The Selectivity for Tumor Cells of Nuclear-Directed Cytotoxic RNases Is Mediated by the Nuclear/Cytoplasmic Distribution of p27KIP1
Abstract
1. Introduction
2. Results
2.1. Selectivity of NLSPE5 In Vitro
2.2. The Effect of NLSPE5 on Proliferation and Viability Differs between Tumor and Non-Tumor Cells
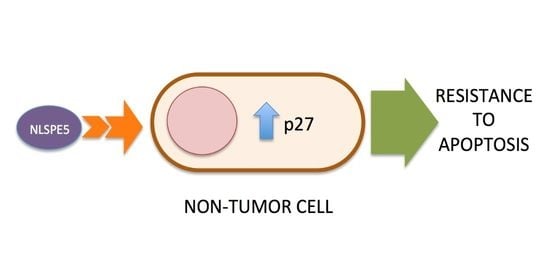
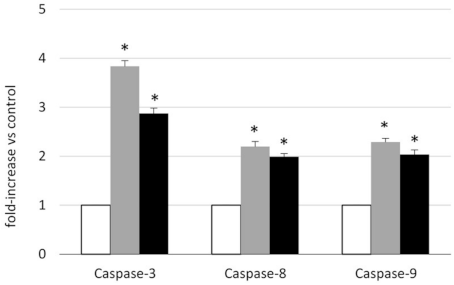
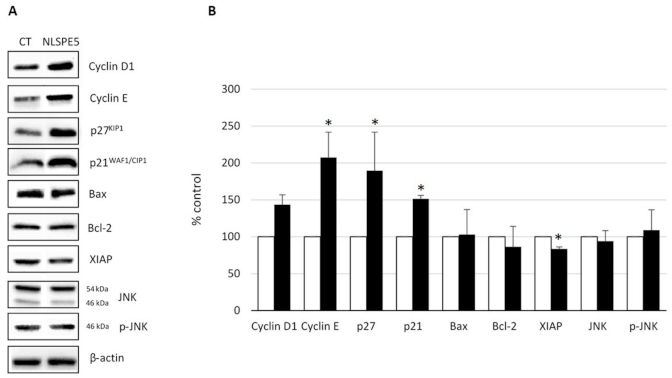
2.3. Analysis of the Mechanism of Cytotoxicity of NLSPE5 on CCD-18Co Cells
3. Discussion
4. Materials and Methods
4.1. RNase Expression and Purification
4.2. Cell Lines and Culture Conditions
4.3. Cell Growth Assays
4.4. Analysis of Cytostatic and Cytotoxic Effects of NLSPE5
4.5. Cell Cycle Phase Analysis
4.6. Procaspase Activation Assay
4.7. Western Blot Analysis
4.8. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Castro, J.; Ribo, M.; Benito, A.; Vilanova, M. Mini-review: Nucleus-targeted ribonucleases as antitumor drugs. Curr. Med. Chem. 2013, 20, 1225–1231. [Google Scholar] [CrossRef] [PubMed]
- Castro, J.; Ribó, M.; Benito, A.; Vilanova, M. Approaches to endow ribonucleases with antitumor activity: Lessons learned from the native cytotoxic ribonucleases. In Anti-Cancer Drugs—Nature, Synthesis and Cell; InTech: London, UK, 2016. [Google Scholar]
- Ribó, M.; Benito, A.; Vilanova, M. Antitumor ribonucleases. In Nucleic Acids and Molecular Biology: Ribonucleases; Springer: Berlin, Germany, 2012; pp. 55–88. ISBN 9783642210785. [Google Scholar]
- Vasandani, V.M.; Wu, Y.N.; Mikulski, S.M.; Youle, R.J.; Sung, C. Molecular determinants in the plasma clearance and tissue distribution of ribonucleases of the ribonuclease A superfamily. Cancer Res. 1996, 56, 4180–4186. [Google Scholar]
- Matoušek, J. Ribonucleases and their antitumor activity. Comp. Biochem. Physiol. C Toxicol. Pharmacol. 2001, 129, 175–191. [Google Scholar] [CrossRef]
- Rodríguez, M.; Moussaoui, M.; Benito, A.; Cuchillo, C.M.; Nogués, M.V.; Vilanova, M. Human pancreatic ribonuclease presents higher endonucleolytic activity than ribonuclease A. Arch. Biochem. Biophys. 2008, 471, 191–197. [Google Scholar] [CrossRef] [PubMed]
- Bosch, M.; Benito, A.; Ribó, M.; Puig, T.; Beaumelle, B.; Vilanova, M. A nuclear localization sequence endows human pancreatic ribonuclease with cytotoxic activity. Biochemistry 2004, 43, 2167–2177. [Google Scholar] [CrossRef] [PubMed]
- Vert, A.; Castro, J.; Ruiz-Martínez, S.; Tubert, P.; Escribano, D.; Ribó, M.; Vilanova, M.; Benito, A. Generation of new cytotoxic human ribonuclease variants directed to the nucleus. Mol. Pharm. 2012, 9, 2894–2902. [Google Scholar] [CrossRef]
