A Comprehensive Analysis Using Colorimetry, Liquid Chromatography-Tandem Mass Spectrometry and Bioassays for the Assessment of Indole Related Compounds Produced by Endophytes of Selected Wheat Cultivars
Abstract
1. Introduction
2. Results
2.1. Identification of Endophytes
2.2. Screening of Production of Indole Related Compounds (IRCs) by Endophytic Isolates—Colorimetric Method
2.3. Determination of Indole-3-Acetic Acid by LC-MS/MS
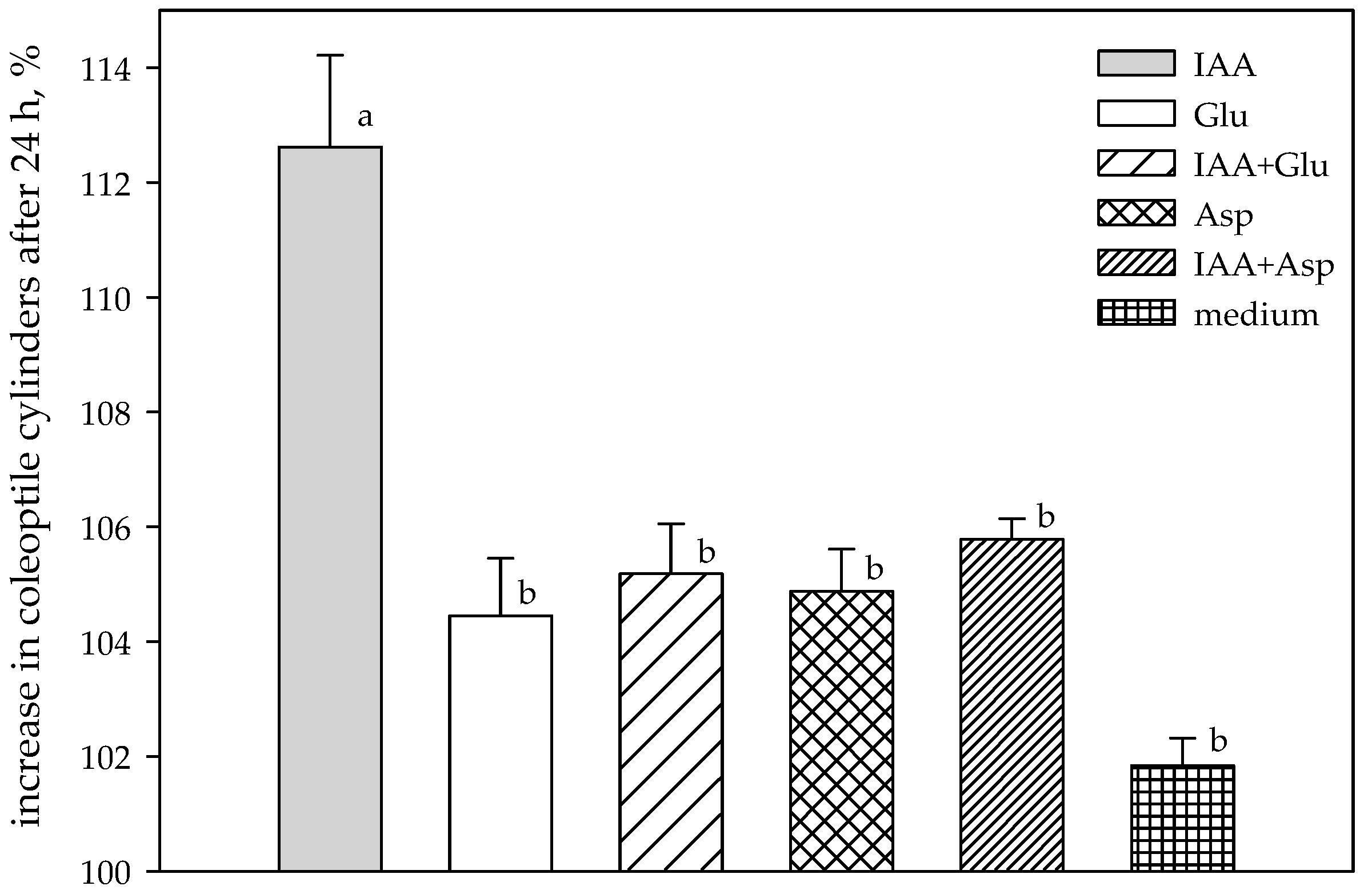
2.4. Biotest of Selected Crude Bacterial Supernatants
2.5. Comparison of the Three Methods for Determination of IRCs
3. Discussion
4. Materials and Methods
4.1. Plant Material
4.2. Isolation and Identification of Endophytes
4.3. Colorimetric Determination of IAA
4.4. Detection and Quantification of Free IAA by LC-MS/MS
4.5. Test of IAA Biological Activity (Biotest)
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Sample Availability
Appendix A
| Bacterial Genus | Culture Collection ID | Plant Part | GenBank Accession Number |
|---|---|---|---|
| Bacillus sp. | HLA 1-7 | Hondia leaf | MT181071 |
| Bacillus sp. | HLA 2-1 | Hondia leaf | MT181072 |
| Serratia sp. | HLA 2-2 | Hondia leaf | MT181073 |
| Bacillus sp. | HLA 2-5 | Hondia leaf | MT181075 |
| Bacillus sp. | HLA 2-7 | Hondia leaf | MT181077 |
| Bacillus sp. | HLB 1-3 | Hondia leaf | MT181080 |
| Bacillus sp. | HLB 2-6 | Hondia leaf | MT181081 |
| Bacillus sp. | HLC 10 | Hondia leaf | MT181084 |
| Serratia sp. | HLC 2 | Hondia leaf | MT181085 |
| Lysinibacillus sp. | HLC 4 | Hondia leaf | MT181087 |
| Bacillus sp. | HLC 7 | Hondia leaf | MT734565 |
| Bacillus sp. | HLC 8 | Hondia leaf | MT181091 |
| Bacillus sp. | HLC 9 | Hondia leaf | MT181095 |
| Bacillus sp. | K 1-2 | Rokosz root | MT181089 |
| Bacillus sp. | K 1-7 | Rokosz root | MT181090 |
| Bacillus sp. | K 2-4 | Rokosz root | MT181092 |
| Bacillus sp. | K 4-3 | Rokosz root | MT181094 |
| Bacillus sp. | K 1-4 | Rokosz root | MT181097 |
| Bacillus sp. | K 4-6 | Rokosz root | MT181098 |
| Bacillus sp. | K 4-7 | Rokosz root | MT734566 |
| Bacillus sp. | KA 1-1 | Hondia root | MT181099 |
| Serratia sp. | KA 1-2 | Hondia root | MT181100 |
| Bacillus sp. | KB 1-6 | Hondia root | MT181107 |
| Lysinibacillus sp. | KC 1-4 | Hondia root | MT181112 |
| Paenibacillus sp. | PC 2-5 | Hondia coleoptile | MT181137 |
| Bacillus sp. | RL 2-2 | Rokosz leaf | MT181139 |
| Bacillus sp. | RL 4-2 | Rokosz leaf | MT181142 |
| Bacillus sp. | RP 2-4 | Rokosz coleoptile | MT181146 |
| Bacillus sp. | RP 4-2 | Rokosz coleoptile | MT181150 |
| Bacillus sp. | TKB 1-4 | Tytanika root | MT181152 |
| Bacillus sp. | TKC 1-6 | Tytanika root | MT181153 |
| Bacillus sp. | TKC 1-7 | Tytanika root | MT181154 |
| Bacillus sp. | TKC 1-8 | Tytanika root | MT181155 |
