Computational Modeling to Explain Why 5,5-Diarylpentadienamides are TRPV1 Antagonists
Abstract
1. Introduction
2. Results
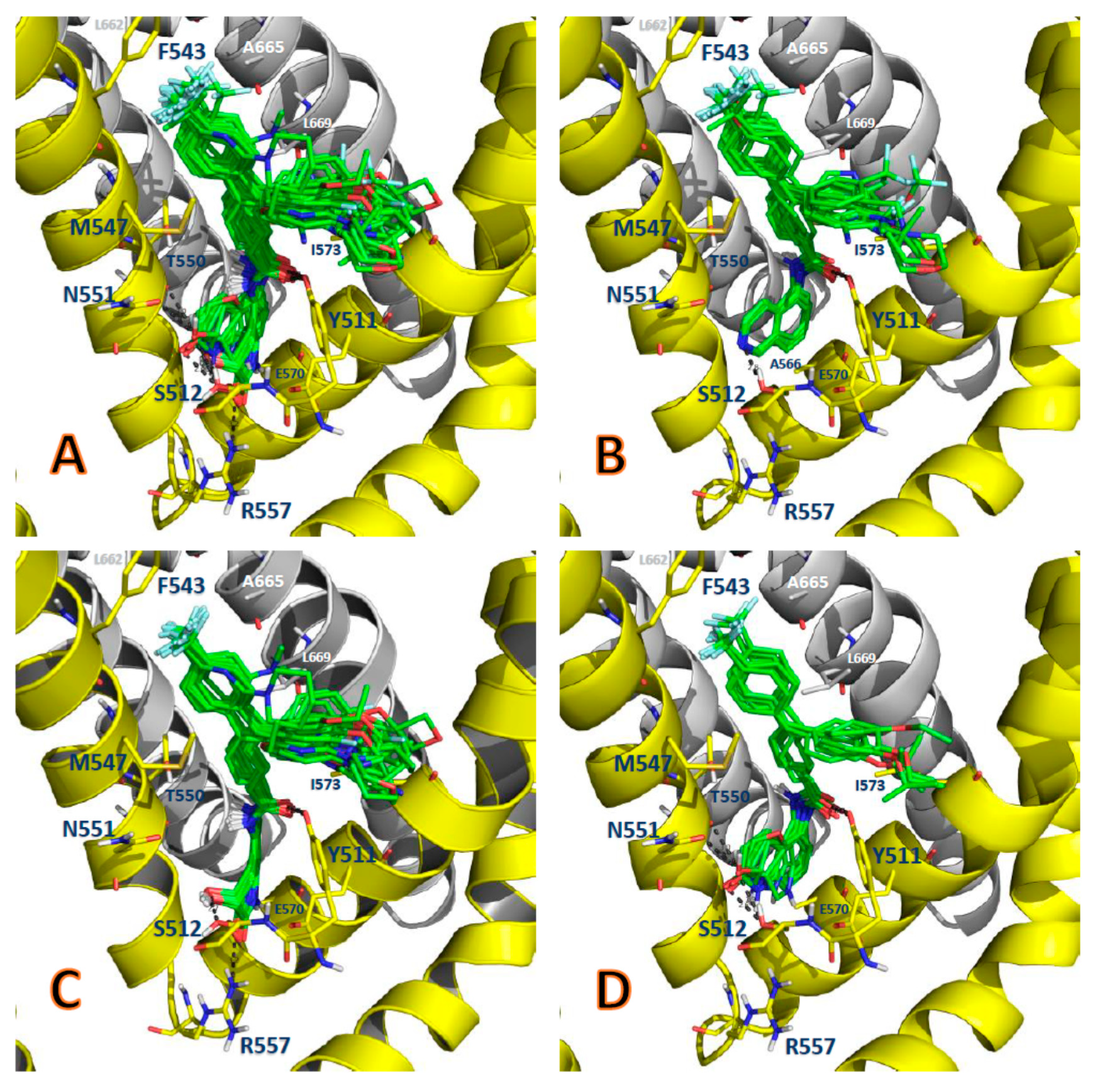
2.1. The Docking Poses
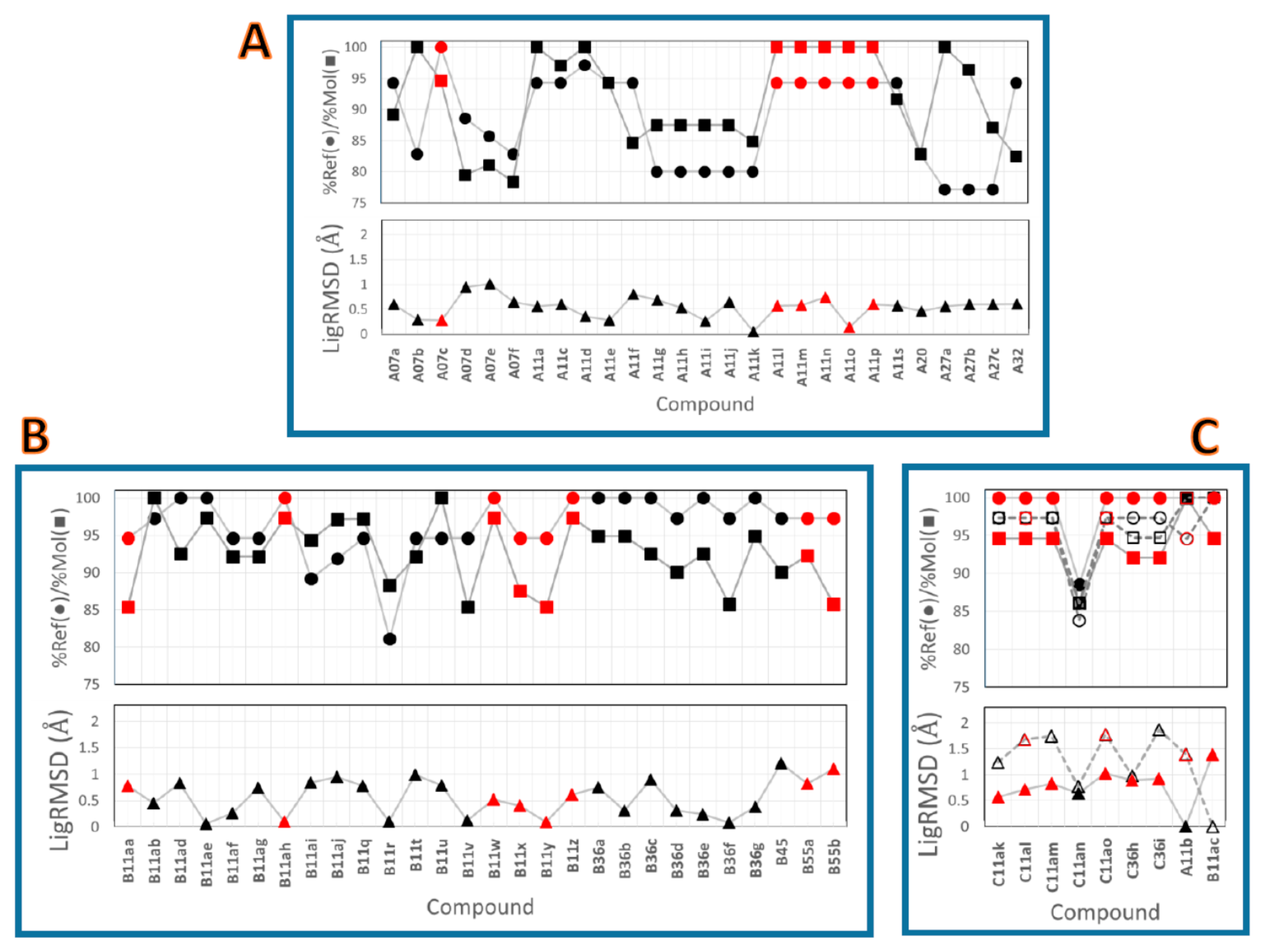
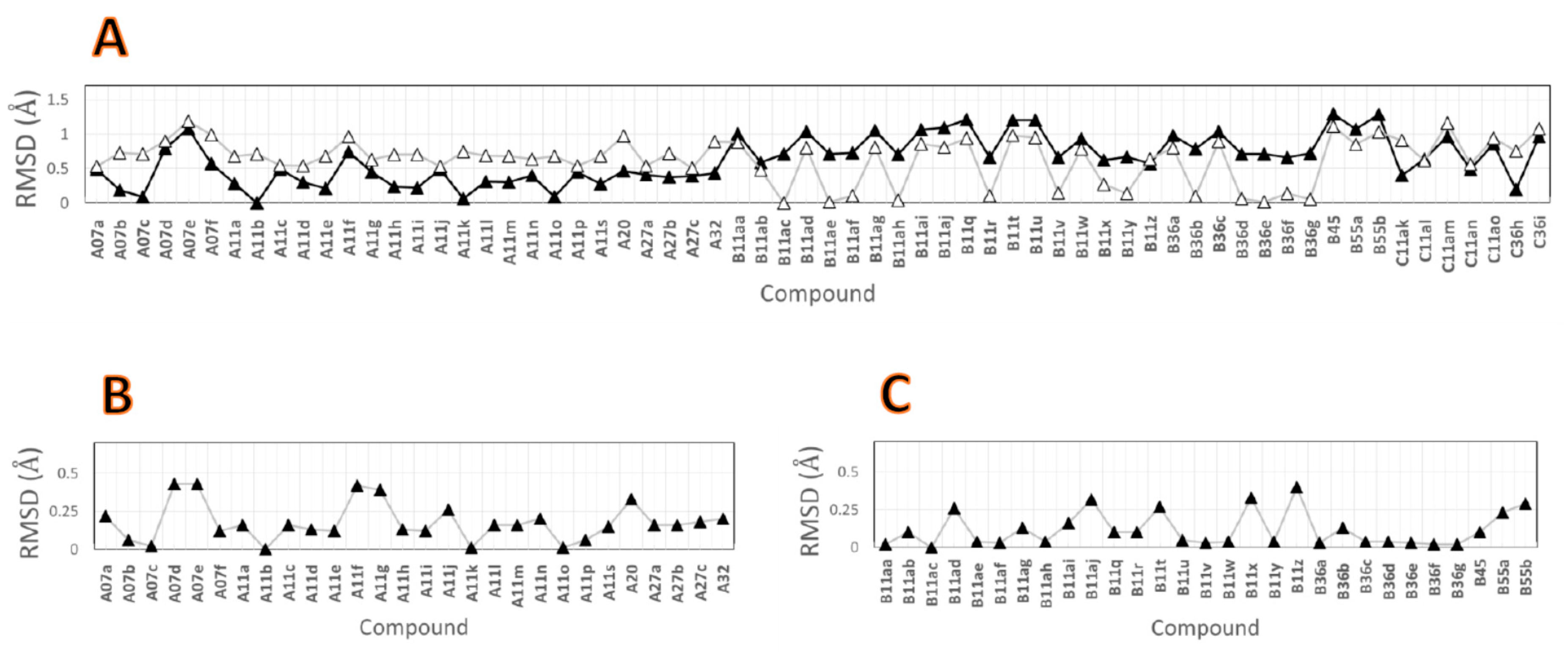
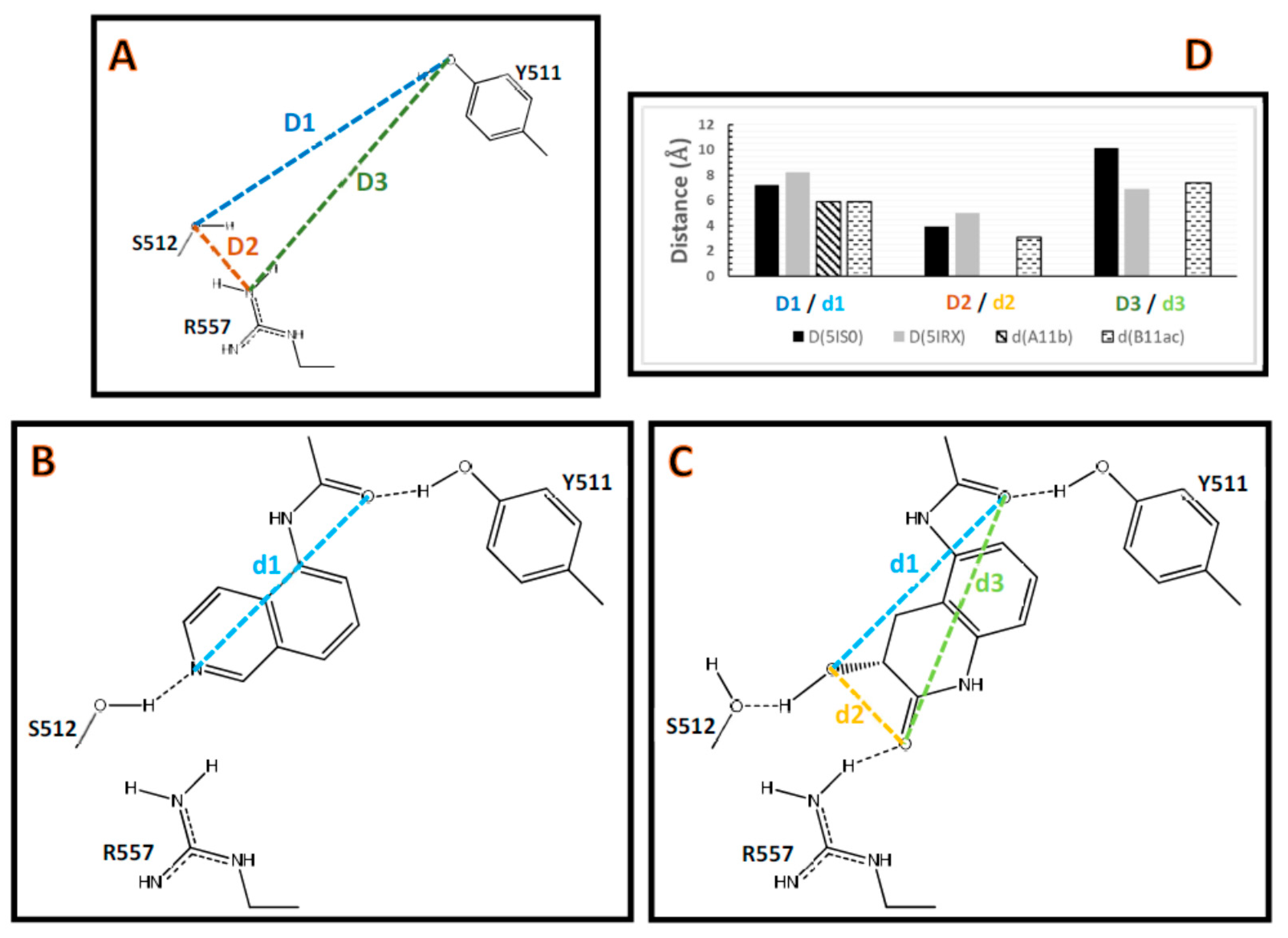
2.2. Comparison between the Poses
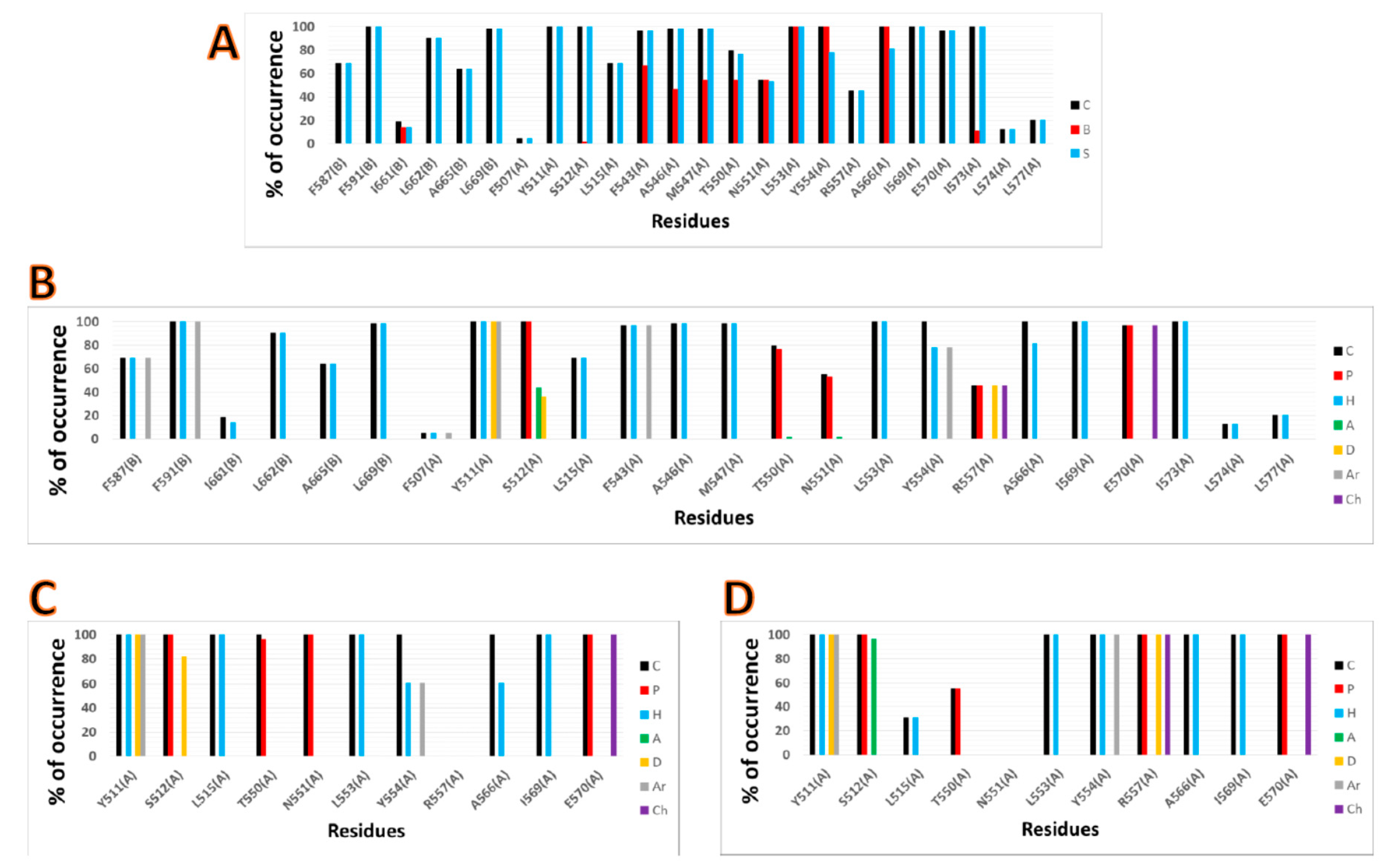
2.3. Interactions with Residues at the TRPV1 Binding Site
2.4. Docking Models Explain Why DPDAs are TRPV1 Antagonists
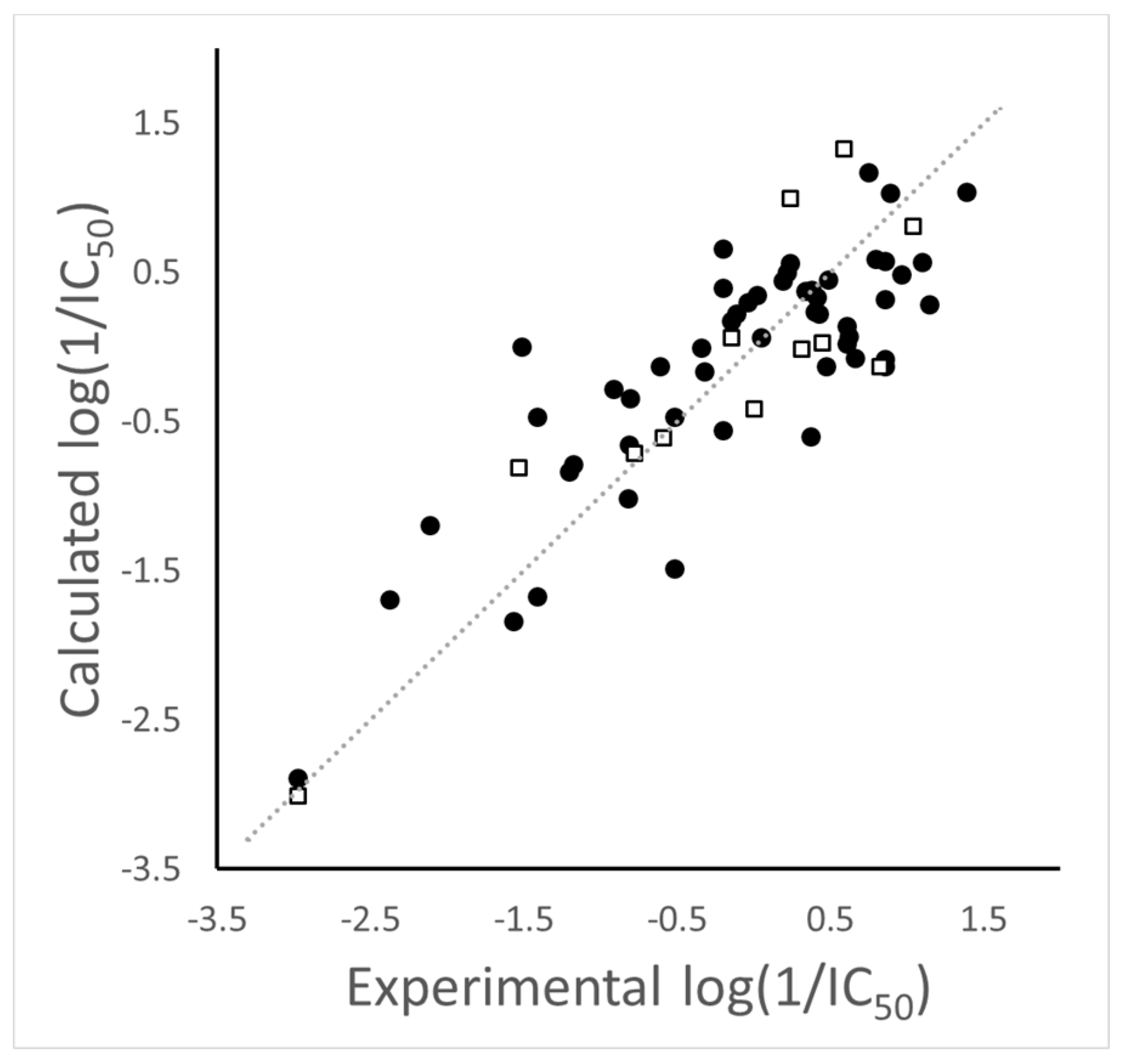
2.5. 2D Autocorrelation Models for Describing Differential Activities
3. Materials and Methods
3.1. Dataset
3.2. Molecular Docking Calculations
3.3. Comparison of the Binding Poses
3.4. IFP Calculations
3.5. 2D Autocorrelation QSAR Modeling
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Sample Availability
Abbreviations
| 3D | Three-dimensional |
| DPDA | 5,5-Diarylpentadienamide |
| GA | Genetic algorithm |
| HB | Hydrogen bond |
| IFP | Interaction fingerprint |
| LOO-CV | Leave-one-out cross-validation |
| PDB | Protein Data Bank |
| RMSD | Root mean square deviation |
| QSAR | Quantitative structure-activity relationship |
| TRPV1 | Transient receptor potential vanilloid 1 |
References
- Montell, C.; Birnbaumer, L.; Flockerzi, V.; Bindels, R.J.; Bruford, E.A.; Caterina, M.J.; Clapham, D.E.; Harteneck, C.; Heller, S.; Julius, D.; et al. A Unified Nomenclature for the Superfamily of TRP Cation Channels. Mol. Cell 2002, 9, 229–231. [Google Scholar] [CrossRef]
- Holzer, P. TRPV1 and the Gut: From a Tasty Receptor for a Painful Vanilloid to a Key Player in Hyperalgesia. Eur. J. Pharm. 2004, 500, 231–241. [Google Scholar] [CrossRef]
- Fischer, M.J.M.; Btesh, J.; McNaughton, P.A. Disrupting Sensitization of Transient Receptor Potential Vanilloid Subtype 1 Inhibits Inflammatory Hyperalgesia. J. Neurosci. 2013, 33, 7407–7414. [Google Scholar] [CrossRef]
- Xiao, Y.; Chen, X.; Zhang, P.-A.; Xu, Q.; Zheng, H.; Xu, G.-Y. TRPV1-Mediated Presynaptic Transmission in Basolateral Amygdala Contributes to Visceral Hypersensitivity in Adult Rats with Neonatal Maternal Deprivation. Sci. Rep. 2016, 6, 29026. [Google Scholar] [CrossRef]
