Coptidis Rhizoma Extract Reverses 5-Fluorouracil Resistance in HCT116 Human Colorectal Cancer Cells via Modulation of Thymidylate Synthase
Abstract
1. Introduction
2. Results
2.1. Compounds Present in CRE
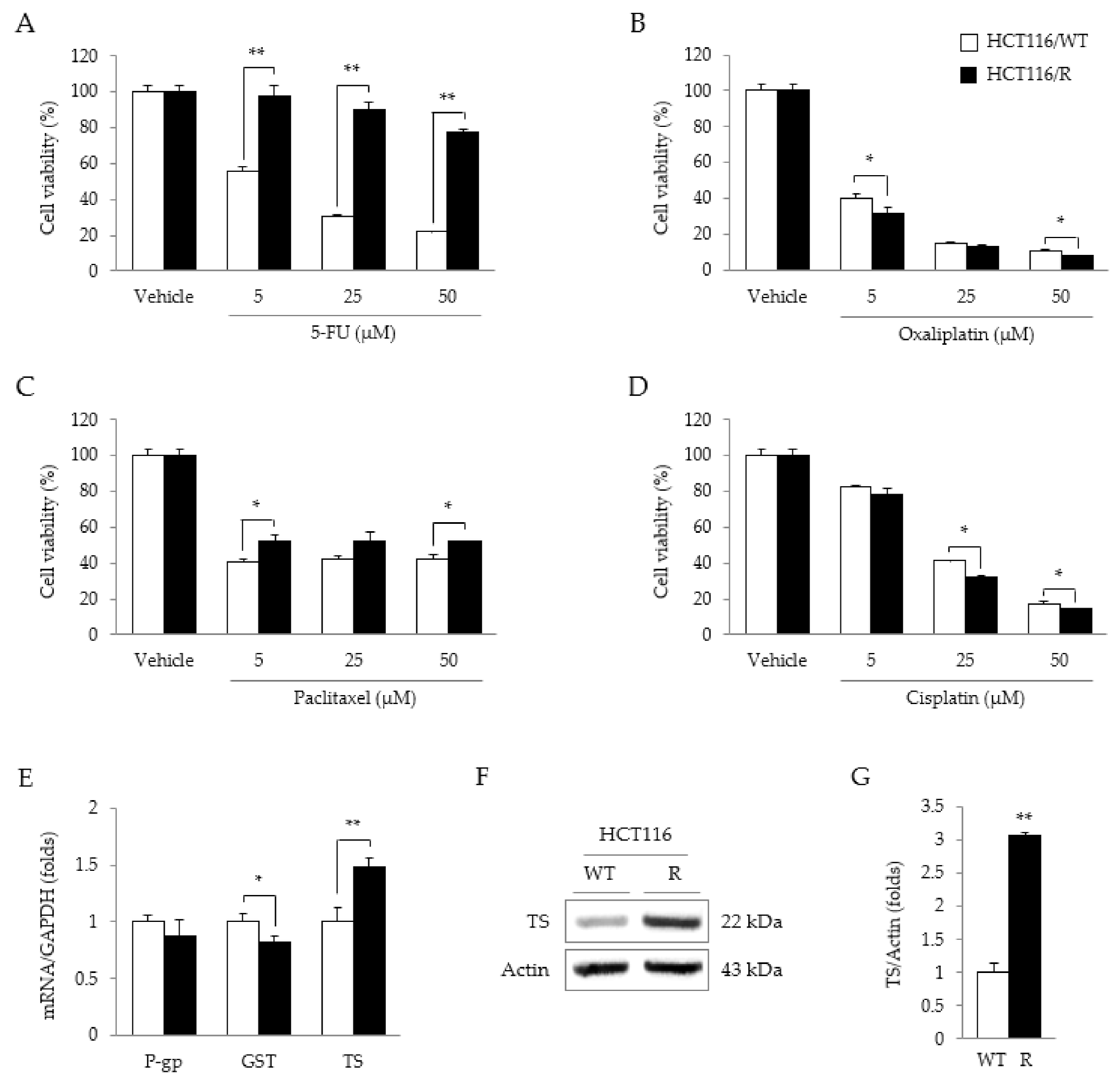
2.2. Characteristics of HCT116/R Cells
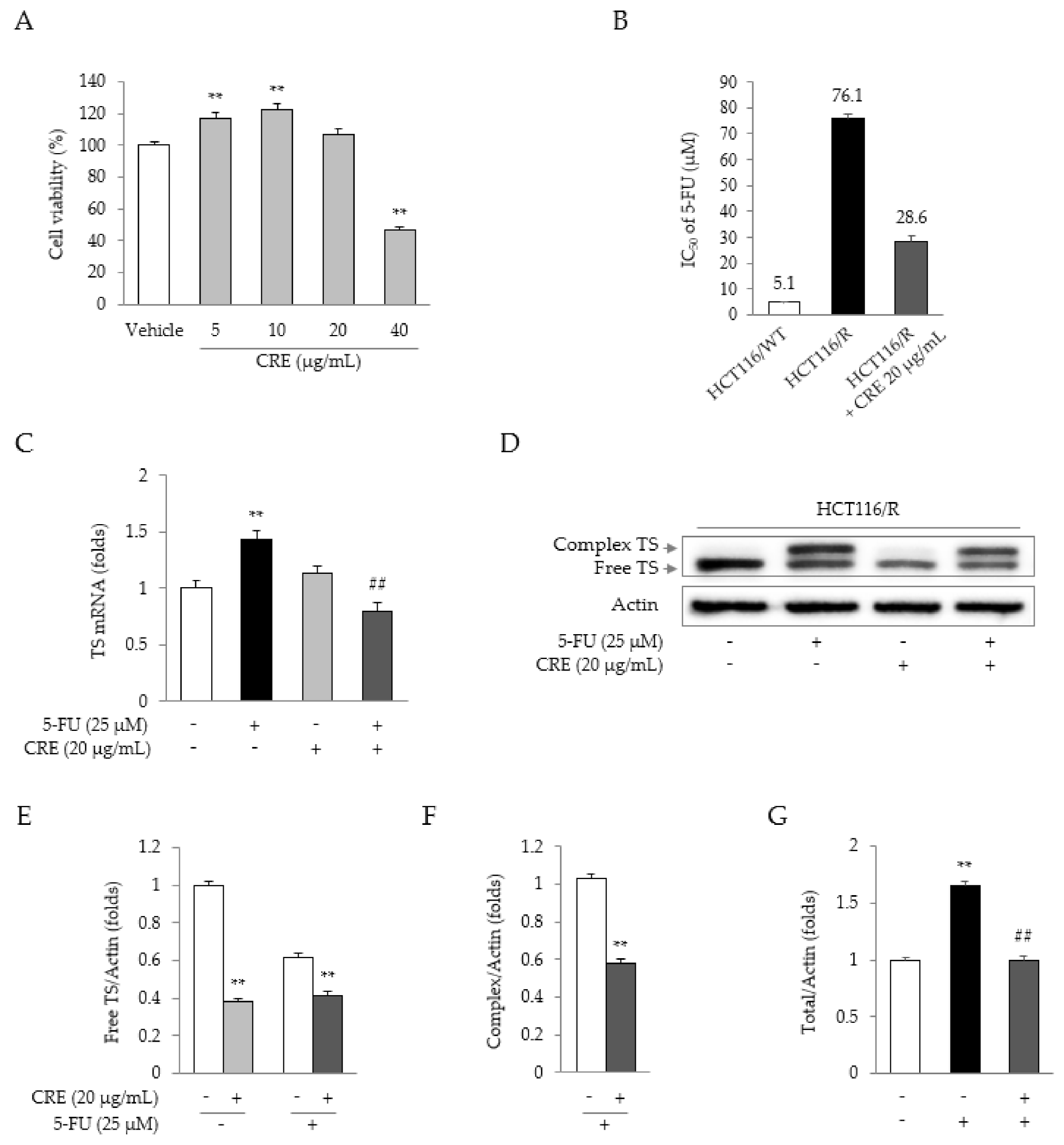
2.3. Synergistic Effects of 5-FU and CRE on Properties Related to 5-FU Resistance
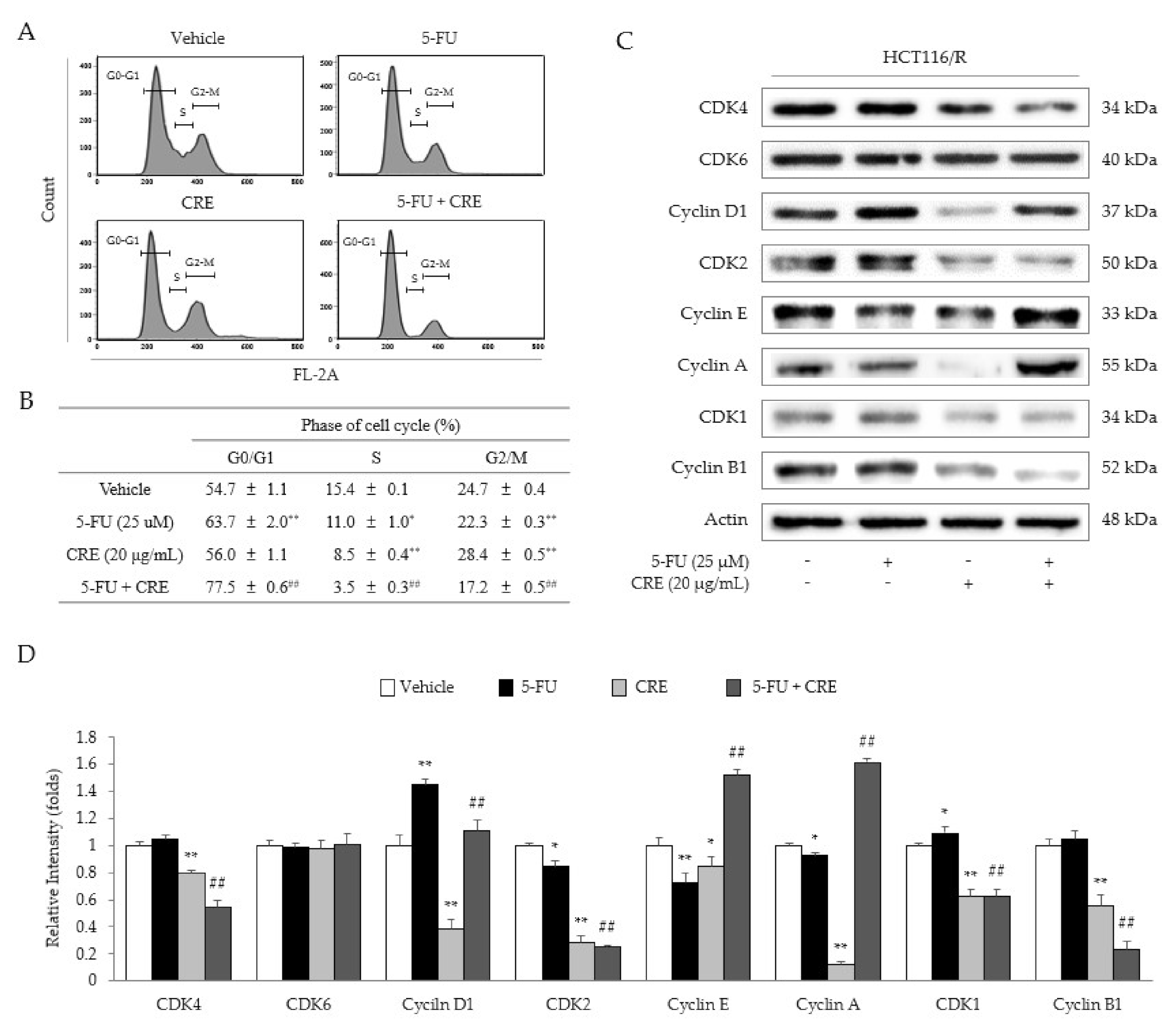
2.4. Synergistic Effects of 5-FU and CRE on Cell Cycle Arrest
2.5. Identification of the Active Compound in CRE
3. Discussion
4. Materials and Methods
4.1. Preparation of C. Rhizoma Extract (CRE)
4.2. Chemicals and Reagents
4.3. Fingerprint Analysis of CRE
4.4. Cell Culture Conditions
4.5. Cell Viability Assay
4.6. Real-Time RT-PCR Analysis
4.7. Western Blot Analysis
4.8. Flow Cytometric Analysis
4.9. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2021, 68, 394–424. [Google Scholar] [CrossRef]
- Karsa, L.; Lignini, T.; Patnick, J.; Lambert, R.; Sauvaget, C. The dimensions of the CRC problem. Best Pr. Res. Clin. Gastroenterol. 2010, 24, 381–396. [Google Scholar] [CrossRef]
- Nizioł, M.; Kostrzewska, B.; Kamińska, D.; Domurat, M.; Zińczuk, J.; Misiura, M.; Guzińska-Ustymowicz, K.; Pryczynicz, A. Symptoms of colorectal cancer contributes to its localization and advancement. Prog. Health Sci. 2019, 1, 76–82. [Google Scholar] [CrossRef]
- Lee, G.-Y.; Lee, J.-S.; Son, C.-G.; Lee, N.-H. Combating Drug Resistance in Colorectal Cancer Using Herbal Medicines. Chin. J. Integr. Med. 2020, 1–10. [Google Scholar] [CrossRef]
