Recent Advances in the Chemical Biology of N-Glycans
Abstract
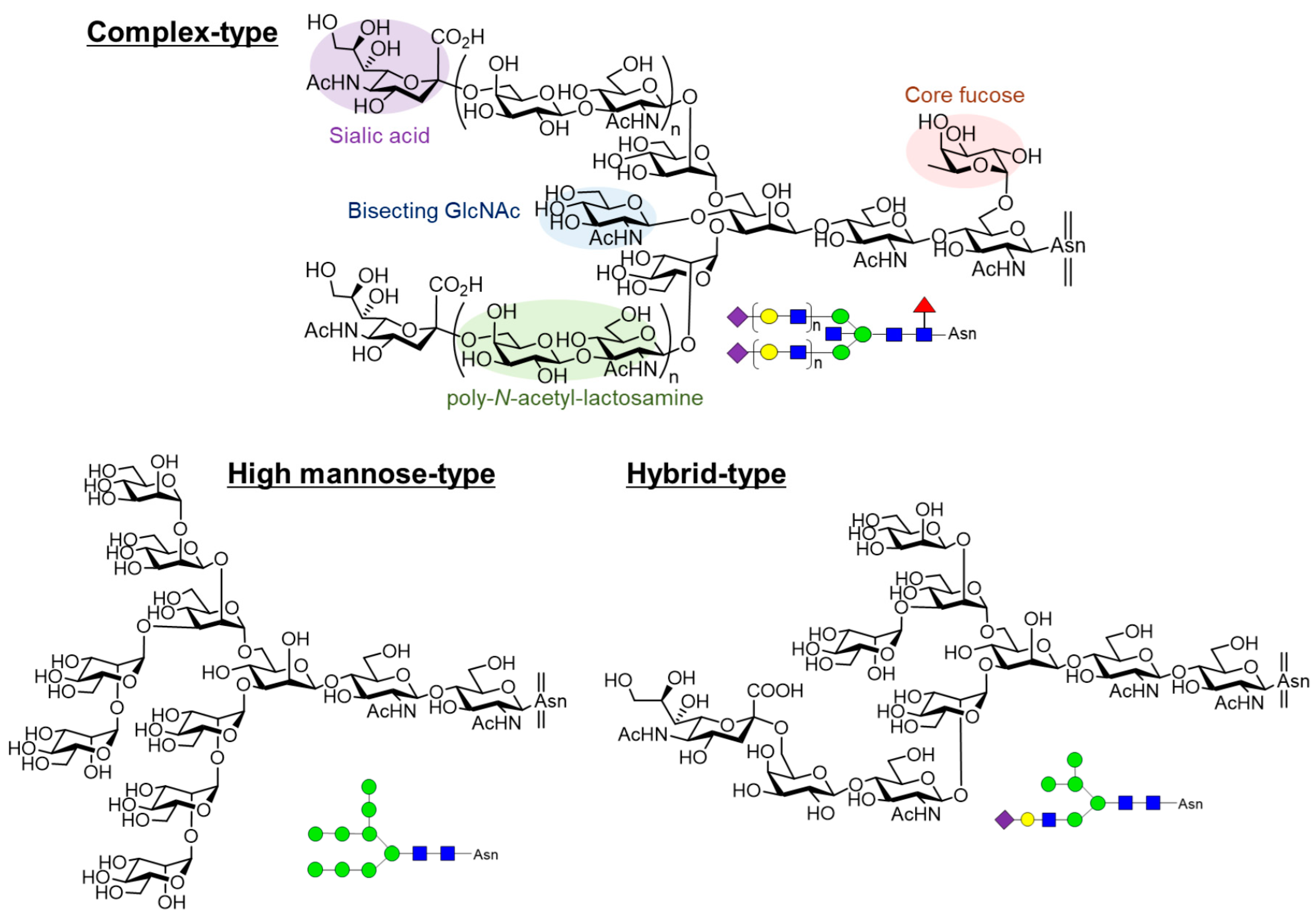
1. Introduction
2. Elucidation of the Molecular Basis of N-Glycan Recognition by Lectins
2.1. Methods for the Glycan‒Lectin Interaction Analysis
2.2. Analysis of Sugar‒Lectin Interactions Using Glycan Arrays
2.3. Analysis Using NMR
3. Functional Analysis of N-glycans on Glycoproteins
4. Use of N-glycans for Drug Development
4.1. Next-Generation Protein/Peptide Drugs Modified with Homogeneous N-Glycans
4.2. Drug Delivery Systems (DDSs) Using N-Glycans
5. Future Perspectives
Author Contributions
Funding
Conflicts of Interest
Sample Availability
References
- Dennis, J.W.; Granovsky, M.; Warren, C.E. Glycoprotein glycosylation and cancer progression. Biochim. Biophys. Acta. Gen. Subj. 1999, 1473, 21–34. [Google Scholar] [CrossRef]
- Crocker, P.R.; Paulson, J.C.; Varki, A. Siglecs and their roles in the immune system. Nat. Rev. Immunol. 2007, 7, 255–266. [Google Scholar] [CrossRef] [PubMed]
- Macauley, M.S.; Crocker, P.R.; Paulson, J.C. Siglec-mediated regulation of immune cell function in disease. Nat. Rev. Immunol. 2014, 14, 653–666. [Google Scholar] [CrossRef]
- Takahashi, M.; Kuroki, Y.; Ohtsubo, K.; Taniguchi, N. Core fucose and bisecting GlcNAc, the direct modifiers of the N-glycan core: Their functions and target proteins. Carbohydr. Res. 2009, 344, 1387–1390. [Google Scholar] [CrossRef]
- Wu, B.; Hua, Z.; Warren, J.D.; Ranganathan, K.; Wan, Q.; Chen, G.; Tan, Z.; Chen, J.; Endo, A.; Danishefsky, S.J. Synthesis of the fucosylated biantennary N-glycan of erythropoietin. Tetrahedron Lett. 2006, 47, 5577–5579. [Google Scholar] [CrossRef]
- Wang, P.; Zhu, J.; Yuan, Y.; Danishefsky, S.J. Total Synthesis of the 2,6-Sialylated Immunoglobulin G Glycopeptide Fragment in Homogeneous Form. J. Am. Chem. Soc. 2009, 131, 16669–16671. [Google Scholar] [CrossRef]
- Walczak, M.A.; Danishefsky, S.J. Solving the Convergence Problem in the Synthesis of Triantennary N-Glycan Relevant to Prostate-Specific Membrane Antigen (PSMA). J. Am. Chem. Soc. 2012, 134, 16430–16433. [Google Scholar] [CrossRef] [PubMed]