- Szymaszkiewicz, A.; Włodarczyk, J.; Wasilewski, A.; Di Marzo, V.; Storr, M.; Fichna, J.; Zielińska, M. Desensitization of Transient Receptor Potential Vanilloid Type-1 (TRPV1) Channel as Promising Therapy of Irritable Bowel Syndrome: Characterization of the Action of Palvanil in the Mouse Gastrointestinal Tract. Naunyn Schmiedebergs Arch. Pharm. 2020, 393, 1357–1364. [Google Scholar] [CrossRef] [PubMed]
- Uchytilova, E.; Spicarova, D.; Palecek, J. TRPV1 Antagonist Attenuates Postoperative Hypersensitivity by Central and Peripheral Mechanisms. Mol. Pain 2014, 10, 67. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Cao, E.; Julius, D.; Cheng, Y. TRPV1 Structures in Nanodiscs Reveal Mechanisms of Ligand and Lipid Action. Nature 2016, 534, 347–351. [Google Scholar] [CrossRef] [PubMed]
- Pretze, M.; Pallavi, P.; Roscher, M.; Klotz, S.; Caballero, J.; Binzen, U.; Greffrath, W.; Treede, R.-D.; Harmsen, M.C.; Hafner, M.; et al. Radiofluorinated N-Octanoyl Dopamine ([18F]F-NOD) as a Tool To Study Tissue Distribution and Elimination of NOD in Vitro and in Vivo. J. Med. Chem. 2016, 59, 9855–9865. [Google Scholar] [CrossRef] [PubMed]
- Pallavi, P.; Pretze, M.; Caballero, J.; Li, Y.; Hofmann, B.B.; Stamellou, E.; Klotz, S.; Wängler, C.; Wängler, B.; Loesel, R.; et al. Analyses of Synthetic N-Acyl Dopamine Derivatives Revealing Different Structural Requirements for Their Anti-Inflammatory and Transient-Receptor-Potential-Channel-of-the-Vanilloid-Receptor-Subfamily-Subtype-1 (TRPV1)-Activating Properties. J. Med. Chem. 2018, 61, 3126–3137. [Google Scholar] [CrossRef]
- Norman, M.H.; Zhu, J.; Fotsch, C.; Bo, Y.; Chen, N.; Chakrabarti, P.; Doherty, E.M.; Gavva, N.R.; Nishimura, N.; Nixey, T.; et al. Novel Vanilloid Receptor-1 Antagonists: 1. Conformationally Restricted Analogues of Trans-Cinnamides. J. Med. Chem. 2007, 50, 3497–3514. [Google Scholar] [CrossRef]
- Ha, T.-H.; Ryu, H.; Kim, S.-E.; Kim, H.S.; Ann, J.; Tran, P.-T.; Hoang, V.-H.; Son, K.; Cui, M.; Choi, S.; et al. TRPV1 Antagonist with High Analgesic Efficacy: 2-Thio Pyridine C-Region Analogues of 2-(3-Fluoro-4-Methylsulfonylaminophenyl)Propanamides. Bioorg. Med. Chem. 2013, 21, 6657–6664. [Google Scholar] [CrossRef] [PubMed]
- Benso, B.; Bustos, D.; Zarraga, M.O.; Gonzalez, W.; Caballero, J.; Brauchi, S. Chalcone Derivatives as Non-Canonical Ligands of TRPV1. Int. J. Biochem. Cell Biol. 2019, 112, 18–23. [Google Scholar] [CrossRef] [PubMed]
