Interactions of Sea Anemone Toxins with Insect Sodium Channel—Insights from Electrophysiology and Molecular Docking Studies
Abstract
1. Introduction
2. Results
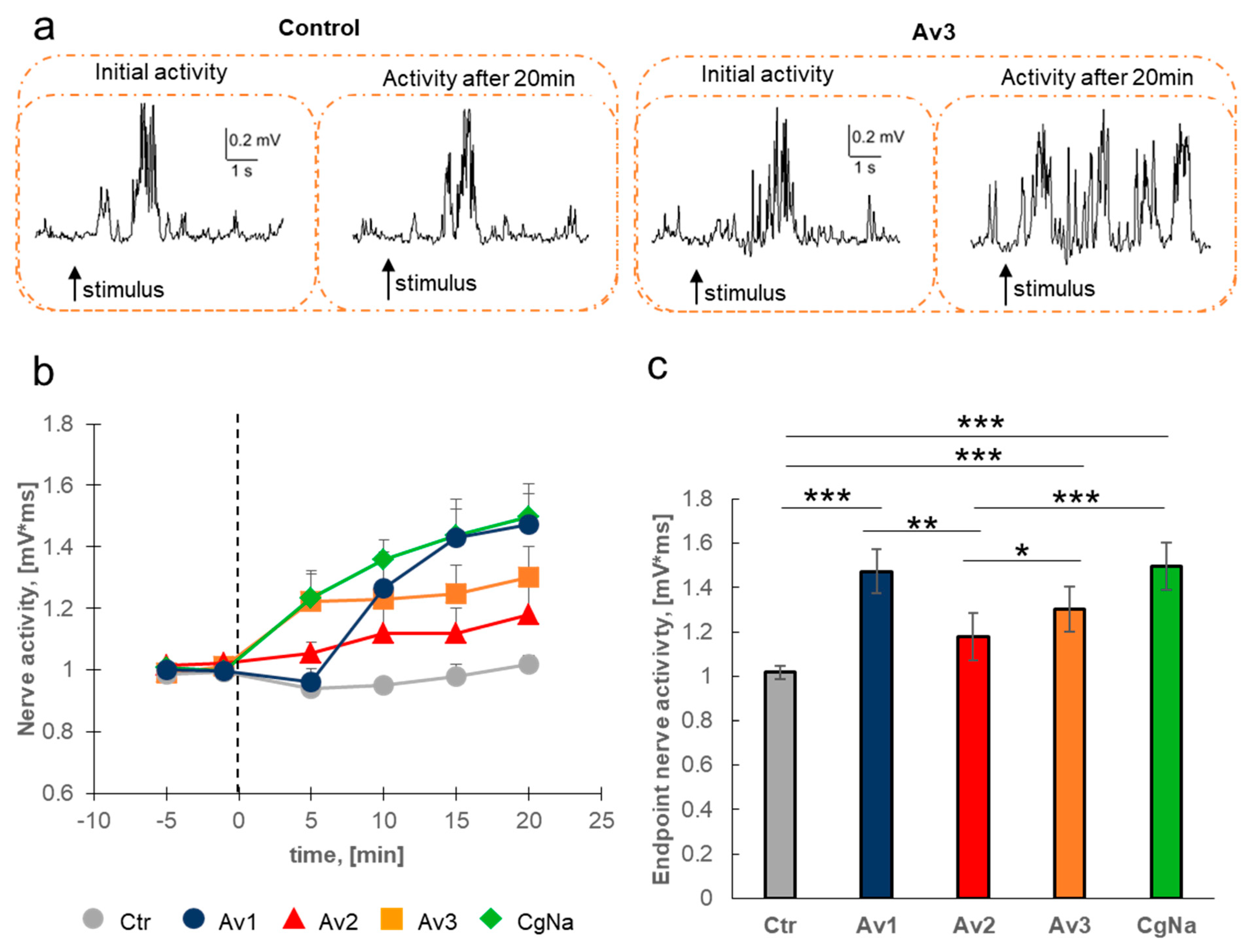
2.1. Electrophysiology
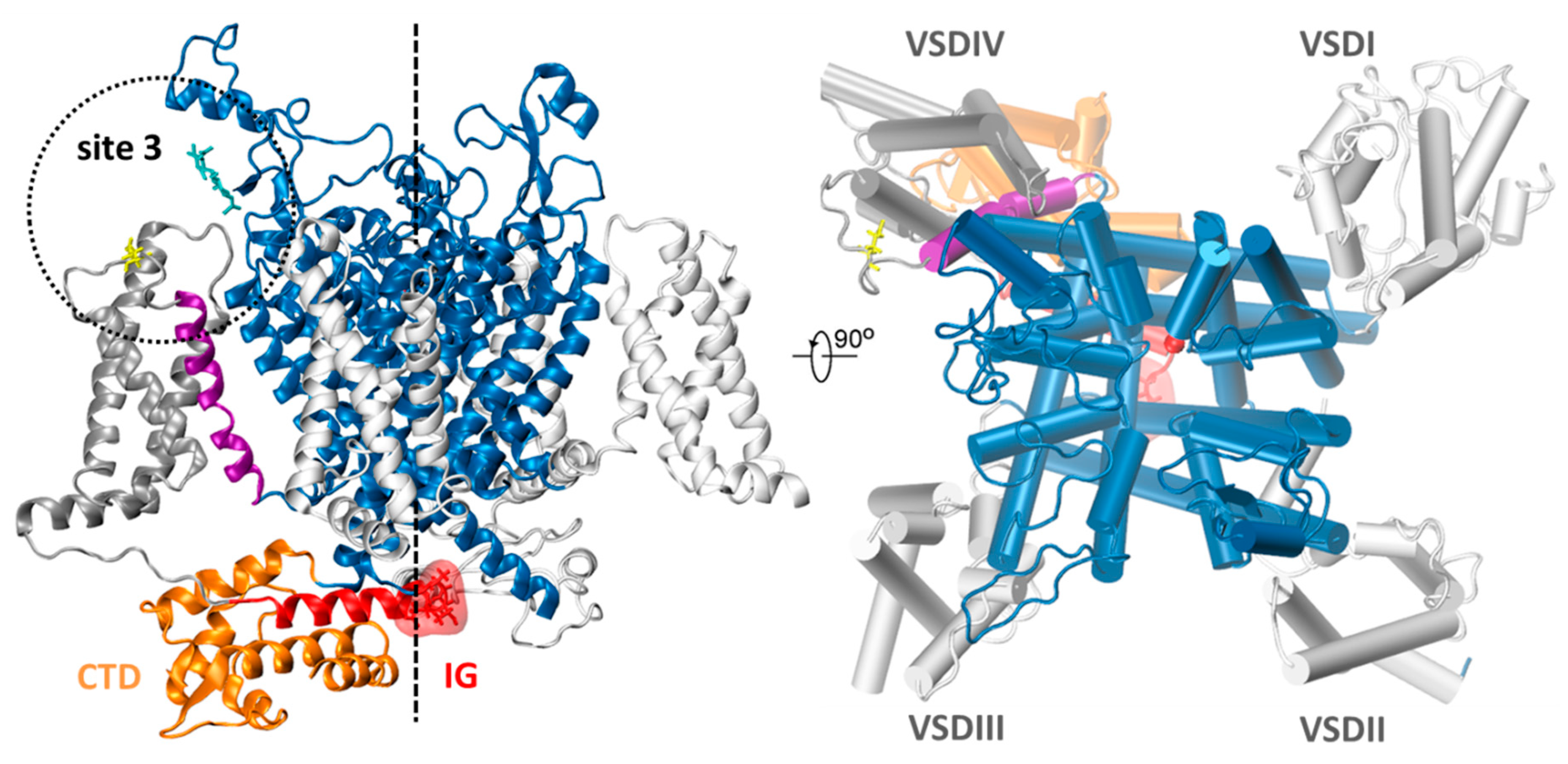
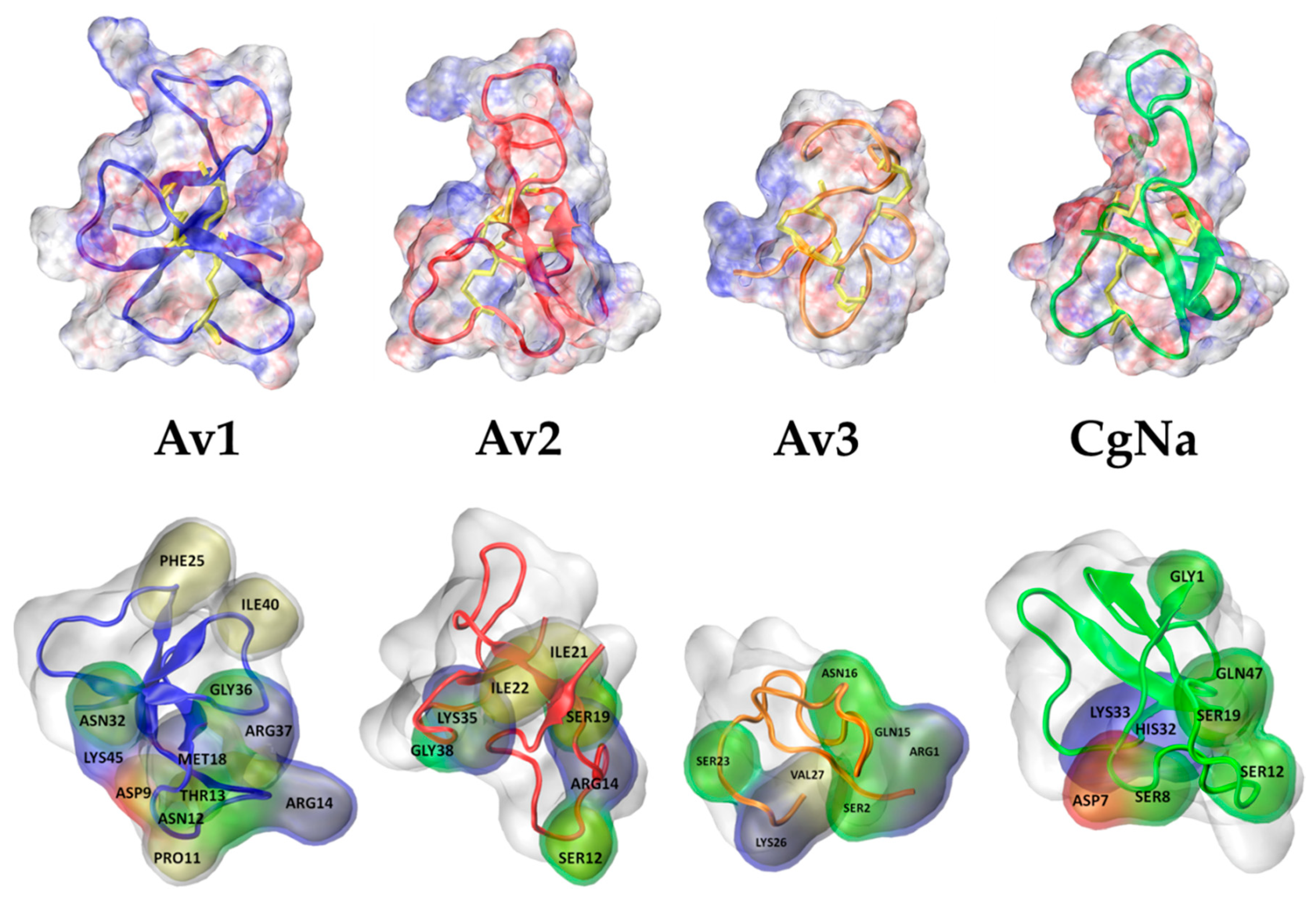
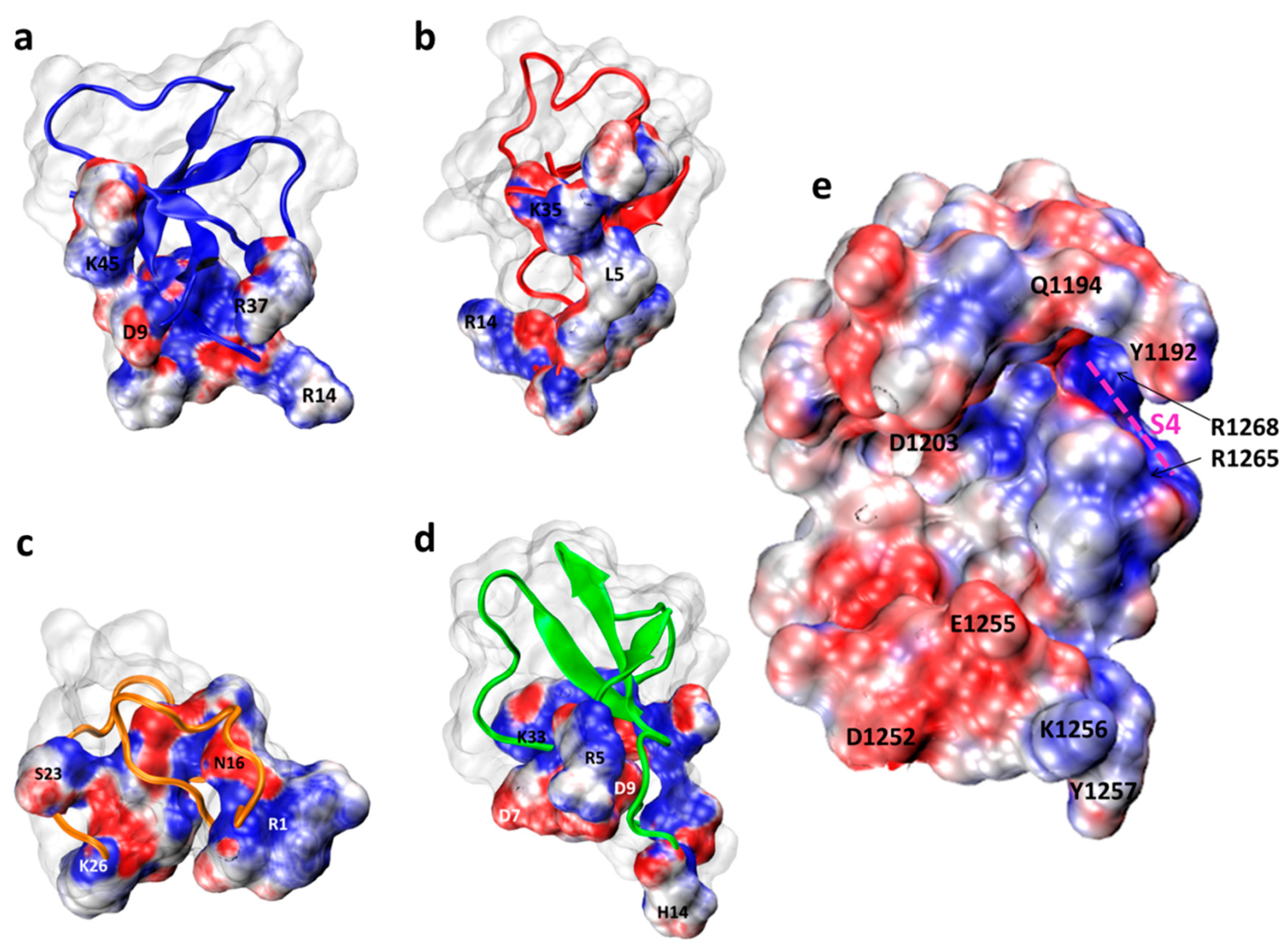
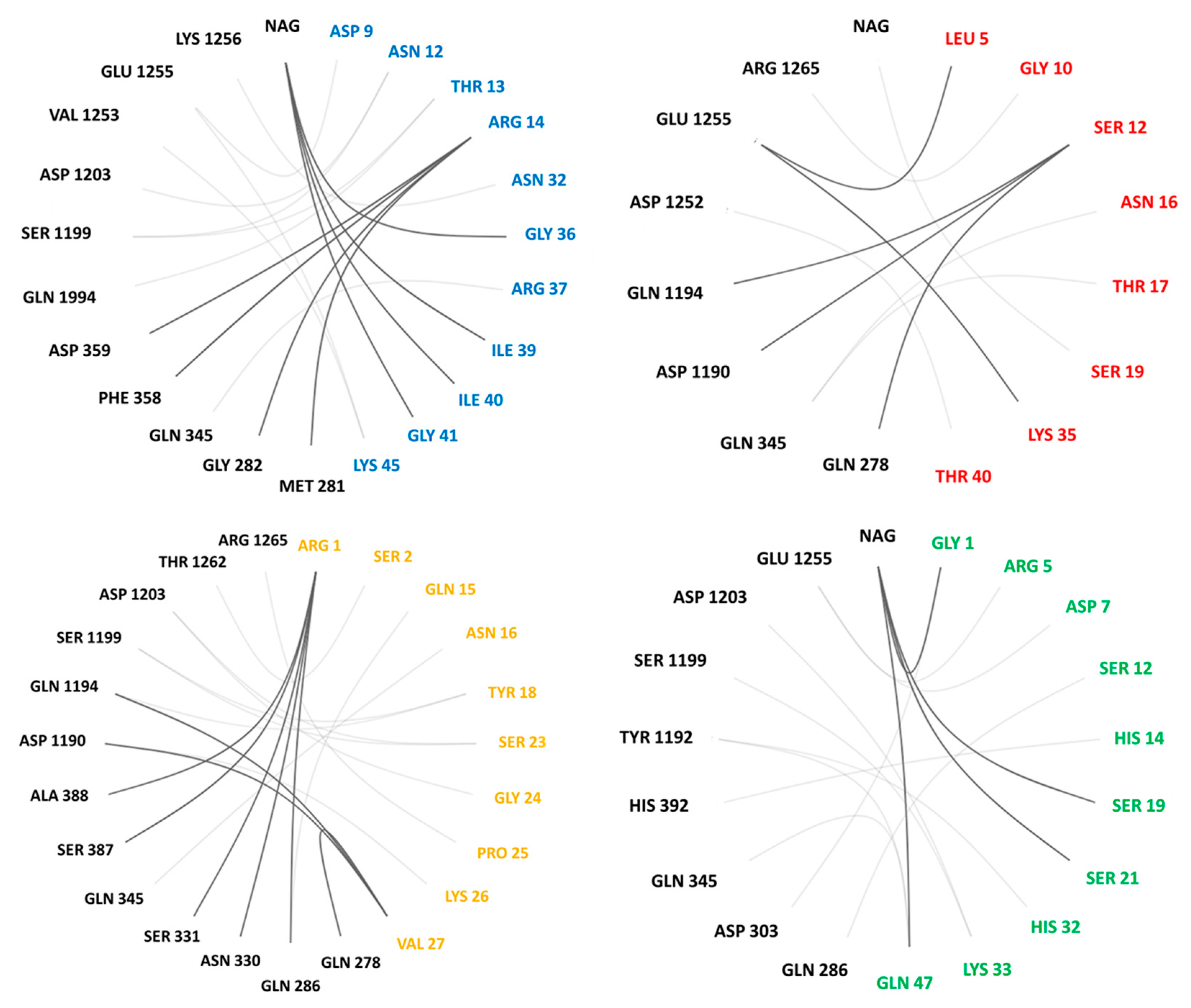
2.2. Molecular Docking
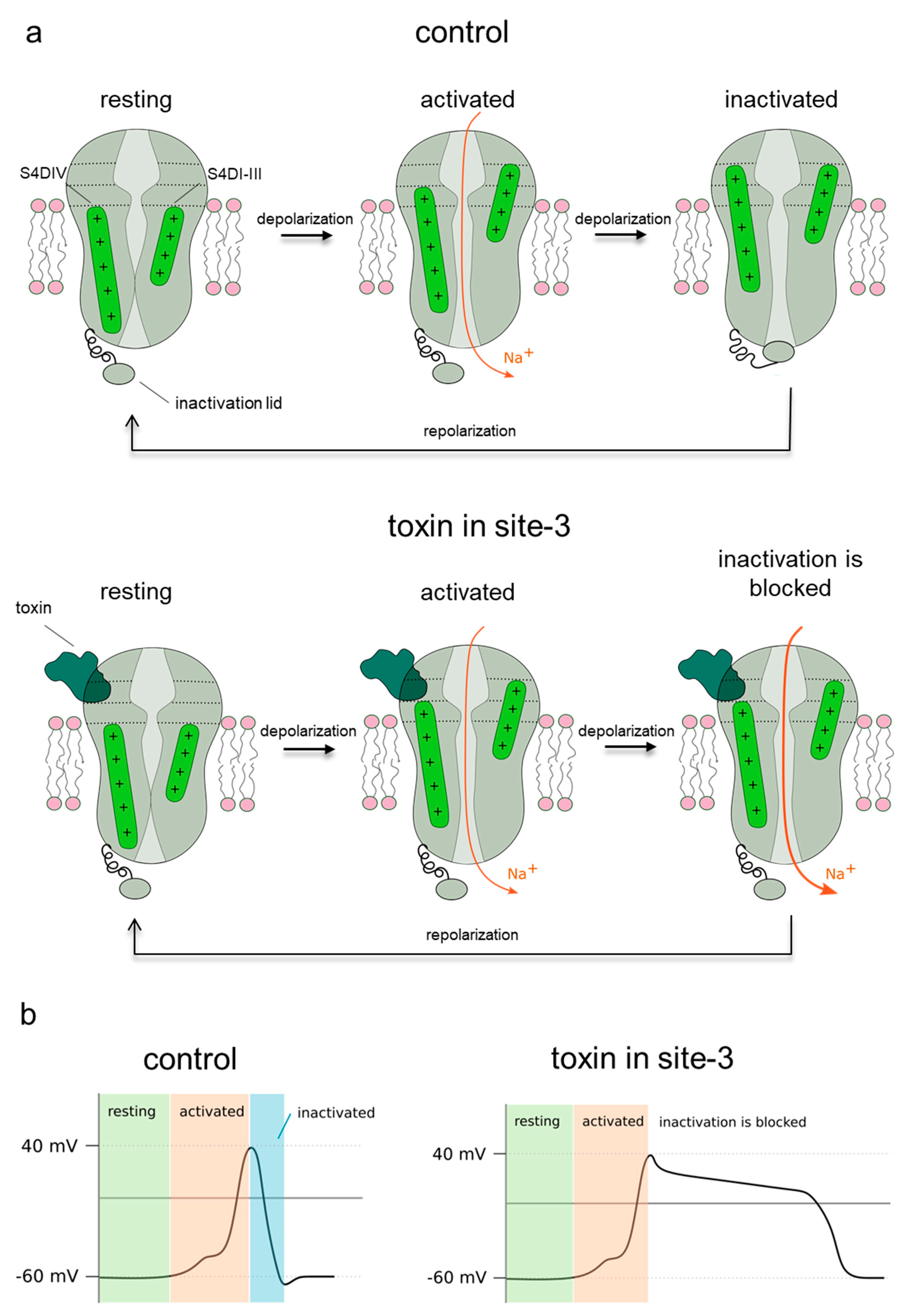
3. Discussion
- Possible Hot Spots in Toxin Binding to Site 3 of Sodium Channels
- Possible NavPaS Residues Affecting Anemone Toxin Binding
- DI: [S5: Gln278, Ile279], [EC3: Met281, Gly282, Val283, Gln286, Phe301, Asp303, Trp306, Phe307, Gly329, Asn330, NAG1601, NAG1602, Ser331, Gln345, Tyr347, Phe358, Ap359], [EC4: Ser387, Ala388, His392]
- DIV: [S1–S2 loop: Asp1190, His1191, Tyr1192, Gly1193, Gln1194, Met1196, Ser1199, Glu1200], [S2: Leu1202, Asp1203, Tyr1204, Asn1206], [S3: Gly1247, Leu1248], [S3–S4 loop: Asp1252, Val1253, Ile1254, Glu1255, Lys1256, Tyr1257, Phe1258, Ile1259, Pro 1261, Thr1262], [S4: Leu1264, Arg1265, Arg1268].
4. Materials and Methods
4.1. Electrophysiology
4.2. Molecular Modeling and Docking
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Catterall, W.A.; Wisedchaisri, G.; Zheng, N. The conformational cycle of a prototypical voltage-gated sodium channel. Nat. Chem. Biol. 2020, 16, 1314–1320. [Google Scholar] [CrossRef] [PubMed]
- Armstrong, C.M. Na channel inactivation from open and closed states. Proc. Natl. Acad. Sci. USA 2006, 103, 17991–17996. [Google Scholar] [CrossRef]
- Vassilev, P.M.; Scheuer, T.; Catterall, W.A. Identification of an intracellular peptide segment involved in sodium channel inactivation. Science 1988, 241, 1658–1661. [Google Scholar] [CrossRef]
- Huang, W.; Liu, M.; Yan, S.F.; Yan, N. Structure-based assessment of disease-related mutations in human voltage-gated sodium channels. Protein Cell 2017, 8, 401–438. [Google Scholar] [CrossRef]
- Ryan, D.P.; Ptáček, L.J. Episodic Neurological Channelopathies. Neuron 2010, 68, 282–292. [Google Scholar] [CrossRef]
- Kasimova, M.; Granata, D.; Carnevale, V. Voltage-gated sodium channels: Evolutionary history and distinctive sequence features. In Current Topics in Membranes; Academic Press: Cambridge, MA, USA, 2016; Volume 78, pp. 261–286. [Google Scholar] [CrossRef]
- Gordon, D.; Karbat, I.; Ilan, N.; Cohen, L.; Kahn, R.; Gilles, N.; Dong, K.; Stühmer, W.; Tytgat, J.; Gurevitz, M. The differential preference of scorpion α-toxins for insect or mammalian sodium channels: Implications for improved insect control. Toxicon 2007, 49, 452–472. [Google Scholar] [CrossRef] [PubMed]