- Furia, A.; Moscato, M.; Calì, G.; Pizzo, E.; Confalone, E.; Amoroso, M.R.; Esposito, F.; Nitsch, L.; D’Alessio, G. The ribonuclease/angiogenin inhibitor is also present in mitochondria and nuclei. FEBS Lett. 2011, 585, 613–617. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez, M.; Benito, A.; Tubert, P.; Castro, J.; Ribó, M.; Beaumelle, B.; Vilanova, M. A cytotoxic ribonuclease variant with a discontinuous nuclear localization signal constituted by basic residues scattered over three areas of the molecule. J. Mol. Biol. 2006, 360, 548–557. [Google Scholar] [CrossRef]
- Tubert, P.; Rodríguez, M.; Ribó, M.; Benito, A.; Vilanova, M. The nuclear transport capacity of a human-pancreatic ribonuclease variant is critical for its cytotoxicity. Invest. New Drugs 2011, 29, 811–817. [Google Scholar] [CrossRef]
- Castro, J.; Ribó, M.; Navarro, S.; Nogués, M.V.; Vilanova, M.; Benito, A. A human ribonuclease induces apoptosis associated with p21WAF1/CIP1 induction and JNK inactivation. BMC Cancer 2011, 11, 9. [Google Scholar] [CrossRef]
- Vert, A.; Castro, J.; Ribó, M.; Benito, A.; Vilanova, M. A nuclear-directed human pancreatic ribonuclease (PE5) targets the metabolic phenotype of cancer cells. Oncotarget 2016, 7, 18309–18324. [Google Scholar] [CrossRef] [PubMed]
- Castro, J.; Ribó, M.; Puig, T.; Colomer, R.; Vilanova, M.; Benito, A. A cytotoxic ribonuclease reduces the expression level of P-glycoprotein in multidrug-resistant cell lines. Investig. New Drugs 2012, 30, 880–888. [Google Scholar] [CrossRef]
- Al Bitar, S.; Gali-Muhtasib, H. The role of the cyclin dependent kinase inhibitor p21cip1/waf1 in targeting cancer: Molecular mechanisms and novel therapeutics. Cancers 2019, 11, 1475. [Google Scholar] [CrossRef] [PubMed]
- Workman, P.; Al-Lazikani, B.; Clarke, P.A. Genome-based cancer therapeutics: Targets, kinase drug resistance and future strategies for precision oncology. Curr. Opin. Pharmacol. 2013, 13, 486–496. [Google Scholar] [CrossRef] [PubMed]
- Ke, X.; Shen, L. Molecular targeted therapy of cancer: The progress and future prospect. Front. Lab. Med. 2017, 1, 69–75. [Google Scholar] [CrossRef]
- Boukamp, P.; Petrussevska, R.T.; Breitkreutz, D.; Hornung, J.; Markham, A.; Fusenig, N.E. Normal keratinization in a spontaneously immortalized aneuploid human keratinocyte cell line. J. Cell Biol. 1988, 106, 761–771. [Google Scholar] [CrossRef]
- Ryle, C.M.; Breitkreutz, D.; Stark, H.-J.; Fusening, N.E.; Leigh, I.M.; Stelnert, P.M.; Roop, D. Density-dependent modulation of synthesis of keratins 1 and 10 in the human keratinocyte line HACAT and in ras-transfected tumorigenic clones. Differentiation 1989, 40, 42–54. [Google Scholar] [CrossRef]
