 16.0 | 1.31 | 16.0 | 0.89 | 11.9 | 0.97 | 16.0 | 0.98 | 16.0 | |
| Glcβ-C19 | 1′ | 6.01 | 96.5 | 6.12 | 96.3 | 6.12 | 96.3 | 5.43 | 94.2 | 5.37 | 95.4 | 5.34 | 95.5 |
| 2′ | 4.09 | 74.4 | 4.13 | 79.8 | 4.17 | 74.4 | 3.39 | 72.6 | 3.36 | 73.8 | 3.35 | 74.0 | |
| 3′ | 4.13 | 79.5 | 4.18 | 78.7 | 3.98 | 79.5 | 3.43 | 77.3 | 3.46 | 78.5 | 3.44 | 78.4 | |
| 4′ | 4.26 | 72.0 | 4.28 | 71.5 | 4.33 | 71.4 | 3.39 | 69.7 | 3.36 | 70.9 | 3.45 | 70.9 | |
| 5′ | 4.02 | 78.8 | 3.97 | 79.2 | 4.22 | 79.9 | 3.39 | 77.3 | 3.36 | 78.2 | 3.45 | 77.5 | |
| 6′ | 4.68; 4.31 | 70.0 | 4.40 | 63.4 | 4.43; 4.57 | 62.6 | 3.85; 3.71 | 61.0 | 3.68; 3.82 | 62.2 | 3.58; 3.61 | 62.1 | |
| Glcβ -C13 | 1″ | 5.03 | 105.9 | 5.08 | 98.8 | 5.19 | 98.3 | 4.56 | 100.5 | 4.59 | 97.2 | 4.61 | 97.4 |
| 2″ | 4.00 | 75.9 | 4.38 | 81.3 | 4.23 | 84.4 | 3.65 | 79.0 | 3.58 | 81.5 | 3.45 | 82.4 | |
| 3″ | 3.88 | 79.1 | 4.15 | 88.6 | 4.27 | 78.6 | 3.71 | 86.0 | 3.55 | 77.9 | 3.56 | 78.1 | |
| 4″ | 4.22 | 77.8 | 3.87 | 71.2 | 4.45 | 71.8 | 3.38 | 68.8 | 3.30 | 71.6 | 3.25 | 72.0 | |
| 5″ | 3.88 | 79.2 | 3.79 | 77.9 | 4.08 | 77.8 | 3.34 | 76.1 | 3.22 | 77.5 | 3.26 | 77.7 | |
| 6″ | 4.57; 4.38 | 63.5 | 4.50; 4.40 | 63.2 | 4.51; 4.78 | 70.3 | 3.91; 3.68 | 61.4 | 3.65; 3.83 | 62.2 | 3.63; 3.85 | 63.0 | |
| Glcβ(1-2) | 1′′′ | 5.15 | 98.7 | 5.58 | 105.3 | 5.30 | 106.8 | 4.81 | 102.2 | 4.61 | 104.3 | 4.57 | 105.1 |
| 2′′′ | 4.18 | 85.4 | 4.20 | 76.8 | 4.11 | 75.7 | 3.19 | 74.6 | 3.28 | 75.6 | 3.27 | 76.2 | |
| 3′′′ | 3.92 | 78.7 | 4.31 | 74.4 | 4.23 | 78.8 | 3.35 | 76.5 | 3.33 | 77.8 | 3.36 | 78.0 | |
| 4′′′ | 4.29 | 78.9 | 4.28 | 72.0 | 4.28 | 72.1 | 3.22 | 70.6 | 3.33 | 71.5 | 3.30 | 71.6 | |
| 5′′′ | 4.03 | 72.8 | 3.97 | 79.0 | 3.91 | 78.3 | 3.33 | 76.6 | 3.44 | 77.5 | 3.25 | 78.4 | |
| 6′′′ | 4.57; 4.22 | 63.2 | 4.40 | 62.5 | 4.22; 4.57 | 63.3 | 3.87; 3.66 | 61.9 | 3.78; 4.10 | 69.3 | 3.63; 3.91 | 63.0 | |
| Sugar*(1-X) | 1′′′′ | 5.30 | 107.6 | 5.33 | 105.3 | 5.14 | 105.9 | 4.65 | 103.1 | 4.54 | 104.0 | 3.61; 3.67 | 63.8 |
| 2′′′′ | 4.22 | 78.8 | 4.08 | 76.8 | 4.06 | 77.2 | 3.28 | 73.9 | 3.19 | 74.8 | -- | 105.1 | |