- Bosmans, F.; Tytgat, J. Animal peptides targeting voltage-activated sodium channels. Curr. Pharm. Des. 2008, 14, 2492–2502. [Google Scholar] [CrossRef]
- Zhu, S.; Gao, B.; Peigneur, S.; Tytgat, J. How a scorpion toxin selectively captures a prey sodium channel: The molecular and evolutionary basis uncovered. Mol. Biol. Evol. 2020, 37, 3149–3164. [Google Scholar] [CrossRef] [PubMed]
- Nicholson, G.M.; Little, M.J.; Birinyi-Strachan, L.C. Structure and function of δ-atracotoxins: Lethal neurotoxins targeting the voltage-gated sodium channel. Toxicon 2004, 43, 587–599. [Google Scholar] [CrossRef]
- Moran, Y.; Gordon, D.; Gurevitz, M. Sea anemone toxins affecting voltage-gated sodium channels–molecular and evolutionary features. Toxicon 2009, 54, 1089–1101. [Google Scholar] [CrossRef] [PubMed]
- Leipold, E.; Hansel, A.; Olivera, B.M.; Terlau, H.; Heinemann, S.H. Molecular interaction of δ-conotoxins with voltage-gated sodium channels. FEBS Lett. 2005, 579, 3881–3884. [Google Scholar] [CrossRef]
- Gilchrist, J.; Olivera, B.M.; Bosmans, F. Animal toxins influence voltage-gated sodium channel function. In Handbook of Experimental Pharmacology; Springer International Publishing: Berlin/Heidelberg, Germany, 2014; Volume 221, pp. 203–229. [Google Scholar] [CrossRef]
- Cardoso, F.C.; Lewis, R.J. Sodium channels and pain: From toxins to therapies. Br. J. Pharmacol. 2018, 175, 2138–2157. [Google Scholar] [CrossRef]
- Shen, H.; Li, Z.; Jiang, Y.; Pan, X.; Wu, J.; Cristofori-Armstrong, B.; Smith, J.J.; Chin, Y.K.Y.; Lei, J.; Zhou, Q.; et al. Structural basis for the modulation of voltage-gated sodium channels by animal toxins. Science 2018, 362, eaau2596. [Google Scholar] [CrossRef]
- Cardoso, F.C. Multi-targeting sodium and calcium channels using venom peptides for the treatment of complex ion channels-related diseases. Biochem. Pharmacol. 2020, 114107. [Google Scholar] [CrossRef] [PubMed]
- Jankowska, M.; Lapied, B.; Jankowski, W.; Stankiewicz, M. The unusual action of essential oil component, menthol, in potentiating the effect of the carbamate insecticide, bendiocarb. Pestic. Biochem. Physiol. 2019, 158, 101–111. [Google Scholar] [CrossRef]
- Pelhate, M.; Laufer, J.; Pichon, Y.; Zlotkin, E. Effects of several sea anemone and scorpion toxins on excitability and ionic currents in the giant axon of the cockroach. J. Physiol. 1984, 79, 309–317. [Google Scholar]