- Schoop, V.M.; Mirancea, N.; Fusenig, N.E. Epidermal organization and differentiation of HaCat keratinocytes in organotypic coculture with human dermal fibroblasts. J. Investig. Dermatol. 1999, 112, 343–353. [Google Scholar] [CrossRef]
- Widder, M.; Lützkendorf, J.; Caysa, H.; Unverzagt, S.; Wickenhauser, C.; Benndorf, R.A.; Schmoll, H.-J.; Müller-Tidow, C.; Müller, T.; Müller, L.P. Multipotent mesenchymal stromal cells promote tumor growth in distinct colorectal cancer cells by a β1-integrin-dependent mechanism. Int. J. Cancer 2016, 138, 964–975. [Google Scholar] [CrossRef]
- Hayashi, Y.; Tsujii, M.; Kodama, T.; Akasaka, T.; Kondo, J.; Hikita, H.; Inoue, T.; Tsujii, Y.; Maekawa, A.; Yoshii, S.; et al. p53 functional deficiency in human colon cancer cells promotes fibroblast-mediated angiogenesis and tumor growth. Carcinogenesis 2016, 37, 972–984. [Google Scholar] [CrossRef]
- Farace, C.; Pisano, A.; Griñan-Lison, C.; Solinas, G.; Jiménez, G.; Serra, M.; Carrillo, E.; Scognamillo, F.; Attene, F.; Montella, A.; et al. Deregulation of cancer-stem-cell-associated miRNAs in tissues and sera of colorectal cancer patients. Oncotarget 2020, 11, 116–130. [Google Scholar] [CrossRef] [PubMed]
- Koh, B.; Jeon, H.; Kim, D.; Kang, D.; Kim, K.R. Effect of fibroblast co-culture on the proliferation, viability and drug response of colon cancer cells. Oncol. Lett. 2019, 17, 2409–2417. [Google Scholar] [CrossRef] [PubMed]
- Debnath, J.; Muthuswamy, S.K.; Brugge, J.S. Morphogenesis and oncogenesis of MCF-10A mammary epithelial acini grown in three-dimensional basement membrane cultures. Methods 2003, 30, 256–268. [Google Scholar] [CrossRef]
- Vidi, P.A.; Bissell, M.J.; Lelièvre, S.A. Three-dimensional culture of human breast epithelial cells: The how and the why. Methods Mol. Biol. 2013, 945, 193–219. [Google Scholar] [CrossRef] [PubMed]
- Abbaszadeh, F.; Clingen, P.H.; Arlett, C.F.; Plowman, P.N.; Bourton, E.C.; Themis, M.; Makarov, E.M.; Newbold, R.F.; Green, M.H.L.; Parris, C.N. A novel splice variant of the DNA-PKcs gene is associated with clinical and cellular radiosensitivity in a patient with xeroderma pigmentosum. J. Med. Genet. 2010, 47, 176–181. [Google Scholar] [CrossRef]
- Parr, C.; Sanders, A.J.; Davies, G.; Martin, T.; Lane, J.; Mason, M.D.; Mansel, R.E.; Jiang, W.G. Matriptase-2 inhibits breast tumor growth and invasion and correlates with favorable prognosis for breast cancer patients. Clin. Cancer Res. 2007, 13, 3568–3576. [Google Scholar] [CrossRef][Green Version]