- Walczak, M.A.; Hayashida, J.; Danishefsky, S.J. Building Biologics by Chemical Synthesis: Practical Preparation of Di- and Triantennary N-Linked Glycoconjugates. J. Am. Chem. Soc. 2013, 135, 4700–4703. [Google Scholar] [CrossRef] [PubMed]
- Schuberth, R.; Unverzagt, C. Synthesis of a N-glycan nonasaccharide of the bisecting type with additional core-fucose. Tetrahedron Lett. 2005, 46, 4201–4204. [Google Scholar] [CrossRef]
- Eller, S.; Schuberth, R.; Gundel, G.; Seifert, J.; Unverzagt, C. Synthesis of Pentaantennary N-Glycans with Bisecting GlcNAc and Core Fucose. Angew. Chem. Int. Ed. 2007, 46, 4173–4175. [Google Scholar] [CrossRef] [PubMed]
- Mönnich, M.; Eller, S.; Karagiannis, T.; Perkams, L.; Luber, T.; Ott, D.; Niemietz, M.; Hoffman, J.; Walcher, J.; Berger, L.; et al. Highly Efficient Synthesis of Multiantennary Bisected N-glycans Based on Imidates. Angew. Chem. Int. Ed. 2016, 55, 10487–10492. [Google Scholar] [CrossRef]
- Luber, T.; Niemietz, M.; Karagiannis, T.; Mönnich, M.; Ott, D.; Perkams, L.; Walcher, J.; Berger, L.; Pischl, M.; Weishaupt, M.; et al. A Single Route to Mammalian N-Glycans Substituted with Core Fucose and Bisecting GlcNAc. Angew. Chem. Int. Ed. 2018, 57, 14543–14549. [Google Scholar] [CrossRef] [PubMed]
- Nagasaki, M.; Manabe, Y.; Minamoto, N.; Tanaka, K.; Silipo, A.; Molinaro, A.; Fukase, K. Chemical Synthesis of a Complex-Type N-Glycan Containing a Core Fucose. J. Org. Chem. 2016, 81, 10600–10616. [Google Scholar] [CrossRef]
- Manabe, Y.; Shomura, H.; Minamoto, N.; Nagasaki, M.; Takakura, Y.; Tanaka, K.; Silipo, A.; Molinaro, A.; Fukase, K. Convergent Synthesis of a Bisecting N-Acetylglucosamine (GlcNAc)-Containing N-Glycan. Chem. Asian J. 2018, 13, 1544–1551. [Google Scholar] [CrossRef] [PubMed]
- Koizumi, A.; Matsuo, I.; Takatani, M.; Seko, A.; Hachisu, M.; Takeda, Y.; Ito, Y. Top-Down Chemoenzymatic Approach to a High-Mannose-Type Glycan Library: Synthesis of a Common Precursor and Its Enzymatic Trimming. Angew. Chem. Int. Ed. 2013, 52, 7426–7431. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Chinoy, Z.S.; Ambre, S.G.; Peng, W.; McBride, R.; de Vries, R.P.; Glushka, J.; Paulson, J.C.; Boons, G.-J. A General Strategy for the Chemoenzymatic Synthesis of Asymmetrically Branched N-Glycans. Science 2013, 341, 379–383. [Google Scholar] [CrossRef]