- Koes, D.R.; Baumgartner, M.P.; Camacho, C.J. Lessons learned in empirical scoring with smina from the CSAR 2011 bench-marking exercise. J. Chem. Inf. Model. 2013, 53, 1893–1904. [Google Scholar] [CrossRef]
- Baker, N.A.; Sept, D.; Joseph, S.; Holst, M.J.; McCammon, J.A. Electrostatics of nanosystems: Application to microtubules and the ribosome. Proc. Natl. Acad. Sci. USA 2001, 98, 10037–10041. [Google Scholar] [CrossRef]
- Humphrey, W.; Dalke, A.; Schulten, K. VMD: Visual molecular dynamics. J. Mol. Graph. 1996, 14, 33–38. [Google Scholar] [CrossRef]
- Venkatakrishnan, A.; Fonseca, R.; Ma, A.K.; Hollingsworth, S.A.; Chemparathy, A.; Hilger, D.; Dror, R.O. Uncovering patterns of atomic interactions in static and dynamic structures of proteins. bioRxiv 2019, 840694. [Google Scholar] [CrossRef]
- Zhou, Q.; Yang, D.; Wu, M.; Guo, Y.; Guo, W.; Zhong, L.; Cai, X.; Dai, A.; Jang, W.; I Shakhnovich, E.; et al. Common activation mechanism of class A GPCRs. eLife 2019, 8, 50279. [Google Scholar] [CrossRef] [PubMed]
- Moran, Y.; Kahn, R.; Cohen, L.; Gur, M.; Karbat, I.; Gordon, D.; Gurevitz, M. Molecular analysis of the sea anemone toxin Av3 reveals selectivity to insects and demonstrates the heterogeneity of receptor site-3 on voltage-gated Na+ channels. Biochem. J. 2007, 406, 41–48. [Google Scholar] [CrossRef]
- Barzilai, M.G.; Kahn, R.; Regev, N.; Gordon, D.; Moran, Y.; Gurevitz, M. The specificity of Av3 sea anemone toxin for arthropods is determined at linker DI/SS2–S6 in the pore module of target sodium channels. Biochem. J. 2014, 463, 271–277. [Google Scholar] [CrossRef] [PubMed]
- Rogers, J.C.; Qu, Y.; Tanada, T.N.; Scheuer, T.; Catterall, W.A. Molecular determinants of high affinity binding of α-scorpion toxin and sea anemone toxin in the S3-S4 extracellular loop in domain IV of the Na+ channel α subunit. J. Biol. Chem. 1996, 271, 15950–15962. [Google Scholar] [CrossRef]
- Shen, H.; Zhou, Q.; Pan, X.; Li, Z.; Wu, J.; Yan, N. Structure of a eukaryotic voltage-gated sodium channel at near-atomic resolution. Science 2017, 355, eaal4326. [Google Scholar] [CrossRef]
- Yan, Z.; Zhou, Q.; Wang, L.; Wu, J.; Zhao, Y.; Huang, G.; Peng, W.; Shen, H.; Lei, J.; Yan, N. Structure of the Nav1. 4-β1 complex from electric eel. Cell 2017, 170, 470–482.e11. [Google Scholar] [CrossRef]
- Madio, B.; King, G.F.; Undheim, E.A.B. Sea anemone toxins: A structural overview. Mar. Drugs 2019, 17, 325. [Google Scholar] [CrossRef]