- Carnevale, V.; Rohacs, T. TRPV1: A Target for Rational Drug Design. Pharmaceuticals 2016, 9, 52. [Google Scholar] [CrossRef] [PubMed]
- Cho, Y.; Kim, M.S.; Kim, H.S.; Ann, J.; Lee, J.; Pearce, L.V.; Pavlyukovets, V.A.; Morgan, M.A.; Blumberg, P.M.; Lee, J. The SAR Analysis of TRPV1 Agonists with the α-Methylated B-Region. Bioorg. Med. Chem. Lett. 2012, 22, 5227–5231. [Google Scholar] [CrossRef] [PubMed]
- Ann, J.; Kim, H.S.; Thorat, S.A.; Kim, H.; Ha, H.-J.; Choi, K.; Kim, Y.-H.; Kim, M.; Hwang, S.W.; Pearce, L.V.; et al. Discovery of Nonpungent Transient Receptor Potential Vanilloid 1 (TRPV1) Agonist as Strong Topical Analgesic. J. Med. Chem. 2020, 63, 418–424. [Google Scholar] [CrossRef]
- Li, J.; Nie, C.; Qiao, Y.; Hu, J.; Li, Q.; Wang, Q.; Pu, X.; Yan, L.; Qian, H. Design, Synthesis and Biological Evaluation of Novel 2,3,4,9-Tetrahydro-1H-Pyrido[3,4-b]Indole Triazole Derivatives as Potent TRPV1 Antagonists. Eur. J. Med. Chem. 2019, 178, 433–445. [Google Scholar] [CrossRef] [PubMed]
- Ahn, S.; Kim, Y.S.; Kim, M.S.; Ann, J.; Ha, H.; Yoo, Y.D.; Kim, Y.H.; Blumberg, P.M.; Frank-Foltyn, R.; Bahrenberg, G.; et al. Discovery of Indane Propanamides as Potent and Selective TRPV1 Antagonists. Bioorg. Med. Chem. Lett. 2020, 30, 126838. [Google Scholar] [CrossRef]
- Saku, O.; Ishida, H.; Atsumi, E.; Sugimoto, Y.; Kodaira, H.; Kato, Y.; Shirakura, S.; Nakasato, Y. Discovery of Novel 5,5-Diarylpentadienamides as Orally Available Transient Receptor Potential Vanilloid 1 (TRPV1) Antagonists. J. Med. Chem. 2012, 55, 3436–3451. [Google Scholar] [CrossRef] [PubMed]
- Yang, F.; Zheng, J. Understand Spiciness: Mechanism of TRPV1 Channel Activation by Capsaicin. Protein Cell 2017, 8, 169–177. [Google Scholar] [CrossRef] [PubMed]
- Pagadala, N.S.; Syed, K.; Tuszynski, J. Software for Molecular Docking: A Review. Biophys. Rev. 2017, 9, 91–102. [Google Scholar] [CrossRef] [PubMed]
- Velázquez-Libera, J.L.; Durán-Verdugo, F.; Valdés-Jiménez, A.; Núñez-Vivanco, G.; Caballero, J. LigRMSD: A Web Server for Automatic Structure Matching and RMSD Calculations among Identical and Similar Compounds in Protein-Ligand Docking. Bioinformatics 2020, 36, 2912–2914. [Google Scholar] [CrossRef]
- Caballero, J.; Morales-Bayuelo, A.; Navarro-Retamal, C. Mycobacterium Tuberculosis Serine/Threonine Protein Kinases: Structural Information for the Design of Their Specific ATP-Competitive Inhibitors. J. Comput. Aided Mol. Des. 2018, 32, 1315–1336. [Google Scholar] [CrossRef] [PubMed]