- Popat, S.; Matakidou, A.; Houlston, R.S. Thymidylate Synthase Expression and Prognosis in Colorectal Cancer: A Systematic Review and Meta-Analysis. J. Clin. Oncol. 2004, 22, 529–536. [Google Scholar] [CrossRef] [PubMed]
- Grothey, A.; Sargent, D. Overall survival of patients with advanced colorectal cancer correlates with availability of fluor-ouracil, irinotecan, and oxaliplatin regardless of whether doublet or single-agent therapy is used first line. J. Clin. Oncol. 2005, 23, 9441–9442. [Google Scholar] [CrossRef] [PubMed]
- Abdel-Rahman, O.; Karachiwala, H. Impact of age on toxicity and efficacy of 5-FU-based combination chemotherapy among patients with metastatic colorectal cancer; a pooled analysis of five randomized trials. Int. J. Color. Dis. 2019, 34, 1741–1747. [Google Scholar] [CrossRef]
- Healey, E.; Stillfried, G.E.; Eckermann, S.; Dawber, J.P.; Clingan, P.R.; Ranson, M. Comparative effectiveness of 5-fluorouracil with and without oxaliplatin in the treatment of colorectal cancer in clinical practice. Anticancer. Res. 2013, 33, 1053–1060. [Google Scholar] [PubMed]
- Folprecht, G.; Cunningham, D.; Glimelius, B.; Dicostanzo, F.; Wils, J.; Scheithauer, W.; Rougier, P.; Aranda, E.; Pabst, U.; Köhne, C. Efficacy of bolus and infusional 5-FU in elderly and non-elderly patients with metastatic colorectal cancer-a pooled analysis of clinical trials. J. Clin. Oncol. 2004, 22, 8045. [Google Scholar] [CrossRef]
- He, L.; Zhu, H.; Zhou, S.; Wu, T.; Wu, H.; Yang, H.; Mao, H.; SekharKathera, C.; Janardhan, A.; Edick, A.M.; et al. Wnt pathway is involved in 5-FU drug resistance of colorectal cancer cells. Exp. Mol. Med. 2018, 50, 1–12. [Google Scholar] [CrossRef]