- Béress, L. Biologically active compounds from coelenterates. Pure Appl. Chem. 1982, 54, 1981–1994. [Google Scholar] [CrossRef]
- Norton, R.S. Structures of sea anemone toxins. Toxicon 2009, 54, 1075–1088. [Google Scholar] [CrossRef] [PubMed]
- Billen, B.; Debaveye, S.; Béress, L.; Garateix, A.; Tytgat, J. Phyla- and subtype-selectivity of CgNa, a Na+ channel toxin from the venom of the giant caribbean sea anemone Condylactis Gigantea. Front. Pharmacol. 2010, 1, 133. [Google Scholar] [CrossRef]
- Manoleras, N.; Norton, R.S. Three-dimensional structure in solution of neurotoxin III from the sea anemone Anemonia sulcata. Biochemistry 1994, 33, 11051–11061. [Google Scholar] [CrossRef]
- Rathmayer, W.; Béress, L. The effect of toxins from Anemonia sulcata (Coelenterata) on neuromuscular transmission and nerve action potentials in the crayfish (Astacus leptodactylus). J. Comp. Physiol. A 1976, 109, 373–382. [Google Scholar] [CrossRef]
- Ravens, U.; Schöllhorn, E. The effects of a toxin (ATX II) from the sea anemone Anemonia sulcata on the electrical and mechanical activity of the denervated hemidiaphragm of the rat. Toxicon 1983, 21, 131–142. [Google Scholar] [CrossRef]
- Kudo, Y.; Shibata, S. The potent excitatory effect of a novel polypeptide, anthopleurin-B, isolated from a sea anemone (Anthopleura xanthogrammica) on the frog spinal cord. J. Pharmacol. Exp. Ther. 1980, 214, 443–448. [Google Scholar]
- Gordon, D.; Zlotkin, E. Binding of an α scorpion toxin to insect sodium channels is not dependent on membrane potential. FEBS Lett. 1993, 315, 125–128. [Google Scholar] [CrossRef]
- Moran, Y.; Cohen, L.; Kahn, R.; Karbat, I.; Gordon, D.; Gurevitz, M. Expression and mutagenesis of the sea anemone toxin Av2 reveals key amino acid residues important for activity on voltage-gated sodium channels. Biochemistry 2006, 45, 8864–8873. [Google Scholar] [CrossRef]
- Chahine, M.; Plante, E.; Kallen, R. Sea anemone toxin (ATX II) modulation of heart and skeletal muscle sodium channel α-subunits expressed in tsA201 cells. J. Membr. Biol. 1996, 152, 39–48. [Google Scholar] [CrossRef]
- Castillo, C.; Piernavieja, C.; Recio-Pinto, E. Interactions between anemone toxin II and veratridine on single neuronal sodium channels. Brain Res. 1996, 733, 243–252. [Google Scholar] [CrossRef]
- Wanke, E.; Zaharenko, A.J.; Redaelli, E.; Schiavon, E. Actions of sea anemone type 1 neurotoxins on voltage-gated sodium channel isoforms. Toxicon 2009, 54, 1102–1111. [Google Scholar] [CrossRef] [PubMed]