| Bacillus sp. | TKC 2-1 | Tytanika root | MT181099 |
| Bacillus sp. | TKC 2-2 | Tytanika root | MT181088 |
| Bacillus sp. | TLA 2-11 | Tytanika leaf | MT181156 |
| Bacillus sp. | TLA 3-2 | Tytanika leaf | MT181157 |
| Pantoea sp. | TLB 1-1 | Tytanika leaf | MT181158 |
| Bacillus sp. | TLB 1-2 | Tytanika leaf | MT181159 |
| Bacillus sp. | TLB 1-3 | Tytanika leaf | MT181160 |
| Bacillus sp. | TLB 2-7 | Tytanika leaf | MT181161 |
| Bacillus sp. | TLB 2-9 | Tytanika leaf | MT181162 |
| Bacillus sp. | TLC 1-8 | Tytanika leaf | MT181163 |
| Bacillus sp. | TLC 2-2 | Tytanika leaf | MT181164 |
| Bacillus sp. | TPA 1-2 | Tytanika coleoptile | MT181165 |
| Bacillus sp. | TPA 1-3 | Tytanika coleoptile | MT181166 |
| Bacillus sp. | TPA 2-6 | Tytanika coleoptile | MT181167 |
| Bacillus sp. | TPA 3-10 | Tytanika coleoptile | MT181168 |
| Bacillus sp. | TPA 3-8 | Tytanika coleoptile | MT181169 |
| Bacillus sp. | TPB 1-1 | Tytanika coleoptile | MT181170 |
| Bacillus sp. | TPB 2-7 | Tytanika coleoptile | MT734569 |
| Bacillus sp. | TPB 2-8 | Tytanika coleoptile | MT181173 |
| Paenibacillus sp. | TPC 1-12 | Tytanika coleoptile | MT181174 |
| Bacillus sp. | TPC 1-2 | Tytanika coleoptile | MT181175 |


| Culture Collection ID | Mean Concentration of DNA (µg·mL−1) | SD | Mean A 260:280 Ratio | SD | Mean A 260:230 Ratio | SD |
|---|---|---|---|---|---|---|
| HLA 1-7 | 1025.00 | 15.00 | 2.38 | 0.03 | 0.59 | 0.01 |
| HLA 2-1 | 9120.00 | 5.00 | 2.08 | 0.01 | 1.47 | 0.00 |
| HLA 2-2 | 355.10 | 1.31 | 2.16 | 0.00 | 0.51 | 0.00 |
| HLA 2-5 | 868.33 | 7.64 | 2.65 | 0.06 | 0.56 | 0.01 |
| HLA 2-7 | 481.67 | 7.64 | 2.96 | 0.16 | 0.34 | 0.00 |
| HLB 1-3 | 2370.00 | 10.00 | 2.10 | 0.02 | 1.35 | 0.02 |
| HLB 2-6 | 343.33 | 2.89 | 6.45 | 1.66 | 0.24 | 0.01 |
| HLC 10 | 41.83 | 0.31 | 1.76 | 0.04 | 0.43 | 0.02 |
| HLC 2 | 748.33 | 18.93 | 2.58 | 0.04 | 0.51 | 0.01 |
| HLC 4 | 96.93 | 0.15 | 2.07 | 0.01 | 1.35 | 0.01 |
| HLC 7 | 71.40 | 0.26 | 2.10 | 0.02 | 0.15 | 0.00 |
| HLC 8 | 1715.00 | 13.23 | 1.93 | 0.04 | 1.32 | 0.01 |
| HLC 9 | 13.30 | 0.10 | 2.88 | 0.13 | 0.01 | 0.00 |
| K 1-1 | 3030.00 | 5.00 | 1.81 | 0.02 | 1.65 | 0.01 |
| K 1-7 | 5763.33 | 10.41 | 2.07 | 0.01 | 1.92 | 0.01 |
| K 2-4 | 6.43 | 0.15 | 2.16 | 0.05 | 1.53 | 0.07 |
| K 4-3 | 1025.17 | 2.04 | 2.19 | 0.00 | 1.01 | 0.01 |
| K 1-4 | 83.50 | 11.32 | 2.25 | 0.02 | 1.66 | 0.02 |
| K 4-6 | 14503.33 | 595.01 | 1.54 | 0.02 | 0.94 | 0.01 |
| K 4-7 | 851.67 | 24.58 | 2.28 | 0.09 | 0.93 | 0.02 |
| KA 1-1 | 2835.00 | 15.00 | 2.14 | 0.02 | 1.91 | 0.00 |
| KA 1-2 | 349.00 | 37.07 | 1.63 | 0.06 | 0.46 | 0.02 |
| KB 1-6 | 676.67 | 2.89 | 2.33 | 0.02 | 1.02 | 0.01 |
| KC 1-4 | 5255.00 | 620.91 | 1.65 | 0.03 | 0.74 | 0.02 |
| PC 2-5 | 6703.33 | 17.56 | 1.98 | 0.01 | 2.28 | 0.01 |