- André, T.; De Gramont, A.A.; Vernerey, D.; Chibaudel, B.B.; Bonnetain, F.; Tijeras-Raballand, A.A.; Scriva, A.A.; Hickish, T.T.; Tabernero, J.; Van Laethem, J.L.; et al. Adjuvant Fluorouracil, Leucovorin, and Oxaliplatin in Stage II to III Colon Cancer: Updated 10-Year Survival and Outcomes According to BRAF Mutation and Mismatch Repair Status of the MOSAIC Study. J. Clin. Oncol. 2015, 33, 4176–4187. [Google Scholar] [CrossRef]
- Francipane, M.G.; Bulanin, D.; Lagasse, E. Establishment and Characterization of 5-Fluorouracil-Resistant Human Colorectal Cancer Stem-Like Cells: Tumor Dynamics under Selection Pressure. Int. J. Mol. Sci. 2019, 20, 1817. [Google Scholar] [CrossRef] [PubMed]
- Thomas, S.A.; Grami, Z.; Mehta, S.; Patel, K. Adverse Effects of 5-fluorouracil: Focus on Rare Side Effects. Cancer Cell Microenviron. 2016, 3. [Google Scholar] [CrossRef]
- Röhrl, K.; Guren, M.G.; Småstuen, M.C.; Rustøen, T. Symptoms during chemotherapy in colorectal cancer patients. Support. Care Cancer 2019, 27, 3007–3017. [Google Scholar] [CrossRef]
- Xiong, L.-R.; Jing, P.-W.; Song, X.-Y.; Wang, Y.-P.; Wang, L. 5-FU-Injured Bone Marrow Stromal Cells Initiate Stress-induced Premature Senescence of Hematopoietic Cells. Zhongguo Shi Yan Xue Ye Xue Za Zhi 2017, 25, 1178–1186. [Google Scholar]
- Colucci, G.; Gebbia, V.; Paoletti, G.; Giuliani, F.; Caruso, M.; Gebbia, N.; Cartenì, G.; Agostara, B.; Pezzella, G.; Manzione, L.; et al. Phase III Randomized Trial of FOLFIRI Versus FOLFOX4 in the Treatment of Advanced Colorectal Cancer: A Multicenter Study of the Gruppo Oncologico Dell’Italia Meridionale. J. Clin. Oncol. 2005, 23, 4866–4875. [Google Scholar] [CrossRef] [PubMed]