- Hartung, K.; Rathmayer, W. Anemonia sulcata toxins modify activation and inactivation of Na+ currents in a crayfish neurone. Pflügers Arch. Eur. J. Physiol. 1985, 404, 119–125. [Google Scholar] [CrossRef]
- Oliveira, J.S.; Redaelli, E.; Zaharenko, A.J.; Cassulini, R.R.; Konno, K.; Pimenta, D.C.; Freitas, J.C.; Clare, J.J.; Wanke, E. Binding specificity of sea anemone toxins to Nav 1.1-1.6 sodium channels. J. Biol. Chem. 2004, 279, 33323–33335. [Google Scholar] [CrossRef] [PubMed]
- Abd-Ella, A.; Stankiewicz, M.; Mikulska, K.; Nowak, W.; Pennetier, C.; Goulu, M.; Fruchart-Gaillard, C.; Licznar, P.; Apaire-Marchais, V.; List, O.; et al. The repellent DEET potentiates carbamate effects via insect muscarinic receptor interactions: An alternative strategy to control insect vector-borne diseases. PLoS ONE 2015, 10, e0126406. [Google Scholar] [CrossRef]
- Moreau, E.; Mikulska-Ruminska, K.; Goulu, M.; Perrier, S.; Deshayes, C.; Stankiewicz, M.; Apaire-Marchais, V.; Nowak, W.; Lapied, B. Orthosteric muscarinic receptor activation by the insect repellent IR3535 opens new prospects in insecticide-based vector control. Sci. Rep. 2020, 10, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Kalina, R.S.; Peigneur, S.; Zelepuga, E.A.; Dmitrenok, P.S.; Kvetkina, A.N.; Kim, N.Y.; Leychenko, E.V.; Tytgat, J.; Kozlovskaya, E.P.; Monastyrnaya, M.M.; et al. New insights into the type II toxins from the sea anemone Heteractis crispa. Toxins 2020, 12, 44. [Google Scholar] [CrossRef] [PubMed]
- Chakrabarti, N.; Ing, C.; Payandeh, J.; Zheng, N.; Catterall, W.A.; Pomès, R. Catalysis of Na+ permeation in the bacterial sodium channel NaVAb. Proc. Natl. Acad. Sci. USA 2013, 110, 11331–11336. [Google Scholar] [CrossRef]
- Flood, E.; Boiteux, C.; Lev, B.; Vorobyov, I.; Allen, T.W. Atomistic simulations of membrane ion channel conduction, gating, and modulation. Chem. Rev. 2019, 119, 7737–7832. [Google Scholar] [CrossRef] [PubMed]
- Vargas, E.; Yarov-Yarovoy, V.; Khalili-Araghi, F.; Catterall, W.A.; Klein, M.L.; Tarek, M.; Roux, B. An emerging consensus on voltage-dependent gating from computational modeling and molecular dynamics simulations. J. Gen. Physiol. 2012, 140, 587–594. [Google Scholar] [CrossRef] [PubMed]
- Demarco, K.R.; Bekker, S.; Vorobyov, I. Challenges and advances in atomistic simulations of potassium and sodium ion channel gating and permeation. J. Physiol. 2019, 597, 679–698. [Google Scholar] [CrossRef]