| 3′′′′ | 4.27 | 71.5 | 4.20 | 74.4 | 4.27 | 78.9 | 3.39 | 76.8 | 3.46 | 77.9 | 4.09 | 78.8 | |
| 4′′′′ | 4.45 | 72.1 | 4.23 | 72.4 | 4.12 | 72.6 | 3.31 | 70.1 | 3.29 | 71.5 | 4.00 | 76.3 | |
| 5′′′′ | 3.97 | 79.4 | 4.05 | 78.8 | 4.03 | 78.5 | 3.37 | 76.8 | 3.30 | 77.8 | 3.72 | 83.6 | |
| 6′′′′ | 4.49; 4.38 | 63.2 | 4.57; 4.32 | 62.8 | 4.22; 4.57 | 63.3 | 3.90; 3.66 | 61.2 | 3.61; 3.86 | 62.6 | 3.72; 3.98 | 61.6 | |
| Moiety | Position | 20 (a) | 21 (a) | 22 (b) | 23 (b) | ||||
|---|---|---|---|---|---|---|---|---|---|
| δH | δC | δH | δC | δH | δC | δH | δC | ||
| Aglycone | 1 | 0.77; 1.78 | 40.8 | 0.75; 1.77 | 40.1 | 0.85; 1.87 | 41.6 | 0.85; 1.88 | 41.5 |
| 2 | 1.41; 2.20 | 19.4 | 1.42; 2.22 | 19.9 | 1.44; 1.93 | 19.8 | 1.41; 1.94 | 19.8 | |
| 3 | 1.00; 2.32 | 38.4 | 1.89; 2.38 | 38.9 | 1.06; 2.17 | 38.7 | 1.05; 2.15 | 38.7 | |
| 4 | -- | 44.0 | -- | 44.4 | -- | 44.3 | -- | 44.8 | |
| 5 | 1.02 | 57.3 | 1.05 | 57.8 | 1.12 | 58.4 | 1.12 | 58.2 | |
| 6 | 1.89; 2.44 | 22.1 | 1.98; 2.40 | 22.6 | 1.83; 2.08 | 22.8 | 1.83; 2.03 | 22.8 | |
| 7 | 1.28 | 41.7 | 1.36; 1.43 | 42.2 | 1.43; 1.55 | 42.5 | 1.44; 1.55 | 42.4 | |
| 8 | -- | 42.5 | -- | 44.7 | -- | 43.2 | -- | 43.3 | |
| 9 | 0.89 | 54.0 | 0.91 | 54.7 | 0.98 | 54.7 | 0.97 | 55.0 | |
| 10 | -- | 39.8 | -- | 40.2 | -- | 38.8 | -- | 40.7 | |
| 11 | 1.66 | 20.6 | 1.68 | 20.9 | 1.66; 1.82 | 21.0 | 1.63; 1.80 | 21.0 | |
| 12 | 1.85; 2.28 | 37.0 | 2.19; 1.89 | 38.4 | 1.46; 2.01 | 37.6 | 1.54; 1.99 | 37.9 | |
| 13 | -- | 86.4 | -- | 87.2 | -- | 87.1 | -- | 88.0 | |
| 14 | 1.79; 2.65 | 44.3 | 1.74; 2.55 | 44.7 | 1.54; 2.27 | 45.0 | 1.51; 2.26 | 45.0 | |
| 15 | 2.03; 2.14 | 47.7 | 2.06; 2.14 | 48.5 | 2.04 | 48.3 | 2.04; 2.14 | 48.3 | |
| 16 | -- | 154.2 | -- | 154.7 | -- | 153.6 | -- | 153.7 | |
| 17 | 4.99; 5.63 | 104.6 | 5.20; 5.60 | 105.4 | 4.82; 5.11 | 104.7 | 4.84; 5.18 | 105.2 | |
| 18 | 1.20 | 28.3 | 1.27 | 29.0 | 1.21 | 28.6 | 1.20 | 28.6 | |
| 19 | -- | 177.0 | -- | 177.3 | -- | 178.2 | -- | 178.5 | |
| 20 | 1.31 | 15.5 | 1.25 | 16.1 | 1.00 | 16.0 | 0.97 | 16.0 | |
| Glcβ-C19 | 1′ | 6.10 | 95.8 | 6.16 | 96.3 | 5.34 | 95.7 | 5.34 | 95.4 |
| 2′ | 4.19 | 73.9 | NA | NA | 3.32 | 73.8 | 3.35 | 73.7 | |