- Caballero, J. Considerations for Docking of Selective Angiotensin-Converting Enzyme Inhibitors. Molecules 2020, 25, 295. [Google Scholar] [CrossRef] [PubMed]
- Doweyko, A.M. 3D-QSAR Illusions. J. Comput. Aided Mol. Des. 2004, 18, 587–596. [Google Scholar] [CrossRef] [PubMed]
- Friesner, R.A.; Banks, J.L.; Murphy, R.B.; Halgren, T.A.; Klicic, J.J.; Mainz, D.T.; Repasky, M.P.; Knoll, E.H.; Shelley, M.; Perry, J.K.; et al. Glide: A New Approach for Rapid, Accurate Docking and Scoring. 1. Method and Assessment of Docking Accuracy. J. Med. Chem. 2004, 47, 1739–1749. [Google Scholar] [CrossRef] [PubMed]
- Banks, J.L.; Beard, H.S.; Cao, Y.; Cho, A.E.; Damm, W.; Farid, R.; Felts, A.K.; Halgren, T.A.; Mainz, D.T.; Maple, J.R.; et al. Integrated Modeling Program, Applied Chemical Theory (IMPACT). J. Comput. Chem. 2005, 26, 1752–1780. [Google Scholar] [CrossRef]
- Harder, E.; Damm, W.; Maple, J.; Wu, C.; Reboul, M.; Xiang, J.Y.; Wang, L.; Lupyan, D.; Dahlgren, M.K.; Knight, J.L.; et al. OPLS3: A Force Field Providing Broad Coverage of Drug-like Small Molecules and Proteins. J. Chem. Theory Comput. 2016, 12, 281–296. [Google Scholar] [CrossRef]
- Quesada-Romero, L.; Mena-Ulecia, K.; Tiznado, W.; Caballero, J. Insights into the Interactions between Maleimide Derivates and GSK3β Combining Molecular Docking and QSAR. PLoS ONE 2014, 9, e102212. [Google Scholar] [CrossRef]
- Quesada-Romero, L.; Caballero, J. Docking and Quantitative Structure—Activity Relationship of Oxadiazole Derivates as Inhibitors of GSK3beta. Mol. Divers. 2014, 18, 149–159. [Google Scholar] [CrossRef]
- Mena-Ulecia, K.; Tiznado, W.; Caballero, J. Study of the Differential Activity of Thrombin Inhibitors Using Docking, QSAR, Molecular Dynamics, and MM-GBSA. PLoS ONE 2015, 10, e0142774. [Google Scholar] [CrossRef]
- Malhotra, S.; Karanicolas, J. When Does Chemical Elaboration Induce a Ligand To Change Its Binding Mode? J. Med. Chem. 2017, 60, 128–145. [Google Scholar] [CrossRef] [PubMed]
- Drwal, M.N.; Jacquemard, C.; Perez, C.; Desaphy, J.; Kellenberger, E. Do Fragments and Crystallization Additives Bind Similarly to Drug-like Ligands? J. Chem. Inf. Model. 2017, 57, 1197–1209. [Google Scholar] [CrossRef]