- Li, T.; Huang, M.; Liu, L.; Wang, S.; Moremen, K.W.; Boons, G.-J. Divergent Chemoenzymatic Synthesis of Asymmetrical-Core-Fucosylated and Core-Unmodified N-Glycans. Chem. Eur. J. 2016, 22, 18742–18746. [Google Scholar] [CrossRef]
- Gagarinov, I.A.; Li, T.; Toraño, J.S.; Caval, T.; Srivastava, A.D.; Kruijtzer, J.A.W.; Heck, A.J.R.; Boons, G.-J. Chemoenzymatic Approach for the Preparation of Asymmetric Bi-, Tri-, and Tetra-Antennary N-Glycans from a Common Precursor. J. Am. Chem. Soc. 2017, 139, 1011–1018. [Google Scholar] [CrossRef]
- Li, L.; Liu, Y.; Ma, C.; Qu, J.; Calderon, A.D.; Wu, B.; Wei, N.; Wang, X.; Guo, Y.; Xiao, Z.; et al. Efficient chemoenzymatic synthesis of an N-glycan isomer library. Chem. Sci. 2015, 6, 5652–5661. [Google Scholar] [CrossRef]
- Shivatare, S.S.; Chang, S.-H.; Tsai, T.-I.; Ren, C.-T.; Chuang, H.-Y.; Hsu, L.; Lin, C.-W.; Li, S.-T.; Wu, C.-Y.; Wong, C.-H. Efficient Convergent Synthesis of Bi-, Tri-, and Tetra-antennary Complex Type N-Glycans and Their HIV-1 Antigenicity. J. Am. Chem. Soc. 2013, 135, 15382–15391. [Google Scholar] [CrossRef]
- Shivatare, S.S.; Chang, S.-H.; Tsai, T.-I.; Tseng, S.Y.; Shivatare, V.S.; Lin, Y.-S.; Cheng, Y.-Y.; Ren, C.-T.; Lee, C.-C.D.; Pawar, S.; et al. Modular synthesis of N-glycans and arrays for the hetero-ligand binding analysis of HIV antibodies. Nat. Chem. 2016, 8, 338–346. [Google Scholar] [CrossRef] [PubMed]
- Chiesa, M.V.; Schmidt, R.R. Synthesis of an Asparagine-Linked Heptasaccharide—Basic Structure of N-Glycans. Eur. J. Org. Chem. 2000, 2000, 3541–3554. [Google Scholar] [CrossRef]
- Kajihara, Y.; Suzuki, Y.; Yamamoto, N.; Sasaki, K.; Sakakibara, T.; Juneja, L.R. Prompt Chemoenzymatic Synthesis of Diverse Complex-Type Oligosaccharides and Its Application to the Solid-Phase Synthesis of a Glycopeptide with Asn-Linked Sialyl-undeca- and Asialo-nonasaccharides. Chem. Eur. J. 2004, 10, 971–985. [Google Scholar] [CrossRef]
- Chao, Q.; Ding, Y.; Chen, Z.-H.; Xiang, M.-H.; Wang, N.; Gao, X.-D. Recent Progress in Chemo-Enzymatic Methods for the Synthesis of N-Glycans. Front. Chem. 2020, 8, 513. [Google Scholar] [CrossRef]