- Soler, M.; González-Bártulos, M.; Figueras, E.; Ribas, X.; Costas, M.; Massaguer, A.; Planas, M.; Feliu, L. Enzyme-triggered delivery of chlorambucil from conjugates based on the cell-penetrating peptide BP16. Org. Biomol. Chem. 2015, 13, 1470–1480. [Google Scholar] [CrossRef]
- Graham, F.L.; Smiley, J.; Russell, W.C.; Nairn, R. Characteristics of a human cell line transformed by DNA from human adenovirus type 5. J. Gen. Virol. 1977, 36, 59–72. [Google Scholar] [CrossRef]
- Kavsan, V.M.; Iershov, A.V.; Balynska, O.V. Immortalized cells and one oncogene in malignant transformation: Old insights on new explanation. BMC Cell Biol. 2011, 12, 23. [Google Scholar] [CrossRef] [PubMed]
- Kannan, N.; Shanmuga Sundar, S.; Balaji, S.; Arul, A.; Anil Kumar, N.; Balasubramanian, N. Physiochemical characterization and cytotoxicity evaluation of mercury-based formulation for the development of anticancer therapeuticals. PLoS ONE 2018, 13, e0195800. [Google Scholar] [CrossRef]
- Jeyarani, S.; Vinita, N.M.; Puja, P.; Senthamilselvi, S.; Devan, U.; Velangani, A.J.; Biruntha, M.; Pugazhendhi, A.; Kumar, P. Biomimetic gold nanoparticles for its cytotoxicity and biocompatibility evidenced by fluorescence-based assays in cancer (MDA-MB-231) and non-cancerous (HEK-293) cells. J. Photochem. Photobiol. B Biol. 2020, 202, 111715. [Google Scholar] [CrossRef] [PubMed]
- Eymin, B.; Haugg, M.; Droin, N.; Sordet, O.; Dimanche-Boitrel, M.T.; Solary, E. p27(Kip1) induces drug resistance by preventing apoptosis upstream of cytochrome c release and procaspase-3 activation in leukemic cells. Oncogene 1999, 18, 1411–1418. [Google Scholar] [CrossRef] [PubMed]
- Ale-Agha, N.; Goy, C.; Jakobs, P.; Spyridopoulos, I.; Gonnissen, S.; Dyballa-Rukes, N.; Aufenvenne, K.; von Ameln, F.; Zurek, M.; Spannbrucker, T.; et al. CDKN1B/p27 is localized in mitochondria and improves respiration-dependent processes in the cardiovascular system—New mode of action for caffeine. PLoS Biol. 2018, 16, e2004408. [Google Scholar] [CrossRef] [PubMed]
- Ophascharoensuk, V.; Fero, M.L.; Hughes, J.; Roberts, J.M.; Shankland, S.J. The cyclin-dependent kinase inhibitor p27(Kip1) safeguards against inflammatory injury. Nat. Med. 1998, 4, 575–580. [Google Scholar] [CrossRef] [PubMed]
- Ribó, M.; Benito, A.; Canals, A.; Nogués, M.V.; Cuchillo, C.M.; Vilanova, M. Purification of engineered human pancreatic ribonuclease. Methods Enzymol. 2001, 341, 221–234. [Google Scholar]
- Ribó, M.; Bosch, M.; Torrent, G.; Benito, A.; Beaumelle, B.; Vilanova, M. Quantitative analysis, using MALDI-TOF mass spectrometry, of the N-terminal hydrolysis and cyclization reactions of the activation process of onconase. Eur. J. Biochem. 2004, 271, 1163–1171. [Google Scholar] [CrossRef]