- Zhang, J.; Mao, W.; Ren, Y.; Sun, R.-N.; Yan, N.; Gong, H. Simulating the ion permeation and ion selection for a eukaryotic voltage-gated sodium channel NaVPaS. Protein Cell 2018, 9, 580–585. [Google Scholar]
- Khera, P.K.; Blumenthal, K.M. Importance of highly conserved anionic residues and electrostatic interactions in the activity and structure of the cardiotonic polypeptide Anthopleurin B. Biochemistry 1996, 35, 3503–3507. [Google Scholar] [CrossRef] [PubMed]
- Gur, M.; Kahn, R.; Karbat, I.; Regev, N.; Wang, J.; Catterall, W.A.; Gordon, D.; Gurevitz, M. Elucidation of the molecular basis of selective recognition uncovers the interaction site for the core domain of scorpion α-toxins on sodium channels*. J. Biol. Chem. 2011, 286, 35209–35217. [Google Scholar] [CrossRef]
- Leipold, E.; Lu, S.; Gordon, D.; Hansel, A.; Heinemann, S.H. Combinatorial interaction of scorpion toxins Lqh-2, Lqh-3, and LqhαIT with sodium channel receptor sites-3. Mol. Pharmacol. 2004, 65, 685–691. [Google Scholar] [CrossRef] [PubMed]
- Rowe, A.H.; Xiao, Y.; Scales, J.; Linse, K.D.; Rowe, M.P.; Cummins, T.R.; Zakon, H.H. Isolation and characterization of CvIV4: A pain inducing α- scorpion toxin. PLoS ONE 2011, 6, e23520. [Google Scholar] [CrossRef]
- Xiao, Y. The tarantula toxins ProTx-II and huwentoxin-IV differentially interact with human Nav1.7 voltage sensors to inhibit channel activation and inactivation. Mol. Pharmacol. 2010, 78, 1124–1134. [Google Scholar] [CrossRef]
- Bosmans, F.; Tytgat, J. Sea anemone venom as a source of insecticidal peptides acting on voltage-gated Na+ channels. Toxicon 2007, 49, 550–560. [Google Scholar] [CrossRef] [PubMed]
- Clairfeuille, T.; Cloake, A.; Infield, D.T.; Llongueras, J.P.; Arthur, C.P.; Li, Z.R.; Jian, Y.; Martin-Eauclaire, M.-F.; Bougis, P.E.; Ciferri, C.; et al. Structural basis of α-scorpion toxin action on Nav channels. Science 2019, 363, eaav8573. [Google Scholar] [CrossRef]
- Lauc, G.; Krištić, J.; Zoldoš, V. Glycans–the third revolution in evolution. Front. Genet. 2014, 5, 145. [Google Scholar] [CrossRef]
- Baycin-Hizal, D.; Gottschalk, A.; Jacobson, E.; Mai, S.; Wolozny, D.; Zhang, H.; Krag, S.S.; Betenbaugh, M.J. Physiologic and pathophysiologic consequences of altered sialylation and glycosylation on ion channel function. Biochem. Biophys. Res. Commun. 2014, 453, 243–253. [Google Scholar] [CrossRef] [PubMed]
- Béress, L.; Béress, R.; Wunderer, G. Purification of three polypeptides with neuro and cardiotoxic activity from the sea anemone Anemonia sulcata. Toxicon 1975, 13, 359–368. [Google Scholar] [CrossRef]