| RL 2-2 | 46.03 | 6.29 | 1.98 | 0.01 | 1.39 | 0.01 |
| RL 2-4 | 2006.67 | 5.77 | 1.80 | 0.01 | 1.80 | 0.02 |
| RP 2-4 | 10273.33 | 270.02 | 1.99 | 0.03 | 1.93 | 0.12 |
| RP 4-2 | 143.37 | 0.57 | 1.94 | 0.02 | 0.41 | 0.00 |
| TKB 1-4 | 1471.67 | 265.58 | 1.83 | 0.02 | 1.11 | 0.01 |
| TKC 1-6 | 262.87 | 6.81 | 1.45 | 0.11 | 1.21 | 0.08 |
| TKC 1-7 | 712.77 | 5.46 | 2.01 | 0.01 | 0.85 | 0.01 |
| TKC 1-8 | 2238.33 | 17.56 | 1.76 | 0.04 | 1.19 | 0.02 |
| TKC 2-1 | 33.00 | 5.00 | 1.68 | 0.13 | 0.36 | 0.00 |
| TKC 2-2 | 226.07 | 3.66 | 2.01 | 0.01 | 0.32 | 0.01 |
| TLA 2-11 | 267.63 | 4.35 | 1.35 | 0.02 | 1.09 | 0.02 |
| TLA 3-2 | 11.80 | 1.41 | 3.22 | 0.40 | 0.55 | 0.04 |
| TLB 1-1 | 388.77 | 11.46 | 1.99 | 0.03 | 0.51 | 0.01 |
| TLB 1-2 | 209.80 | 0.72 | 1.54 | 0.01 | 0.47 | 0.01 |
| TLB 1-3 | 862.40 | 1.49 | 1.98 | 0.01 | 1.23 | 0.01 |
| TLB 2-7 | 8923.33 | 12.58 | 1.90 | 0.01 | 1.89 | 0.01 |
| TLB 2-9 | 3908.33 | 5.77 | 1.90 | 0.01 | 1.57 | 0.01 |
| TLC 1-8 | 1383.33 | 89.63 | 1.49 | 0.02 | 0.46 | 0.01 |
| TLC 2-2 | 1835.00 | 10.00 | 1.83 | 0.01 | 0.81 | 0.01 |
| TPA 1-2 | 27.17 | 2.28 | 1.61 | 0.02 | 0.74 | 0.04 |
| TPA 1-3 | 1214.97 | 2.37 | 2.02 | 0.01 | 1.75 | 0.01 |
| TPA 2-6 | 427.73 | 34.40 | 1.67 | 0.07 | 0.82 | 0.06 |
| TPA 3-10 | 374.47 | 8.12 | 2.03 | 0.02 | 0.71 | 0.01 |
| TPA 3-8 | 2078.33 | 10.41 | 1.82 | 0.03 | 0.98 | 0.01 |
| TPB 1-1 | 80.53 | 0.25 | 1.92 | 0.01 | 0.11 | 0.00 |
| TPB 2-7 | 243.83 | 6.17 | 1.97 | 0.02 | 0.31 | 0.01 |
| TPB 2-8 | 236.00 | 7.21 | 1.58 | 0.01 | 0.39 | 0.00 |
| TPC 1-12 | 879.23 | 10.06 | 1.97 | 0.00 | 1.05 | 0.01 |
| TPC 1-2 | 4595.00 | 21.79 | 1.79 | 0.00 | 1.68 | 0.01 |
References
- Augustyniak, A.; Roszak, M. Zastosowanie mikrobiologii w nowoczesnym rolnictwie. In Application of Microbiology in Modern Agriculture; Młodzi Naukowcy: Poznań, Poland, 2017; pp. 7–12. ISBN 9788365362483. [Google Scholar]
- Shahzad, R.; Khan, A.L.; Waqas, M.; Ullah, I.; Bilal, S.; Kim, Y.-H.; Asaf, S.; Kang, S.-M.; Lee, I.-J. Metabolic and proteomic alteration in phytohormone-producing endophytic Bacillus amyloliquefaciens RWL-1 during methanol utilization. Metabolomics 2019, 15, 16. [Google Scholar] [CrossRef]
- White, J.F.; Kingsley, K.L.; Zhang, Q.; Verma, R.; Obi, N.; Dvinskikh, S.; Elmore, M.T.; Verma, S.K.; Gond, S.K.; Kowalski, K.P. Review: Endophytic microbes and their potential applications in crop management. Pest Manag. Sci. 2019, 75, 2558–2565. [Google Scholar] [CrossRef]
- Fadiji, A.E.; Babalola, O.O. Exploring the potentialities of beneficial endophytes for improved plant growth. Saudi J. Biol. Sci. 2020, 27, 3622–3633. [Google Scholar] [CrossRef]
- Kuźniar, A.; Włodarczyk, K.; Wolińska, A. Agricultural and Other Biotechnological Applications Resulting from Trophic Plant-Endophyte Interactions. Agronomy 2019, 9, 779. [Google Scholar] [CrossRef]