- Muñoz-Gutierrez, C.; Adasme-Carreño, F.; Fuentes, E.; Palomo, I.; Caballero, J. Computational Study of the Binding Orientation and Affinity of PPARγ Agonists: Inclusion of Ligand-Induced Fit by Cross-Docking. RSC. Adv. 2016, 6, 64756–64768. [Google Scholar] [CrossRef]
- Ramírez, D.; Caballero, J. Is It Reliable to Take the Molecular Docking Top Scoring Position as the Best Solution without Considering Available Structural Data? Molecules 2018, 23, 1038. [Google Scholar] [CrossRef] [PubMed]
- Deng, Z.; Chuaqui, C.; Singh, J. Structural Interaction Fingerprint (SIFt): A Novel Method for Analyzing Three-Dimensional Protein-Ligand Binding Interactions. J. Med. Chem. 2004, 47, 337–344. [Google Scholar] [CrossRef] [PubMed]
- Moreau, G.; Broto, P. Autocorrelation of Molecular Structures: Application to SAR Studies. Nouv. J. Chem. 1980, 4, 757–764. [Google Scholar]
- Moran, P. Notes on Continuous Stochastic Processes. Biometrika 1950, 37, 17–23. [Google Scholar] [CrossRef]
- Geary, R.C. The Contiguity Ratio and Statistical Mapping. Inc. Stat. 1954, 5, 115–146. [Google Scholar] [CrossRef]
- Fernández, M.; Tundidor-Camba, A.; Caballero, J.M. 2D Autocorrelation Modeling of the Activity of Trihalobenzocycloheptapyridine Analogues as Farnesyl Protein Transferase Inhibitors. Mol. Simul. 2005, 31, 575–584. [Google Scholar] [CrossRef]
- González, M.; Caballero, J.; Helguera, A.; Garriga, M.; González, G.; Fernández, M. 2D Autocorrelation Modelling of the Inhibitory Activity of Cytokinin-Derived Cyclin-Dependent Kinase Inhibitors. Bull. Math. Biol. 2006, 68, 735–751. [Google Scholar] [CrossRef]
- Fernández, M.; Caballero, J. QSAR Modeling of Matrix Metalloproteinase Inhibition by N-Hydroxy-[Alpha]-Phenylsulfonylacetamide Derivatives. Bioorg. Med. Chem. 2007, 15, 6298–6310. [Google Scholar] [CrossRef]
- Caballero, J.; Fernández, M.; González-Nilo, F.D. Structural Requirements of Pyrido[2,3-d]Pyrimidin-7-One as CDK4/D Inhibitors: 2D Autocorrelation, CoMFA and CoMSIA Analyses. Bioorg. Med. Chem. 2008, 16, 6103–6115. [Google Scholar] [CrossRef] [PubMed]
- Caballero, J.; Fernández, M.; Saavedra, M.; González-Nilo, F.D. 2D Autocorrelation, CoMFA, and CoMSIA Modeling of Protein Tyrosine Kinases’ Inhibition by Substituted Pyrido[2,3-d]Pyrimidine Derivatives. Bioorg. Med. Chem. 2008, 16, 810–821. [Google Scholar] [CrossRef] [PubMed]