- Ständker, L.; Béress, L.; Garateix, A.; Christ, T.; Ravens, U.; Salceda, E.; Soto, E.; John, H.; Forssmann, W.-G.; Aneiros, A. A new toxin from the sea anemone Condylactis gigantea with effect on sodium channel inactivation. Toxicon 2006, 48, 211–220. [Google Scholar] [CrossRef] [PubMed]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef] [PubMed]
- Waterhouse, A.; Bertoni, M.; Bienert, S.; Studer, G.; Tauriello, G.; Gumienny, R.; Heer, F.T.; de Beer, T.A.P.; Rempfer, C.; Bordoli, L.; et al. SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 2018, 46, W296–W303. [Google Scholar] [CrossRef]
- Jo, S.; Kim, T.; Iyer, V.G.; Im, W. CHARMM-GUI: A web-based graphical user interface for CHARMM. J. Comput. Chem. 2008, 29, 1859–1865. [Google Scholar] [CrossRef] [PubMed]
- Krissinel, E.; Henrick, K. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 2007, 372, 774–797. [Google Scholar] [CrossRef] [PubMed]
- Dolinsky, T.J.; Nielsen, J.E.; McCammon, J.A.; Baker, N.A. PDB2PQR: An automated pipeline for the setup of Poisson-Boltzmann electrostatics calculations. Nucleic Acids Res. 2004, 32, W665–W667. [Google Scholar] [CrossRef]

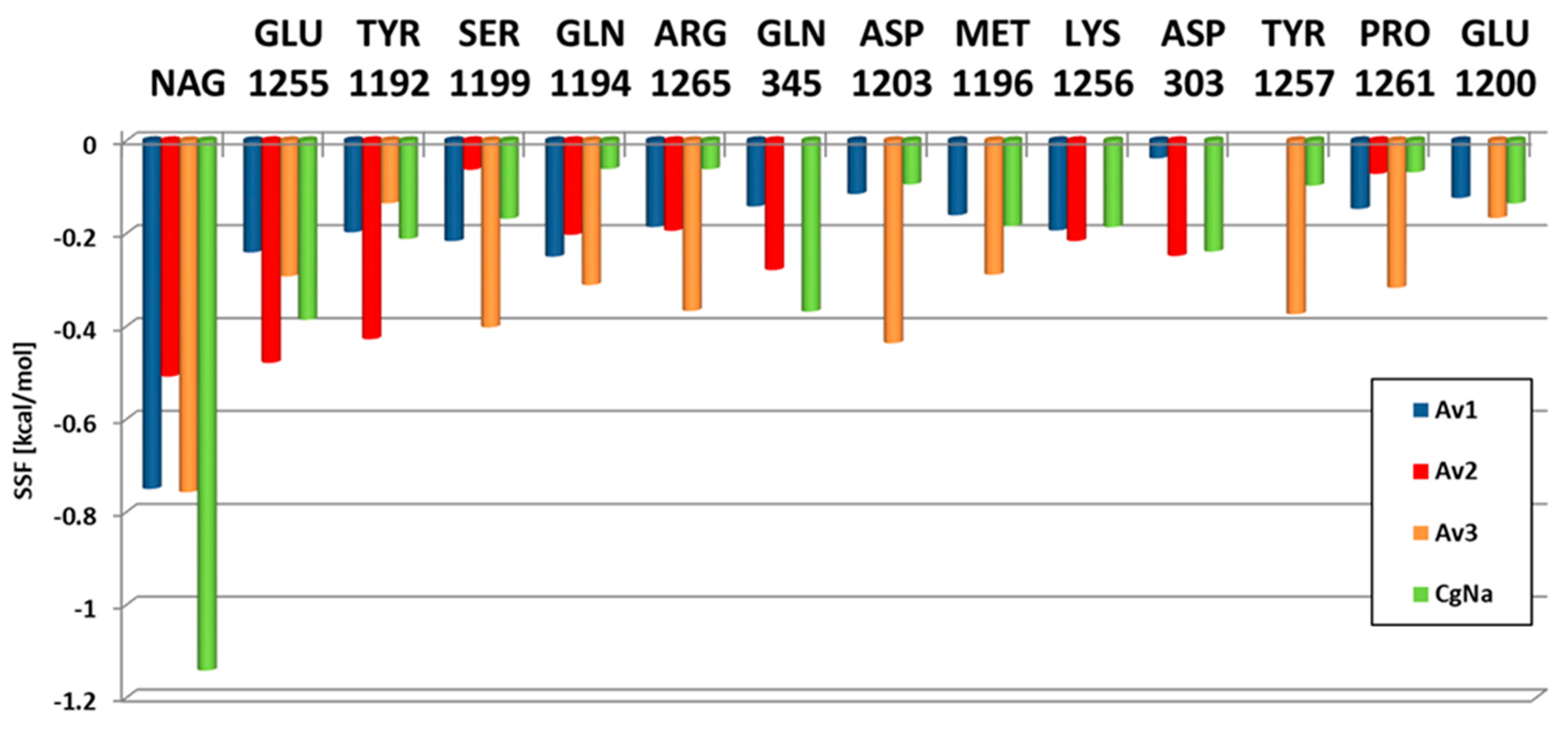
| Parameter * | Av1 | Av2 | Av3 | Av3’ | CgNa |
|---|---|---|---|---|---|
| Activity in experiment | $$$ | $ | $$ | $$ | $$$ |
| SSF (kcal/mol) | −4.89 | −4.98 | −9.72 | −8.85 | −5.53 |
| Total RRCS (AA) | 122.10 | 86.00 | 152.80 | 145.90 | 119.13 |
| Total RRCS for NAG | 26.63 | 19.90 | 13.90 | 2.53 | 38.08 |
| RRCS S4 only | 14.80 | 7.52 | 21.84 | 6.51 | 10.14 |
| Toxin AA in interface (%) | 61 | 55 | 85 | 78 | 70 |
| NAG atoms in interface (%) | 68 | 75 | 50 | 46 | 86 |
| ASA_TI (%) | 34 | 22 | 46 | 45 | 37 |
| ASA_NavI (%) | 1.40 | 1.20 | 1.40 | 1.40 | 1.70 |
| % BSA Glu1255 | 98 | 74 | 43 | 89 | 72 |
| % BSA Arg1265 (S4) | 77 | 54 | 94 | 31 | 35 |
| % BSA Arg1268 (S4) | 27 | 0 | 83 | 22 | 0 |
| % BSA NAG | 35 | 42 | 25 | 20 | 42 |
| Interaction | Av1 | Av2 | Av3 | Av3’ | CgNa |
|---|---|---|---|---|---|
| Type | |||||
| salt bridge | ASP 359-ARG 14 | GLU 1255-LYS 35 | ASP 1190-LYS 26 * | ASP 1203-LYS 33 | |
| GLU 1255-LYS 45 * | |||||
| π-cation | TYR 1192-ARG 1 | HIS 392-HIS 14 | |||
| TYR 1204-LYS 26 | TYR 1192-HIS 32 | ||||
| H bond | sb MET 281-ARG 14 | ss GLN 278-SER 12 | ss GLN 286-ARG 1 | sb ILE 279-TRP 13 | ss GLN 286-SER 12 |
| s—side chain | sb GLY 282-ARG 14 | ss GLN 345-ASN 16 | ss GLN 286-GLN 15 | sb HIS 1191-ARG 1 | sb ASP 303-GLY 1 |