- De Aquino, J.P.A.; Junior, F.B.D.M.; Antunes, J.E.L.; Figueiredo, M.D.V.B.; Neto, F.D.A.; De Araujo, A.S.F. Plant growth-promoting endophytic bacteria on maize and sorghum1. Agropecuária Trop. 2019, 49, e56241. [Google Scholar] [CrossRef]
- Napora, A.; Kacprzak, M.; Nowak, K.; Grobelak, A. Wpływ Bakterii Endofitycznych Na Promowanie Wzrostu Roślin w Warunkach Stresowych/Influence Endophytic Bacteria to Promote Plants Growth in Stress Conditions. Postępy Biochem. Adv. Biochem. 2015, 61, 398–402. [Google Scholar]
- Emami, S.; Alikhani, H.A.; Pourbabaei, A.A.; Etesami, H.; Sarmadian, F.; Motessharezadeh, B. Effect of rhizospheric and endophytic bacteria with multiple plant growth promoting traits on wheat growth. Environ. Sci. Pollut. Res. 2019, 26, 19804–19813. [Google Scholar] [CrossRef]
- Mateo-Bonmatí, E.; Casanova-Sáez, R.; Ljung, K. Epigenetic Regulation of Auxin Homeostasis. Biomolecules 2019, 9, 623. [Google Scholar] [CrossRef]
- Goswami, D.; Thakker, J.N.; Dhandhukia, P.C. Portraying mechanics of plant growth promoting rhizobacteria (PGPR): A review. Cogent Food Agric. 2016, 2, 1127500. [Google Scholar] [CrossRef]
- Korasick, D.A.; Enders, T.A.; Strader, L.C. Auxin biosynthesis and storage forms. J. Exp. Bot. 2013, 64, 2541–2555. [Google Scholar] [CrossRef]
- Yu, P.; Hegeman, A.D.; Cohen, J.D. A facile means for the identification of indolic compounds from plant tissues. Plant J. 2014, 79, 1065–1075. [Google Scholar] [CrossRef]
- Mano, Y.; Nemoto, K. The pathway of auxin biosynthesis in plants. J. Exp. Bot. 2012, 63, 2853–2872. [Google Scholar] [CrossRef]
- Bajguz, A.; Piotrowska, A. Conjugates of auxin and cytokinin. Phytochemistry 2009, 70, 957–969. [Google Scholar] [CrossRef]
- Sun, Z.; Liu, K.; Zhang, J.; Zhang, Y.; Xu, K.; Yu, D.; Wang, J.; Hu, L.; Chen, L.; Li, C. IAA producing Bacillus altitudinis alleviates iron stress in Triticum aestivum L. seedling by both bioleaching of iron and up-regulation of genes encoding ferritins. Plant Soil 2017, 419, 1–11. [Google Scholar] [CrossRef]
- Rana, K.L.; Kour, D.; Kaur, T.; Sheikh, I.; Yadav, A.N.; Kumar, V.; Suman, A.; Dhaliwal, H.S. Endophytic Microbes from Diverse Wheat Genotypes and Their Potential Biotechnological Applications in Plant Growth Promotion and Nutrient Uptake. Proc. Natl. Acad. Sci. India Sect. B Boil. Sci. 2020, 90, 969–979. [Google Scholar] [CrossRef]
- Kiani, T.; Khan, S.A.; Noureen, N.; Yasmin, T.; Zakria, M.; Ahmed, H.; Mehboob, F.; Farrakh, S. Isolation and characterization of culturable endophytic bacterial community of stripe rust–resistant and stripe rust–susceptible Pakistani wheat cultivars. Int. Microbiol. 2019, 22, 191–201. [Google Scholar] [CrossRef]