- Sui, H.; Liu, X.; Jin, B.-H.; Pan, S.-F.; Zhou, L.-H.; Yu, N.A.; Wu, J.; Cai, J.-F.; Fan, Z.-Z.; Zhu, H.-R.; et al. Zuo Jin Wan, a Traditional Chinese Herbal Formula, Reverses P-gp-Mediated MDRIn VitroandIn Vivo. Evid.-Based Complement. Altern. Med. 2013, 2013, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Wang, L.; Lou, G.-H.; Zeng, H.-R.; Hu, J.; Huang, Q.-W.; Peng, W.; Yang, X.-B. Coptidis Rhizoma: A comprehensive review of its traditional uses, botany, phytochemistry, pharmacology and toxicology. Pharm. Biol. 2019, 57, 193–225. [Google Scholar] [CrossRef]
- Huang, T.; Xiao, Y.; Yi, L.; Li, L.; Wang, M.; Tian, C.; Ma, H.; He, K.; Wang, Y.; Han, B.; et al. Coptisine from Rhizoma Coptidis Suppresses HCT-116 Cells-related Tumor Growth in vitro and in vivo. Sci. Rep. 2017, 7, srep38524. [Google Scholar] [CrossRef]
- Wang, W. A review on pharmacologic effects of effective ingredients in huanglian. Clin. J. Chin. Med. 2016, 8, 147–148. [Google Scholar]
- Dan, L.; Guangshang, C.; Xixi, S.; Qianqian, C.; Hongsheng, S. An overview of the antiarrhythmic study of alkaloids in Coptidis Rhizoma. Shandong J. Trad Chin. Med. 2017, 2017, 164–166. [Google Scholar]
- Mou, S.-J.; Yang, P.-F.; Liu, Y.-P.; Xu, N.; Jiang, W.-W.; Yue, W.-J. BCLAF1 promotes cell proliferation, invasion and drug-resistance though targeting lncRNA NEAT1 in hepatocellular carcinoma. Life Sci. 2020, 242, 117177. [Google Scholar] [CrossRef]
- Satapathy, S.R.; Sjölander, A. Cysteinyl leukotriene receptor 1 promotes 5-fluorouracil resistance and resistance-derived stemness in colon cancer cells. Cancer Lett. 2020, 488, 50–62. [Google Scholar] [CrossRef]