- Caballero, J.; Tundidor-Camba, A.; Fernández, M. Modeling of the Inhibition Constant (Ki) of Some Cruzain Ketone-Based Inhibitors Using 2D Spatial Autocorrelation Vectors and Data-Diverse Ensembles of Bayesian-Regularized Genetic Neural Networks. QSAR Comb. Sci. 2007, 26, 27–40. [Google Scholar] [CrossRef]
- de Oliveira, D.B.; Gaudio, A.C. BuildQSAR: A New Computer Program for QSAR Analysis. Quant. Struct. Act. Relat. 2000, 19, 599–601. [Google Scholar] [CrossRef]
| Series A | 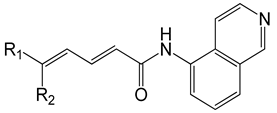 | ||||
| Compound | R1 | R2 | Experimental log(1/IC50) | Predicted log(1/IC50) | Glide XP Score (kcal/mol) |
| A07a | 4-(CF3)-phenyl | 4-(CF3)-phenyl | 0.377 | 0.382 | −7.75 |
| A07b | Phenyl | Phenyl | −1.568 | −1.837 | −9.10 |
| A07c | 6-(CF3)-pyridin-3-yl | 6-(CF3)-pyridin-3-yl | −2.114 | −1.192 | −10.84 |
| A07d | 4-(OCF3)-phenyl) | 4-(OCF3)-phenyl | −0.820 | −1.015 | −6.83 |
| A07e | 4-(tBu)-phenyl | 4-(tBu)-phenyl | 0.745 | 1.169 | −6.83 |
| A07f | 3-(CF3)-phenyl | 3-(CF3)-phenyl | −0.204 | −0.557 | −7.71 |
| A11a | 4-(CF3)-phenyl | Phenyl | 0.854 | −0.127 | −10.85 |
| A11b 1 | 4-(CF3)-phenyl | 4-(OMe)-phenyl | 0.824 | −0.132 | −10.79 |
| A11c | 4-(CF3)-phenyl | 4-(F)-phenyl | 1.143 | 0.285 | −10.68 |
| A11d | 4-(CF3)-phenyl | 4-(OH)-phenyl | −0.914 | −0.279 | −9.33 |
| A11e | 4-(CF3)-phenyl | 3-(CN)-phenyl | −0.322 | −0.164 | −7.88 |
| A11f | 4-(CF3)-phenyl | 4-Morpholinophenyl | 0.237 | 0.560 | −7.97 |
| A11g | 4-(CF3)-phenyl | Thiophen-2-yl | 0.018 | 0.350 | −10.28 |
| A11h 1 | 4-(CF3)-phenyl | Thiophen-3-yl | 0.310 | −0.012 | −7.74 |
| A11i | 4-(CF3)-phenyl | Furan-2-yl | −0.519 | −0.470 | −7.36 |
| A11j | 4-(CF3)-phenyl | Furan-3-yl | −0.519 | −1.487 | −10.04 |
| A11k | 4-(CF3)-phenyl | 5-(Me)-furan-2-yl | 0.469 | −0.128 | −10.59 |
| A11l 1 | 4-(CF3)-phenyl | Pyridin-3-yl | −1.531 | −0.810 | −10.70 |
| A11m | 4-(CF3)-phenyl | Pyridin-4-yl | −1.204 | −0.837 | −10.81 |
| A11n | 4-(CF3)-phenyl | Pyrimidin-5-yl | −2.380 | −1.695 | −10.48 |
| A11o | 4-(CF3)-phenyl | Cyclohex-1-en-1-yl | 0.481 | 0.452 | −10.55 |
| A11p | 4-(CF3)-phenyl | 3,6-Dihydro-2H-pyran-4-yl | −1.415 | −0.467 | −9.54 |
| A11s 1 | 4-(CF3)-phenyl | 4-(NMe2)-phenyl | 0.444 | 0.027 | −11.05 |
| A20 | 4-(CF3)-phenyl | 4-(CF3)-phenyl | −0.613 | −0.127 | −9.04 |
| A27a 1 | 4-(CF3)-phenyl | H | −2.973 | −3.007 | −8.52 |