- Matsuda, F.; Miyazawa, H.; Wakasa, K.; Miyagawa, H. Quantification of Indole-3-Acetic Acid and Amino Acid Conjugates in Rice by Liquid Chromatography–Electrospray Ionization–Tandem Mass Spectrometry. Biosci. Biotechnol. Biochem. 2005, 69, 778–783. [Google Scholar] [CrossRef]
- Gordon, S.A.; Weber, R.P. Colorimetric estimation of indoleacetic acid. Plant Physiol. 1951, 26, 192–195. [Google Scholar] [CrossRef]
- Gilbert, S.; Xu, J.; Acosta, K.; Poulev, A.; Lebeis, S.; Lam, E. Bacterial Production of Indole Related Compounds Reveals Their Role in Association Between Duckweeds and Endophytes. Front. Chem. 2018, 6, 265. [Google Scholar] [CrossRef]
- Glickmann, E.; Dessaux, Y. A critical examination of the specificity of the salkowski reagent for indolic compounds produced by phytopathogenic bacteria. Appl. Environ. Microbiol. 1995, 61, 793–796. [Google Scholar] [CrossRef]
- Zhang, L.-N.; Wang, D.-C.; Hu, Q.; Dai, X.-Q.; Xie, Y.-S.; Li, Q.; Liu, H.-M.; Guo, J.-H. Consortium of Plant Growth-Promoting Rhizobacteria Strains Suppresses Sweet Pepper Disease by Altering the Rhizosphere Microbiota. Front. Microbiol. 2019, 10, 1668. [Google Scholar] [CrossRef] [PubMed]
- Nawaz, A.; Shahbaz, M.; Asadullah; Imran, A.; Marghoob, M.U.; Imtiaz, M.; Mubeen, F. Potential of Salt Tolerant PGPR in Growth and Yield Augmentation of Wheat (Triticum aestivum L.) Under Saline Conditions. Front. Microbiol. 2020, 11, 2019. [Google Scholar] [CrossRef]
- Djaya, L.; Hersanti; Istifadah, N.; Hartati, S.; Joni, I. In vitro study of plant growth promoting rhizobacteria (PGPR) and endophytic bacteria antagonistic to Ralstonia solanacearum formulated with graphite and silica nano particles as a biocontrol delivery system (BDS). Biocatal. Agric. Biotechnol. 2019, 19, 101153. [Google Scholar] [CrossRef]
- Sheirdil, R.A.; Hayat, R.; Zhang, X.-X.; Abbasi, N.A.; Ali, S.; Ahmed, M.; Khattak, J.Z.K.; Ahmad, S. Exploring Potential Soil Bacteria for Sustainable Wheat (Triticum aestivum L.) Production. Sustainability 2019, 11, 3361. [Google Scholar] [CrossRef]
- Cheng, D.; Tian, Z.; Feng, L.; Xu, L.; Wang, H. Diversity analysis of the rhizospheric and endophytic bacterial communities of Senecio vulgaris L. (Asteraceae) in an invasive range. PeerJ 2019, 6, e6162. [Google Scholar] [CrossRef]
- Kuźniar, A.; Włodarczyk, K.; Grządziel, J.; Goraj, W.; Gałązka, A.; Wolińska, A. Culture-independent analysis of an endophytic core microbiome in two species of wheat: Triticum aestivum L. (cv. ‘Hondia’) and the first report of microbiota in Triticum spelta L. (cv. ‘Rokosz’). Syst. Appl. Microbiol. 2020, 43, 126025. [Google Scholar] [CrossRef]