- Li, T.; Liu, L.; Wei, N.; Yang, J.-Y.; Chapla, D.G.; Moremen, K.W.; Boons, G.-J. An automated platform for the enzyme-mediated assembly of complex oligosaccharides. Nat. Chem. 2019, 11, 229–236. [Google Scholar] [CrossRef]
- Paulson, J.C.; Blixt, O.; Collins, B.E. Sweet spots in functional glycomics. Nat. Chem. Biol. 2006, 2, 238–248. [Google Scholar] [CrossRef]
- Dam, T.K.; Talaga, M.L.; Fan, N.; Brewer, C.F. Chapter Four—Measuring Multivalent Binding Interactions by Isothermal Titration Calorimetry. In Methods Enzymol; Feig, A.L., Ed.; Academic Press: Cambridge, MA, USA, 2016; Volume 567, pp. 71–95. [Google Scholar]
- Duverger, E.; Frison, N.; Roche, A.-C.; Monsigny, M. Carbohydrate-lectin interactions assessed by surface plasmon resonance. Biochimie 2003, 85, 167–179. [Google Scholar] [CrossRef]
- Safina, G. Application of surface plasmon resonance for the detection of carbohydrates, glycoconjugates, and measurement of the carbohydrate-specific interactions: A comparison with conventional analytical techniques. A critical review. Anal. Chim. Acta 2012, 712, 9–29. [Google Scholar] [CrossRef] [PubMed]
- Kakehi, K.; Oda, Y.; Kinoshita, M. Fluorescence Polarization: Analysis of Carbohydrate–Protein Interaction. Anal. Biochem. 2001, 297, 111–116. [Google Scholar] [CrossRef]
- Nagae, M.; Yamaguchi, Y. Function and 3D Structure of the N-Glycans on Glycoproteins. Int. J. Mol. Sci. 2012, 13, 8398–8429. [Google Scholar] [CrossRef] [PubMed]
- Feizi, T.; Fazio, F.; Chai, W.; Wong, C.-H. Carbohydrate microarrays—A new set of technologies at the frontiers of glycomics. Curr. Opin. Struct. Biol. 2003, 13, 637–645. [Google Scholar] [CrossRef] [PubMed]
- Liang, P.-H.; Wu, C.-Y.; Greenberg, W.A.; Wong, C.-H. Glycan arrays: Biological and medical applications. Curr. Opin. Chem. Biol. 2008, 12, 86–92. [Google Scholar] [CrossRef]
- Rillahan, C.D.; Paulson, J.C. Glycan Microarrays for Decoding the Glycome. Annu. Rev. Biochem 2011, 80, 797–823. [Google Scholar] [CrossRef]
- Analysis of Glycans, Polysaccharide Functional Properties & Biochemistry of Glycoconjugate Glycans, Carbohydrate-Mediated Interactions: Comprehensive Glycoscience: From Chemistry to Systems Biology; Elsevier: Amsterdam, The Netherlands, 2007; Volumes II–III.