- Das, D.; Preet, R.; Mohapatra, P.; Satapathy, S.R.; Kundu, C.N. 1,3-Bis(2-chloroethyl)-1-nitrosourea enhances the inhibitory effect of Resveratrol on 5-fluorouracil sensitive/resistant colon cancer cells. World J. Gastroenterol. 2013, 19, 7374–7388. [Google Scholar] [CrossRef] [PubMed]
- Longley, D.B.; Allen, W.L.; Johnston, P.G. Drug resistance, predictive markers and pharmacogenomics in colorectal cancer. Biochim. Biophys. Acta (BBA) Bioenerg. 2006, 1766, 184–196. [Google Scholar] [CrossRef]
- Huang, L.; Zhang, S.; Zhou, J.; Li, X. Effect of resveratrol on drug resistance in colon cancer chemotherapy. RSC Adv. 2019, 9, 2572–2580. [Google Scholar] [CrossRef]
- Housman, G.; Byler, S.; Heerboth, S.; Lapinska, K.; Longacre, M.; Snyder, N.; Sarkar, S. Drug resistance in cancer: An over-view. Cancers 2014, 6, 1769–1792. [Google Scholar] [CrossRef]
- Yokogawa, T.; Yano, W.; Tsukioka, S.; Osada, A.; Wakasa, T.; Ueno, H.; Hoshino, T.; Yamamura, K.; Fujioka, A.; Fukuoka, M.; et al. dUTPase inhibition confers susceptibility to a thymidylate synthase inhibitor in DNA-repair-defective human cancer cells. Cancer Sci. 2021, 112, 422–432. [Google Scholar] [CrossRef] [PubMed]
- Showalter, S.L.; Showalter, T.N.; Witkiewicz, A.; Havens, R.; Kennedy, E.P.; Hucl, T.; Kern, S.E.; Yeo, C.J.; Brody, J.R. Eval-uating the drug-target relationship between thymidylate synthase expression and tumor response to 5-fluorouracil: Is it time to move forward? Cancer Biol. Ther. 2008, 7, 986–994. [Google Scholar] [CrossRef] [PubMed]
- Chong, D.; Ma, L.; Liu, F.; Zhang, Z.; Zhao, S.; Huo, Q.; Zhang, P.; Zheng, H.; Liu, H. Synergistic antitumor effect of 3-bromopyruvate and 5-fluorouracil against human colorectal cancer through cell cycle arrest and induction of apoptosis. Anti-Cancer Drugs 2017, 28, 831–840. [Google Scholar] [CrossRef]