| 3′ | 4.10 | 79.2 | NA | NA | 3.44 | 78.4 | 3.42 | 78.2 | |
| 4′ | 4.24 | 71.0 | NA | NA | 3.34 | 70.9 | 3.39 | 70.9 | |
| 5′ | 3.93 | 78.5 | NA | NA | 3.36 | 78.2 | 3.52 | 77.6 | |
| 6′ | 4.39; 4.30 | 62.1 | NA | NA | 3.60; 3.82 | 62.5 | 3.76; 4.06 | 68.8 | |
| Xyl(1-6) | 1″ | -- | -- | -- | -- | -- | -- | 4.30 | 104.6 |
| 2″ | -- | -- | -- | -- | -- | -- | 3.58 | 72.2 | |
| 3″ | -- | -- | -- | -- | -- | -- | 3.53 | 74.0 | |
| 4″ | -- | -- | -- | -- | -- | -- | 3.79 | 69.1 | |
| 5″ | -- | -- | -- | -- | -- | -- | 3.50; 3.84 | 66.3 | |
| 6″ | -- | -- | -- | -- | -- | -- | -- | -- | |
| Sugar*-C13 | 1′′′ | 5.02 | 97.9 | 4.96 | 98.7 | 4.59 | 97.8 | 4.60 | 97.4 |
| 2′′′ | 4.22 | 80.7 | NA | NA | 3.59 | 81.5 | 3.45 | 82.4 | |
| 3′′′ | 4.06 | 88.3 | NA | NA | 3.68 | 87.3 | 3.54 | 77.9 | |
| 4′′′ | 3.83 | 70.6 | NA | NA | 3.34 | 70.9 | 3.25 | 71.6 | |
| 5′′′ | 3.72 | 77.3 | NA | NA | 3.30 | 77.2 | 3.25 | 77.9 | |
| 6′′′ | 4.42; 4.02 | 62.6 | -- | -- | 3.60; 3.82 | 62.3 | 3.62; 3.83 | 62.6 | |
| Sugar**(1-2) | 1′′′′ | 5.42 | 105.4 | 5.54 | 105.0 | 4.63 | 104.0 | 4.58 | 105.1 |
| 2′′′′ | 4.10 | 75.9 | NA | NA | 3.25 | 73.4 | 3.27 | 77.8 | |
| 3′′′′ | 4.12 | 78.6 | NA | NA | 3.42 | 78.6 | 3.36 | 77.9 | |
| 4′′′′ | 4.23 | 71.2 | NA | NA | 3.32 | 71.1 | 3.29 | 71.4 | |
| 5′′′′ | 4.35; 3.65 | 67.5 | NA | NA | 3.36 | 78.4 | 3.24 | 77.7 | |
| 6′′′′ | -- | -- | NA | NA | 3.62; 3.80 | 62.7 | 3.62; 3.83 | 62.6 | |
| Sugar***(1-3) | 1′′′′′ | 5.26 | 104.8 | 5.33 | 105.1 | 4.61 | 105.3 | -- | -- |
| 2′′′′′ | 4.01 | 75.2 | NA | NA | 3.54 | 73.0 | -- | -- | |
| 3′′′′′ | 4.17 | 79.0 | NA | NA | 3.50 | 74.3 | -- | -- | |
| 4′′′′′ | 4.13 | 71.5 | NA | NA | 3.46 | 69.9 | -- | -- | |
| 5′′′′′ | 4.02 | 78.6 | NA | NA | 3.62; 3.86 | 67.6 | -- | -- | |
| 6′′′′′ | 4.51; 4.26 | 62.3 | NA | NA | -- | -- | -- | -- | |
| Moiety | Position | 24 (b) | 25 (b) | 26 (b) | 27 (a) | ||||
|---|---|---|---|---|---|---|---|---|---|
| δH | δC | δH | δC | δH | δC | δH | δC | ||
| Aglycone | 1 | 0.76; 1.66 | 41.1 | 0.83; 1.85 | 40.9 | 0.85; 1.86 | 41.6 | NA | 40.8 |
| 2 | 1.70; 2.11 | 20.3 | 1.40; 1.92 | 19.2 | 1.40; 1.94 | 19.8 | NA | 19.4 | |