| b—backbone | ss GLN 345-ARG 37 | ss GLN 345-THR 17 | ss ASN 330-ARG 1 | sb GLY 1993-ARG 1 | ss TYR 1192-GLN 47 |
| sb PHE 358-ARG 14 | ss ASP 1190-SER 12 | sb SER 331-ARG 1 | sb GLN 1994-ARG 1 | ss SER 1199-LYS 33 | |
| sb ASP 359-ARG 14 | sb GLN 1194-SER 12 | ss SER 331-ARG 1 | sb GLU 1200-GLY 24 | ss ASP 1203-LYS 33 | |
| ss ASP 359-ARG 14 | ss ASP 1252-THR 40 | sb GLN 345-ASN 16 | sb ASP 1203-LYS 26 | sb GLU 1255-ARG 5 | |
| ss GLN 1194-THR 13 | sb GLU 1255-LEU 5 | sb SER 387-ARG 1 | ss ASP 1203-VAL 27 | sb GLU 1255-ASP 7 | |
| ss SER 1199-ASN 12 | ss GLU 1255-LYS 35 | bb ALA 388-ARG 1 | sb LEU 1248-TYR 7 | b NAG-GLY 1 | |
| sb SER 1199-THR 13 | sb ARG 1265-GLY 10 | ss ASP 1190-LYS 26 | sb GLU 1255-GLY 9 | s NAG-HYP 3 | |
| ss ASP 1203-ASN 12 | s NAG-SER 19 | ss GLN 1194-TYR 18 | s NAG-THR 21 | ||
| Av3’ | sb VAL 1253-LYS 45 | sb GLN 1194-VAL 27 | VAL 283-TRP 13 | s NAG-GLN 47 | |
| hydrophobic | ss GLU 1255-ASP 9 | ss SER 1199-TYR 18 | ALA 388-TRP 13 | ||
| ss GLU 1255-LYS 45 | sb SER 1199-SER 23 | PHE 1258-TRP 8 | |||
| ss LYS 1256-ASN 32 | ss ASP 1203-SER 23 | ILE 1259-TRP 8 | |||
| b NAG-GLY 36 | sb ASP 1203-GLY 24 | PRO 1261-TRP 8 | |||
| b NAG-ILE 39 | sb THR 1262-SER 2 | ||||
| b NAG-ILE 40 | ss THR 1262-SER 2 | ||||
| b NAG-GLY 41 | sb ARG 1265-PRO 25 | ||||
| s NAG-ASN 16 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Niklas, B.; Jankowska, M.; Gordon, D.; Béress, L.; Stankiewicz, M.; Nowak, W. Interactions of Sea Anemone Toxins with Insect Sodium Channel—Insights from Electrophysiology and Molecular Docking Studies. Molecules 2021, 26, 1302. https://doi.org/10.3390/molecules26051302
Niklas B, Jankowska M, Gordon D, Béress L, Stankiewicz M, Nowak W. Interactions of Sea Anemone Toxins with Insect Sodium Channel—Insights from Electrophysiology and Molecular Docking Studies. Molecules. 2021; 26(5):1302. https://doi.org/10.3390/molecules26051302
Chicago/Turabian StyleNiklas, Beata, Milena Jankowska, Dalia Gordon, László Béress, Maria Stankiewicz, and Wieslaw Nowak. 2021. "Interactions of Sea Anemone Toxins with Insect Sodium Channel—Insights from Electrophysiology and Molecular Docking Studies" Molecules 26, no. 5: 1302. https://doi.org/10.3390/molecules26051302
APA StyleNiklas, B., Jankowska, M., Gordon, D., Béress, L., Stankiewicz, M., & Nowak, W. (2021). Interactions of Sea Anemone Toxins with Insect Sodium Channel—Insights from Electrophysiology and Molecular Docking Studies. Molecules, 26(5), 1302. https://doi.org/10.3390/molecules26051302