- Chang, H.-X.; Haudenshield, J.S.; Bowen, C.R.; Hartman, G.L. Metagenome-Wide Association Study and Machine Learning Prediction of Bulk Soil Microbiome and Crop Productivity. Front. Microbiol. 2017, 8, 519. [Google Scholar] [CrossRef]
- Tsukanova, K.; Сhеbоtаr, V.; Meyer, J.; Bibikova, T. Effect of plant growth-promoting Rhizobacteria on plant hormone homeostasis. S. Afr. J. Bot. 2017, 113, 91–102. [Google Scholar] [CrossRef]
- Kramer, E.M.; Ackelsberg, E.M. Auxin metabolism rates and implications for plant development. Front. Plant Sci. 2015, 6, 150. [Google Scholar] [CrossRef]
- Vyas, P.; Kaur, R. Culturable Stress-Tolerant Plant Growth-Promoting Bacterial Endophytes Associated with Adhatoda vasica. J. Soil Sci. Plant Nutr. 2019, 19, 290–298. [Google Scholar] [CrossRef]
- Susilowati, A.; Puspita, A.A.; Yunus, A. Drought resistant of bacteria producing exopolysaccharide and IAA in rhizosphere of soybean plant (Glycine max) in Wonogiri Regency Central Java Indonesia. IOP Conf. Ser. Earth Environ. Sci. 2018, 142, 012058. [Google Scholar] [CrossRef]
- Ferchichi, N.; Toukabri, W.; Boularess, M.; Smaoui, A.; Mhamdi, R.; Trabelsi, D. Isolation, identification and plant growth promotion ability of endophytic bacteria associated with lupine root nodule grown in Tunisian soil. Arch. Microbiol. 2019, 201, 1333–1349. [Google Scholar] [CrossRef] [PubMed]
- Govarthanan, M.; Mythili, R.; Selvankumar, T.; Kamala-Kannan, S.; Rajasekar, A.; Chang, Y.-C. Bioremediation of heavy metals using an endophytic bacterium Paenibacillus sp. RM isolated from the roots of Tridax procumbens. 3 Biotech 2016, 6, 1–7. [Google Scholar] [CrossRef]
- Banach, A.M.; Kuźniar, A.; Grządziel, J.; Wolińska, A. Azolla filiculoides L. as a source of metal-tolerant microorganisms. PLoS ONE 2020, 15, e0232699. [Google Scholar] [CrossRef]
- Salkowski, E. Ueber das Verhalten der Skatolcarbonsa üre im Organismus. Z. Physiol. Chem. 1885, 9, 23–33. [Google Scholar]
- Lin, G.-H.; Chang, C.-Y.; Lin, H.-R. Systematic profiling of indole-3-acetic acid biosynthesis in bacteria using LC–MS/MS. J. Chromatogr. B 2015, 988, 53–58. [Google Scholar] [CrossRef]
- Lane, D.J. 16S/23S rRNA sequencing. In Nucleic Acid Techniques in Bacterial Systematic; Stackebrandt, E., Goodfellow, M., Eds.; John Wiley and Sons: New York, NY, USA, 1991; pp. 115–175. [Google Scholar]
- Turner, S.; Pryer, K.M.; Miao, V.P.W.; Palmer, J.D. Investigating Deep Phylogenetic Relationships among Cyanobacteria and Plastids by Small Subunit rRNA Sequence Analysis. J. Eukaryot. Microbiol. 1999, 46, 327–338. [Google Scholar] [CrossRef]
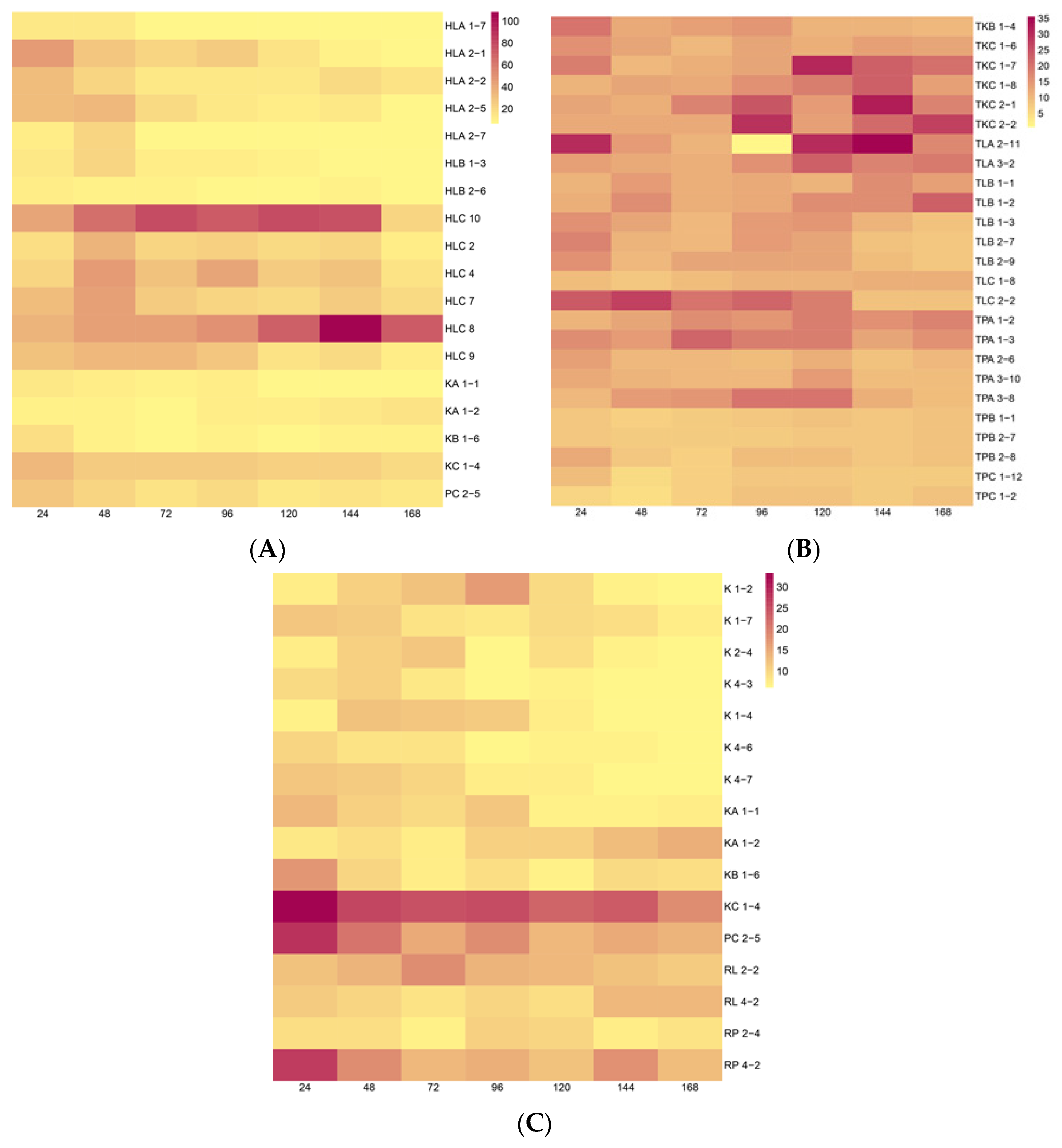
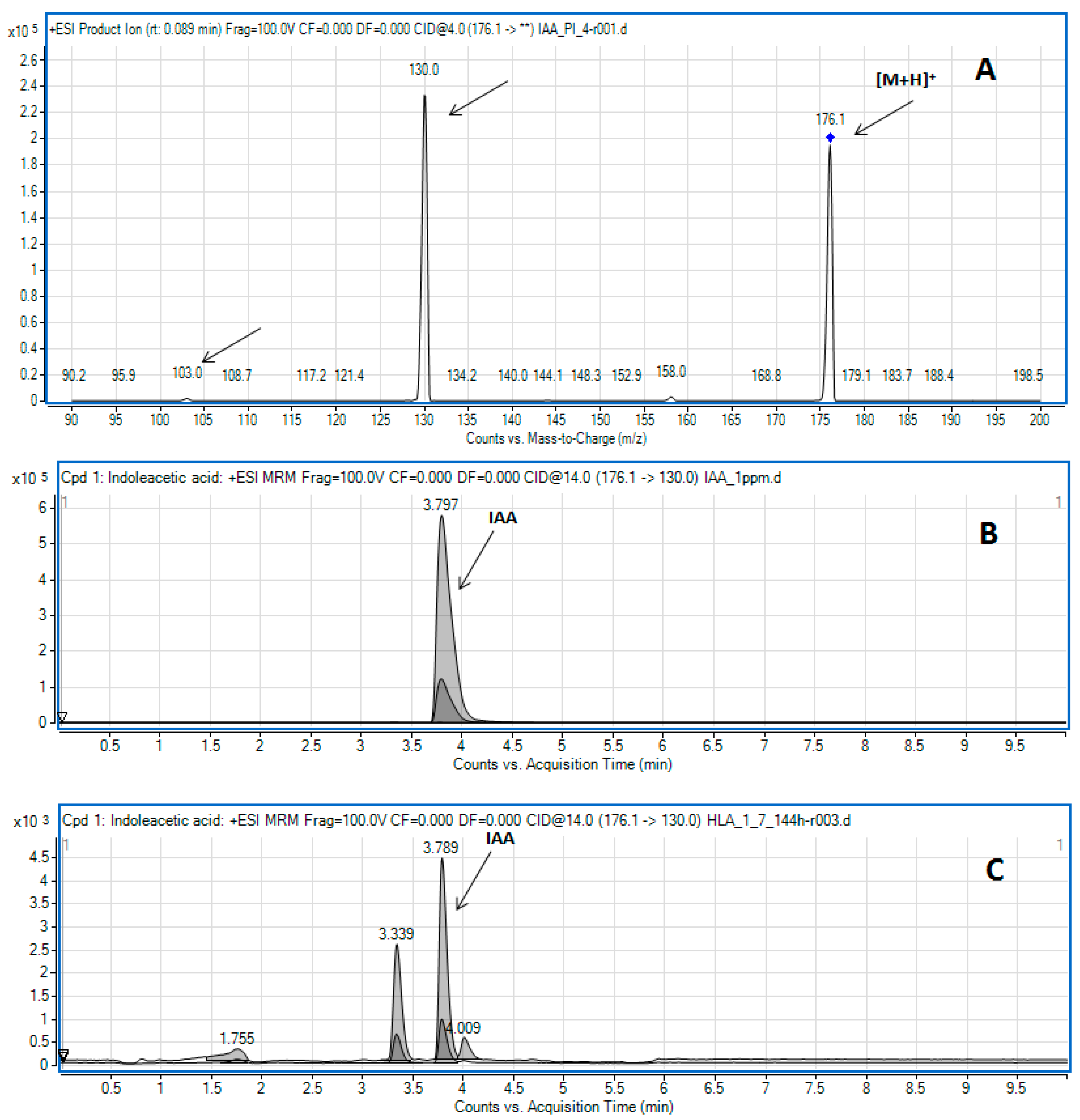
| Strain Name | Results (µg∙mL−1) | ||||||
|---|---|---|---|---|---|---|---|
| Time of Cultivation (h) | |||||||
| 24 | 48 | 72 | 96 | 120 | 144 | 168 | |