| 3 | 1.91; 2.15 | 38.1 | 1.55; 1.96 | 37.0 | 1.05; 2.14 | 38.9 | NA | 38.5 | |
| 4 | -- | 44.8 | -- | 43.5 | -- | 44.8 | NA | 43.9 | |
| 5 | 1.00 | 58.8 | 1.07 | 57.5 | 1.12 | 58.3 | NA | 57.5 | |
| 6 | 1.86; 2.11 | 22.5 | 1.86; 1.90 | 21.8 | 1.84; 2.02 | 22.7 | NA | 22.0 | |
| 7 | 1.32; 1.62 | 42.1 | 1.43; 1.54 | 41.7 | 1.41; 1.54 | 42.5 | NA | 41.8 | |
| 8 | -- | 43.1 | -- | 54.0 | - | 43.2 | NA | 42.2 | |
| 9 | 0.91 | 54.4 | 0.98 | 54.1 | 0.97 | 55.0 | NA | 54.1 | |
| 10 | -- | 40.2 | -- | 39.0 | -- | NA | NA | 39.9 | |
| 11 | 1.70 | 21.1 | 1.63; 1.80 | 19.8 | 1.63; 1.77 | 21.0 | NA | 20.6 | |
| 12 | 2.68; 1.15 | 38.1 | 1.07; 2.27 | 37.5 | 1.51; 1.97 | 38.1 | NA | 38.5 | |
| 13 | -- | 86.6 | -- | 87.6 | - | 88.7 | NA | 87.3 | |
| 14 | 1.70; 2.50 | 45.1 | 1.50; 2.24 | 44.6 | 1.52; 2.24 | 44.9 | NA | 43.2 | |
| 15 | 2.10 | 48.3 | 2.04; 2.13 | 47.7 | 2.03; 2.14 | 48.5 | NA | 48.7 | |
| 16 | -- | 155.1 | -- | 152.5 | -- | 153.2 | NA | 152.9 | |
| 17 | 5.10; 5.72 | 105.6 | 4.84; 2.13 | 104.7 | 4.83; 5.18 | 105.1 | NA | 106.1 | |
| 18 | 1.50 | 29.6 | 1.25 | 28.4 | 1.21 | 28.6 | NA | 28.2 | |
| 19 | -- | 176.0 | -- | 177.3 | -- | 178.4 | NA | 177.1 | |
| 20 | 1.14 | 17.3 | 0.93 | 16.1 | 0.99 | 16.1 | NA | 15.2 | |
| Glcβ-C19 | 1′ | 6.24 | 94.1 | 5.60 | 93.5 | 5.38 | 95.5 | 5.98 | 96.0 |
| 2′ | NA | NA | 3.60 | 77.7 | 3.34 | 73.6 | NA | NA | |
| 3′ | NA | NA | 3.57 | 78.2 | 3.44 | 78.4 | NA | NA | |
| 4′ | NA | NA | 3.40 | 70.1 | 3.34 | 70.9 | NA | NA | |
| 5′ | NA | NA | 3.39 | 77.4 | 3.36 | 78.2 | NA | NA | |
| 6′ | NA | NA | 3.70; 3.79 | 62.3 | 3.60; 3.82 | 62.3 | NA | NA | |
| Rha(1-2) | 1″ | 6.40 | 102.0 | 5.30 | 100.9 | -- | -- | -- | -- |
| 2″ | NA | NA | 3.90 | 71.0 | -- | -- | -- | -- | |
| 3″ | NA | NA | 3.61 | 71.1 | -- | -- | -- | -- | |
| 4″ | NA | NA | 3.37 | 72.7 | -- | -- | -- | -- | |
| 5″ | NA | NA | 3.75 | 69.3 | -- | -- | -- | -- | |
| 6″ | NA | NA | 1.24 | 17.1 | -- | -- | -- | -- | |
| Glcβ-C13 | 1′′′ | 5.10 | 98.5 | 4.61 | 96.5 | 4.56 | 97.7 | 5.04 | 97.7 |
| 2′′′ | NA | NA | 3.47 | 81.2 | 3.54 | 80.9 | NA | NA | |
| 3′′′ | NA | NA | 3.57 | 79.2 | 3.67 | 87.5 | NA | NA | |