- Otto, T.; Sicinski, P. Cell cycle proteins as promising targets in cancer therapy. Nat. Rev. Cancer 2017, 17, 93. [Google Scholar] [CrossRef]
- Zhang, J.; Zhou, L.; Zhao, S.; Dicker, D.T.; El-Deiry, W.S. The CDK4/6 inhibitor palbociclib synergizes with irinotecan to promote colorectal cancer cell death under hypoxia. Cell Cycle 2017, 16, 1193–1200. [Google Scholar] [CrossRef]
- Chen, J.; Wang, F.; Liu, J.; Lee, F.S.-C.; Wang, X.; Yang, H. Analysis of alkaloids in Coptis chinensis Franch by accelerated solvent extraction combined with ultra performance liquid chromatographic analysis with photodiode array and tandem mass spec-trometry detections. Anal. Chim. Acta 2008, 613, 184–195. [Google Scholar] [CrossRef] [PubMed]
- Ma, B.-L.; Ma, Y.-M.; Shi, R.; Wang, T.-M.; Zhang, N.; Wang, C.-H.; Yang, Y. Identification of the toxic constituents in Rhizoma Coptidis. J. Ethnopharmacol. 2010, 128, 357–364. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Zhang, Z.; Li, S.; Ye, X.; Li, X.; He, K. Synergy effects of herb extracts: Pharmacokinetics and pharmacodynamic basis. Fitoterapia 2014, 92, 133–147. [Google Scholar] [CrossRef] [PubMed]
- Longley, D.B.; Johnston, P.G. Molecular mechanisms of drug resistance. J. Pathol. 2005, 205, 275–292. [Google Scholar] [CrossRef] [PubMed]
- Zahreddine, H.; Borden, K.L.B. Mechanisms and insights into drug resistance in cancer. Front. Pharmacol. 2013, 4, 28. [Google Scholar] [CrossRef] [PubMed]
- Blondy, S.; David, V.; Verdier, M.; Mathonnet, M.; Perraud, A.; Christou, N. 5-Fluorouracil resistance mechanisms in colorectal cancer: From classical pathways to promising processes. Cancer Sci. 2020, 111, 3142–3154. [Google Scholar] [CrossRef]