| A27b | 4-(CF3)-phenyl | Me | −2.978 | −2.888 | −10.09 |
| A27c 1 | 4-(CF3)-phenyl | nBu | −0.778 | −0.711 | −10.88 |
| A32 | 4-(CF3)-phenyl | 4-(Morpholinomethyl)-phenyl | −0.204 | 0.663 | −4.83 |
| Series B |  | ||||
| Compound | R1 | R2 | Experimental log(1/IC50) | Predicted log(1/IC50) | Glide XP Score (kcal/mol) |
| B11aa | 4-(CF3)-phenyl | 2-(Piperidin-1-yl)-pyrimidin-5-yl) | 0.367 | −0.600 | −10.41 |
| B11ab | 4-(CF3)-phenyl | 4-(OH)-phenyl | −1.519 | 0.006 | −11.32 |
| B11ac | 4-(CF3)-phenyl | 4-(OMe)-phenyl | 0.602 | 0.024 | −11.85 |
| B11ad | 4-(CF3)-phenyl | 4-(OCF3)-phenyl | 0.854 | −0.081 | −10.94 |
| B11ae | 4-(CF3)-phenyl | 4-(OEt)-phenyl | 0.409 | 0.335 | −12.02 |
| B11af | 4-(CF3)-phenyl | 2-(OEt)-phenyl | −0.204 | 0.399 | −11.85 |
| B11ag | 4-(CF3)-phenyl | 3-(OEt)-phenyl | 0.046 | 0.062 | −11.26 |
| B11ah 1 | 4-(CF3)-phenyl | 6-(OEt)-pyridin-3-yl | −0.591 | −0.607 | −11.98 |
| B11ai | 4-(Cl)-phenyl | 4-(OEt)-phenyl | 0.620 | 0.071 | −9.34 |
| B11aj | 4-(Me)-phenyl | 4-(OEt)-phenyl | 0.398 | 0.237 | −8.96 |
| B11q | 4-(CF3)-phenyl | 4-(F)-phenyl | 0.959 | 0.488 | −10.90 |
| B11r | 4-(CF3)-phenyl | Furan-2-yl | −0.806 | −0.342 | −11.42 |
| B11t | 4-(CF3)-phenyl | 4-(NMe2)-phenyl | −0.146 | 0.174 | −10.17 |
| B11u | 4-(CF3)-phenyl | Phenyl | 0.420 | 0.222 | −10.61 |
| B11v 1 | 4-(CF3)-phenyl | 4-(Piperidin-1-yl)-phenyl | 0.585 | 1.327 | −12.59 |
| B11w | 4-(CF3)-phenyl | 6-(NMe2)-pyridin-3-yl | −0.813 | −0.656 | −10.22 |
| B11x | 4-(CF3)-phenyl | 6-(pyrrolidin-1-yl)-pyridin-3-yl | −0.342 | −0.006 | −11.63 |
| B11y | 4-(CF3)-phenyl | 6-(piperidin-1-yl)-pyridin-3-yl) | 0.187 | 0.447 | −12.38 |
| B11z | 4-(CF3)-phenyl | 2-(NMe2)-pyrimidin-5-yl | −1.415 | −1.671 | −11.18 |
| B36a | 4-(CF3)-phenyl | 4-(O-n-Pr)-phenyl | 1.097 | 0.573 | −11.86 |
| B36b | 4-(CF3)-phenyl | 4-(O-i-Pr)-phenyl | 0.854 | 0.581 | −11.62 |
| B36c 1 | 4-(CF3)-phenyl | 4-(O-t-Bu)-phenyl | 1.041 | 0.806 | −10.05 |
| B36d | 4-(CF3)-phenyl | 4-Cyclobutoxyphenyl | 1.387 | 1.041 | −12.30 |
| B36e | 4-(CF3)-phenyl | 4-(Cyclopropylmethoxy)phenyl | 0.886 | 1.034 | −12.12 |
| B36f 1 | 4-(CF3)-phenyl | 4-((Tetrahydro-2H-pyran-4-yl)oxy)-phenyl | 0.237 | 0.998 | −12.36 |
| B36g 1 | 4-(CF3)-phenyl | 4-(Cyanomethoxy)-phenyl | −0.146 | 0.060 | −12.02 |
| B45 | 4-(CF3)-phenyl | 4-(Oxetan-3-yloxy)-phenyl | −0.041 | 0.300 | −9.27 |