| 4′′′ | NA | NA | 3.31 | 70.7 | 3.34 | 70.9 | NA | NA | |
| 5′′′ | NA | NA | 3.25 | 77.2 | 3.30 | 77.2 | NA | NA | |
| 6′′′ | NA | NA | 3.63; 3.83 | 62.0 | 3.60; 3.82 | 62.3 | NA | NA | |
| Sugar*(1-2) | 1′′′′ | 5.22 | 107.0 | 4.60 | 104.4 | 4.71 | 104.0 | 6.34 | 104.2 |
| 2′′′′ | NA | NA | 3.24 | 75.0 | 3.22 | 75.8 | NA | NA | |
| 3′′′′ | NA | NA | 3.35 | 77.7 | 3.30 | 74.0 | NA | NA | |
| 4′′′′ | NA | NA | 3.21 | 70.7 | 3.02 | 76.9 | NA | NA | |
| 5′′′′ | NA | NA | 3.21 | 76.5 | 3.26 | 81.2 | NA | NA | |
| 6′′′′ | NA | NA | 3.64; 3.84 | 61.8 | 1.25 | 18.0 | NA | 18.8 | |
| Glcβ(1-3) | 1′′′′′′ | -- | -- | -- | -- | 4.63 | 104.2 | 4.94 | 102.1 |
| 2′′′′′′ | -- | -- | -- | -- | 3.24 | 73.4 | NA | NA | |
| 3′′′′′′ | -- | -- | -- | -- | 3.42 | 78.6 | NA | NA | |
| 4′′′′′′ | -- | -- | -- | -- | 3.32 | 71.1 | NA | NA | |
| 5′′′′′′ | -- | -- | -- | -- | 3.36 | 78.4 | NA | NA | |
| 6′′′′′′ | -- | -- | -- | -- | 3.62; 3.80 | 62.7 | NA | NA | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Perera, W.H.; McChesney, J.D. Approaches toward the Separation, Modification, Identification and Scale up Purification of Tetracyclic Diterpene Glycosides from Stevia rebaudiana (Bertoni) Bertoni. Molecules 2021, 26, 1915. https://doi.org/10.3390/molecules26071915
Perera WH, McChesney JD. Approaches toward the Separation, Modification, Identification and Scale up Purification of Tetracyclic Diterpene Glycosides from Stevia rebaudiana (Bertoni) Bertoni. Molecules. 2021; 26(7):1915. https://doi.org/10.3390/molecules26071915
Chicago/Turabian StylePerera, Wilmer H., and James D. McChesney. 2021. "Approaches toward the Separation, Modification, Identification and Scale up Purification of Tetracyclic Diterpene Glycosides from Stevia rebaudiana (Bertoni) Bertoni" Molecules 26, no. 7: 1915. https://doi.org/10.3390/molecules26071915
APA StylePerera, W. H., & McChesney, J. D. (2021). Approaches toward the Separation, Modification, Identification and Scale up Purification of Tetracyclic Diterpene Glycosides from Stevia rebaudiana (Bertoni) Bertoni. Molecules, 26(7), 1915. https://doi.org/10.3390/molecules26071915