| HLA 1-7 | 1.836 ± 0.067 | na | na | na | 1.810 ± 0.013 | 1.776 ± 0.018 | na |
| HLA 2-7 | 0.344 ± 0.009 | na | 0.702 ± 0.002 | 0.668 ± 0.003 | na | na | na |
| HLB 1-3 | 0.519 ± 0.009 | na | 0.720 ± 0.014 | 0.686 ± 0.009 | na | na | na |
| HLB 2-6 | 1.716 ± 0.021 | 3.958 ± 0.030 | 4.643 ± 0.045 | na | na | na | na |
| K 1-4 | 0.718 ± 0.013 | na | na | 0.731 ± 0.017 | 0.711 ± 0.014 | na | na |
| KC 1-4 | 2.789 ± 0.310 | na | na | na | na | na | 6.649 ± 0.032 |
| PC 2-5 | 11.770 ± 0.094 | na | na | na | 7.178 ± 0.043 | 7.181 ± 0.062 | na |
| RL 2-2 | na | 0.357 ± 0.006 | na | na | 0.301 ± 0.011 | 0.360 ± 0.013 | na |
| RP 2-4 | na | na | 0.650 ± 0.004 | 0.702 ± 0.008 | 0.500 ± 0.019 | na | na |
| TLB 2-9 | na | na | 0.982 ± 0.002 | 0.965 ± 0.075 | 1.207 ± 0.027 | na | na |
| TPA 1-3 | na | na | 2.397 ± 0.038 | 2.260 ± 0.142 | 2.226 ± 0.022 | na | na |
| TKC 2-1 | na | na | na | na | 0.724 ± 0.013 | 0.558 ± 0.024 | 0.557 ± 0.006 |
| Probes | The Growth of Wheat Coleoptile Segments Treated with Post-Culture Medium [% of Control] Time of Cultivation (h) | |||
|---|---|---|---|---|
| 24 | 48 | 72 | 96 | |
| Control | 101.998 ± 1.718 | 113.164 ± 1.779 | 105.158 ± 1.365 | 108.532 ± 1.142 |
| HLA 1-7 | 106.111 ± 2.110 | 101.622 ± 0.957 | 102.145 ± 1.365 | 107.46 ± 1.083 |
| HLA 2-7 | 105.682 ± 1.688 | 101.622 ± 0.957 | 104.283 ± 1.079 | 106.740 ± 0.993 |
| KC 1-4 | 117.941 ± 1.077 | 121.452 ± 1.304 | 126.178 ± 0.981 | 113.860 ± 1.051 |
| PC 2-5 | 121.086 ± 1.802 | 119.046 ± 487 | 113.067 ± 0.872 | 115.532 ± 1.228 |
| HLB 2-6 | 104.011 ± 1.215 | 109.016 ± 1.140 | 104.770 ± 0.867 | 106.248 ± 458 |
| Strain Name | Methods | Results | ||||||
|---|---|---|---|---|---|---|---|---|
| Time of Cultivation (h) | ||||||||
| 24 | 48 | 72 | 96 | 120 | 144 | 168 | ||
| HLA 1-7 | LC-MS/MS | 1.836 | na | na | na | 1.810 | 1.776 | na |
| Colorimetric method | 14.220 | 12.673 | 7.970 | 7.256 | 7.286 | nd | nd | |
| Biotest | - | - | - | - | - | na | na | |
| HLA 2-7 | LC-MS/MS | 0.344 | na | 0.702 | 0.668 | na | na | na |
| Colorimetric method | 10.708 | 21.839 | 7.792 | nd | nd | nd | nd | |
| Biotest | - | - | - | - | - | na | na | |
| HLB 1-3 | LC-MS/MS | 0.519 | na | 0.720 | 0.686 | na | na | na |
| Colorimetric method | 13.536 | 20.9762 | 10.232 | nd | nd | nd | nd | |
| HLB 2-6 | LC-MS/MS | 1.716 | 3.958 | 4.643 | na | na | na | na |
| Colorimetric method | 12.077 | 10.024 | nd | nd | nd | nd | nd | |
| Biotest | - | - | - | - | na | na | na | |
| K 1-4 | LC-MS/MS | 0.718 | na | na | 0.731 | 0.711 | na | na |
| Colorimetric method | nd | 12.375 | 11.899 | 11.214 | nd | nd | nd | |
| KC 1-4 | LC-MS/MS | 2.789 | na | na | na | na | na | 6.649 |
| Colorimetric method | 33.298 | 25.977 | 24.607 | 25.500 | 22.851 | 24.012 | 18.446 | |