- Pace, C.N.; Vajdos, F.; Fee, L.; Grimsley, G.; Gray, T. How to measure and predict the molar absorption coefficient of a protein. Protein Sci. 1995, 4, 2411–2423. [Google Scholar] [CrossRef]


| Cell Line | Origin | ONC IC50 (μM) | NLSPE5 IC50 (μM) | |
|---|---|---|---|---|
| Tumor Cell Lines | NCI-H460 | Large cell lung carcinoma | 0.40 ± 0.06 | 0.15 ± 0.05 |
| OVCAR-8 | Ovarian serous adenocarcinoma | 0.54 ± 0.08 | 0.24 ± 0.04 | |
| NCI-H460/R | NCI-H460 doxorubicin resistant | 0.50 ± 0.10 | 0.40 ± 0.10 | |
| Sk-Br-3 | Breast cancer derived from metastatic pleural effusion | 4.20 ± 0.60 | 0.36 ± 0.09 | |
| HeLa | Cervix carcinoma | 0.15 ± 0.02 | 0.26 ± 0.04 | |
| Non-Tumor Cell Lines | HaCaT | Spontaneously transformed keratinocyte | 0.70 ± 0.09 | 0.92 ± 0.14 |
| CCD-18Co | Colon fibroblasts | 0.56 ± 0.10 | 7.10 ± 1.04 | |
| HEK-293 | Embryonic kidney cells | 0.28 ± 0.05 | 0.74 ± 0.18 | |
| 1BR3G | Skin transformed fibroblasts | 0.50 ± 0.13 | 0.75 ± 0.10 | |
| MCF10A | Epithelial mammary gland | 1.07 ± 0.07 | 1.50 ± 0.09 |
| CCD-18Co | NCI-H460/R | |||
|---|---|---|---|---|
| Control | NLSPE5 | Control | NLSPE5 | |
| G0/G1 | 74.7 ± 1.0 | 70.8 ± 2.8 | 61.2 ± 6.5 | 26.9 ± 5.2 |
| S | 15.4 ± 0.3 | 11.7 ± 0.4 | 20.4 ± 3.5 | 33.4 ± 2.7 |
| G2/M | 5.1 ± 0.4 | 6.4 ± 0.5 | 10.7 ± 2.2 | 16.0 ± 2.2 |
| subG1 | 1.4 ± 0.5 | 10.6 ± 3.0 | 3.7 ± 0.8 | 18.9 ± 6.2 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
García-Galindo, G.; Castro, J.; Matés, J.; Bravo, M.; Ribó, M.; Vilanova, M.; Benito, A. The Selectivity for Tumor Cells of Nuclear-Directed Cytotoxic RNases Is Mediated by the Nuclear/Cytoplasmic Distribution of p27KIP1. Molecules 2021, 26, 1319. https://doi.org/10.3390/molecules26051319
García-Galindo G, Castro J, Matés J, Bravo M, Ribó M, Vilanova M, Benito A. The Selectivity for Tumor Cells of Nuclear-Directed Cytotoxic RNases Is Mediated by the Nuclear/Cytoplasmic Distribution of p27KIP1. Molecules. 2021; 26(5):1319. https://doi.org/10.3390/molecules26051319
Chicago/Turabian StyleGarcía-Galindo, Glòria, Jessica Castro, Jesús Matés, Marlon Bravo, Marc Ribó, Maria Vilanova, and Antoni Benito. 2021. "The Selectivity for Tumor Cells of Nuclear-Directed Cytotoxic RNases Is Mediated by the Nuclear/Cytoplasmic Distribution of p27KIP1" Molecules 26, no. 5: 1319. https://doi.org/10.3390/molecules26051319
APA StyleGarcía-Galindo, G., Castro, J., Matés, J., Bravo, M., Ribó, M., Vilanova, M., & Benito, A. (2021). The Selectivity for Tumor Cells of Nuclear-Directed Cytotoxic RNases Is Mediated by the Nuclear/Cytoplasmic Distribution of p27KIP1. Molecules, 26(5), 1319. https://doi.org/10.3390/molecules26051319