| B55a 1 | 2-(NMe2)-6-(CF3)-pyridin-3-yl | 4-(F)-phenyl | 0.000 | −0.416 | −10.65 |
| B55b | 2-(Piperidin-1-yl)-6-(CF3)-pyridin-3-yl | 4-(F)-phenyl | 0.215 | 0.498 | −9.96 |
| Series C | 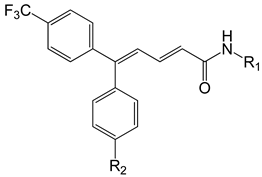 | ||||
| Compound | R1 | R2 | Experimental log(1/IC50) | Predicted log(1/IC50) | Glide XP Score (kcal/mol) |
| C11ak | 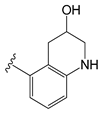 | OEt | 0.854 | 0.321 | −11.18 |
| C11al |  | OEt | 0.658 | −0.076 | −10.76 |
| C11am |  | OEt | 0.602 | 0.139 | −10.66 |
| C11an | 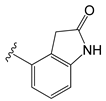 | OEt | −0.114 | 0.224 | −11.56 |
| C11ao | 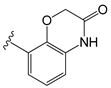 | OEt | −1.176 | −0.786 | −8.27 |
| C36h | 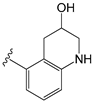 | O−i−Pr | 0.796 | 0.593 | −11.74 |
| C36i | 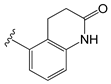 | O−i−Pr | 0.337 | 0.380 | −10.38 |
| Compound | Head Group | Residues and Their Role in an HB 1 |
|---|---|---|
| Series A | Isoquinoline | Y511 (donor); S512 (donor) |
| Series B | 3-Hydroxy-3,4-dihydroquinolin-2(1H)-one | Y511 (donor); S512 (acceptor); R557 (donor). |
| C11ak | 1,2,3,4-Tetrahydroquinolin-3-ol | Y511 (donor); S512 (donor); N551 (acceptor). 2 |
| C11al | 2-Oxo-1,2-dihydro-quinoline | Y511 (donor); S512 (donor). |
| C11am and C36i | 3,4-Dihydroquinolin-2(1H)-one | Y511 (donor); S512 (donor). |
| C11an | Indolin-2-one | Y511 (donor); S512 (donor). |
| C11ao | 2H-benzo[b]-[1,4] oxazin-3(4H)-one | Y511 (donor); S512 (donor). |
| C36h | 1,2,3,4-Tetrahydroquinolin-3-ol | Y511 (donor); T550 (acceptor). 3 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Caballero, J. Computational Modeling to Explain Why 5,5-Diarylpentadienamides are TRPV1 Antagonists. Molecules 2021, 26, 1765. https://doi.org/10.3390/molecules26061765
Caballero J. Computational Modeling to Explain Why 5,5-Diarylpentadienamides are TRPV1 Antagonists. Molecules. 2021; 26(6):1765. https://doi.org/10.3390/molecules26061765
Chicago/Turabian StyleCaballero, Julio. 2021. "Computational Modeling to Explain Why 5,5-Diarylpentadienamides are TRPV1 Antagonists" Molecules 26, no. 6: 1765. https://doi.org/10.3390/molecules26061765
APA StyleCaballero, J. (2021). Computational Modeling to Explain Why 5,5-Diarylpentadienamides are TRPV1 Antagonists. Molecules, 26(6), 1765. https://doi.org/10.3390/molecules26061765