| Biotest | + | + | + | + | na | na | na | |
| PC 2-5 | LC-MS/MS | 11.770 | na | na | na | 7.178 | 7.181 | na |
| Colorimetric method | 28.327 | 20.917 | 15.054 | 18.506 | 13.625 | nd | nd | |
| Biotest | + | + | + | + | + | na | na | |
| RL 2-2 | LC-MS/MS | na | 0.357 | na | na | 0.301 | 0.360 | na |
| Colorimetric method | nd | nd | 18.536 | 13.893 | 13.506 | nd | nd | |
| RP 2-4 | LC-MS/MS | na | na | 0.650 | 0.702 | 0.500 | na | na |
| Colorimetric method | nd | nd | nd | 10.976 | nd | nd | nd | |
| TLB 2-9 | LC-MS/MS | na | na | 0.982 | 0.965 | 1.207 | na | na |
| Colorimetric method | 15.679 | 10.202 | 13.119 | nd | 12.762 | nd | nd | |
| TPA 1-3 | LC-MS/MS | na | na | 2.397 | 2.260 | 2.226 | na | na |
| Colorimetric method | 16.512 | 13.893 | 21.839 | nd | 18.506 | 12.792 | 15.351 | |
| TKC 2-1 | LC-MS/MS | na | na | na | na | 0.724 | 0.558 | 0.557 |
| Colorimetric method | nd | nd | nd | nd | nd | 31.869 | nd | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kuźniar, A.; Włodarczyk, K.; Sadok, I.; Staniszewska, M.; Woźniak, M.; Furtak, K.; Grządziel, J.; Gałązka, A.; Skórzyńska-Polit, E.; Wolińska, A. A Comprehensive Analysis Using Colorimetry, Liquid Chromatography-Tandem Mass Spectrometry and Bioassays for the Assessment of Indole Related Compounds Produced by Endophytes of Selected Wheat Cultivars. Molecules 2021, 26, 1394. https://doi.org/10.3390/molecules26051394
Kuźniar A, Włodarczyk K, Sadok I, Staniszewska M, Woźniak M, Furtak K, Grządziel J, Gałązka A, Skórzyńska-Polit E, Wolińska A. A Comprehensive Analysis Using Colorimetry, Liquid Chromatography-Tandem Mass Spectrometry and Bioassays for the Assessment of Indole Related Compounds Produced by Endophytes of Selected Wheat Cultivars. Molecules. 2021; 26(5):1394. https://doi.org/10.3390/molecules26051394
Chicago/Turabian StyleKuźniar, Agnieszka, Kinga Włodarczyk, Ilona Sadok, Magdalena Staniszewska, Małgorzata Woźniak, Karolina Furtak, Jarosław Grządziel, Anna Gałązka, Ewa Skórzyńska-Polit, and Agnieszka Wolińska. 2021. "A Comprehensive Analysis Using Colorimetry, Liquid Chromatography-Tandem Mass Spectrometry and Bioassays for the Assessment of Indole Related Compounds Produced by Endophytes of Selected Wheat Cultivars" Molecules 26, no. 5: 1394. https://doi.org/10.3390/molecules26051394
APA StyleKuźniar, A., Włodarczyk, K., Sadok, I., Staniszewska, M., Woźniak, M., Furtak, K., Grządziel, J., Gałązka, A., Skórzyńska-Polit, E., & Wolińska, A. (2021). A Comprehensive Analysis Using Colorimetry, Liquid Chromatography-Tandem Mass Spectrometry and Bioassays for the Assessment of Indole Related Compounds Produced by Endophytes of Selected Wheat Cultivars. Molecules, 26(5), 1394. https://doi.org/10.3390/molecules26051394