| Gene Name | Primer Sequence (Forword and Reverse, 5′→3′) |
|---|---|
| 1 P-gp | AAG GCC TAA TGC CGA ACA CA TCC AGG CTC AGT CCC TGA AG |
| 2 GST | TTC CTG TGG CAT AAT GTG AT CTG ATT CAA AGG CAA ATC TC |
| 3 TS | ACC GAG CTC CCG AGA CTT TTT GGA CAG CCT ACC AAG CTT AAG AAT CCT GAG CTT TGG GAA |
| 4 GAPDH | CAT GGC CTT CCG TGT TCC T CCT GCT TCA CCA CCT TCT TGA |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kang, Y.-H.; Lee, J.-S.; Lee, N.-H.; Kim, S.-H.; Seo, C.-S.; Son, C.-G. Coptidis Rhizoma Extract Reverses 5-Fluorouracil Resistance in HCT116 Human Colorectal Cancer Cells via Modulation of Thymidylate Synthase. Molecules 2021, 26, 1856. https://doi.org/10.3390/molecules26071856
Kang Y-H, Lee J-S, Lee N-H, Kim S-H, Seo C-S, Son C-G. Coptidis Rhizoma Extract Reverses 5-Fluorouracil Resistance in HCT116 Human Colorectal Cancer Cells via Modulation of Thymidylate Synthase. Molecules. 2021; 26(7):1856. https://doi.org/10.3390/molecules26071856
Chicago/Turabian StyleKang, Yong-Hwi, Jin-Seok Lee, Nam-Hun Lee, Seung-Hyung Kim, Chang-Seob Seo, and Chang-Gue Son. 2021. "Coptidis Rhizoma Extract Reverses 5-Fluorouracil Resistance in HCT116 Human Colorectal Cancer Cells via Modulation of Thymidylate Synthase" Molecules 26, no. 7: 1856. https://doi.org/10.3390/molecules26071856
APA StyleKang, Y.-H., Lee, J.-S., Lee, N.-H., Kim, S.-H., Seo, C.-S., & Son, C.-G. (2021). Coptidis Rhizoma Extract Reverses 5-Fluorouracil Resistance in HCT116 Human Colorectal Cancer Cells via Modulation of Thymidylate Synthase. Molecules, 26(7), 1856. https://doi.org/10.3390/molecules26071856