- Wang, D.; Liu, S.; Trummer, B.J.; Deng, C.; Wang, A. Carbohydrate microarrays for the recognition of cross-reactive molecular markers of microbes and host cells. Nat. Biotechnol. 2002, 20, 275–281. [Google Scholar] [CrossRef]
- Blixt, O.; Head, S.; Mondala, T.; Scanlan, C.; Huflejt, M.E.; Alvarez, R.; Bryan, M.C.; Fazio, F.; Calarese, D.; Stevens, J.; et al. Printed covalent glycan array for ligand profiling of diverse glycan binding proteins. Proc. Natl. Acad. Sci. USA 2004, 101, 17033–17038. [Google Scholar] [CrossRef] [PubMed]
- Marchetti, R.; Perez, S.; Arda, A.; Imberty, A.; Jimenez-Barbero, J.; Silipo, A.; Molinaro, A. “Rules of Engagement” of Protein–Glycoconjugate Interactions: A Molecular View Achievable by using NMR Spectroscopy and Molecular Modeling. ChemistryOpen 2016, 5, 274–296. [Google Scholar] [CrossRef]
- Gimeno, A.; Valverde, P.; Ardá, A.; Jiménez-Barbero, J. Glycan structures and their interactions with proteins. A NMR view. Curr. Opin. Struct. Biol. 2020, 62, 22–30. [Google Scholar] [CrossRef]
- Mayer, M.; Meyer, B. Group Epitope Mapping by Saturation Transfer Difference NMR To Identify Segments of a Ligand in Direct Contact with a Protein Receptor. J. Am. Chem. Soc. 2001, 123, 6108–6117. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Delbianco, M. Conformational Studies of Oligosaccharides. Chem. Eur. J. 2020, 26, 9814–9825. [Google Scholar] [CrossRef]
- Zhang, Y.; Yamaguchi, T.; Kato, K. New NMR Tools for Characterizing the Dynamic Conformations and Interactions of Oligosaccharides. Chem. Lett. 2013, 42, 1455–1462. [Google Scholar] [CrossRef]
- Yamaguchi, T.; Sakae, Y.; Zhang, Y.; Yamamoto, S.; Okamoto, Y.; Kato, K. Exploration of Conformational Spaces of High-Mannose-Type Oligosaccharides by an NMR-Validated Simulation. Angew. Chem. Int. Ed. 2014, 53, 10941–10944. [Google Scholar] [CrossRef] [PubMed]
- Canales, A.; Boos, I.; Perkams, L.; Karst, L.; Luber, T.; Karagiannis, T.; Domínguez, G.; Cañada, F.J.; Pérez-Castells, J.; Häussinger, D.; et al. Breaking the Limits in Analyzing Carbohydrate Recognition by NMR Spectroscopy: Resolving Branch-Selective Interaction of a Tetra-Antennary N-Glycan with Lectins. Angew. Chem. Int. Ed. 2017, 56, 14987–14991. [Google Scholar] [CrossRef] [PubMed]
- Fernández de Toro, B.; Peng, W.; Thompson, A.J.; Domínguez, G.; Cañada, F.J.; Pérez-Castells, J.; Paulson, J.C.; Jiménez-Barbero, J.; Canales, Á. Avenues to Characterize the Interactions of Extended N-Glycans with Proteins by NMR Spectroscopy: The Influenza Hemagglutinin Case. Angew. Chem. Int. Ed. 2018, 57, 15051–15055. [Google Scholar] [CrossRef]
- Canales, A.; Mallagaray, A.; Pérez-Castells, J.; Boos, I.; Unverzagt, C.; André, S.; Gabius, H.-J.; Cañada, F.J.; Jiménez-Barbero, J. Breaking Pseudo-Symmetry in Multiantennary Complex N-Glycans Using Lanthanide-Binding Tags and NMR Pseudo-Contact Shifts. Angew. Chem. Int. Ed. 2013, 52, 13789–13793. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, S.; Yamaguchi, T.; Erdélyi, M.; Griesinger, C.; Kato, K. Paramagnetic Lanthanide Tagging for NMR Conformational Analyses of N-Linked Oligosaccharides. Chem. Eur. J. 2011, 17, 9280–9282. [Google Scholar] [CrossRef]
- Weiss, M.; Ott, D.; Karagiannis, T.; Weishaupt, M.; Niemietz, M.; Eller, S.; Lott, M.; Martínez-Orts, M.; Canales, Á.; Razi, N.; et al. Efficient Chemoenzymatic Synthesis of N-Glycans with a β1,4-Galactosylated Bisecting GlcNAc Motif. ChemBioChem 2020, 21, 3212–3215. [Google Scholar] [CrossRef] [PubMed]
- Brzezicka, K.; Echeverria, B.; Serna, S.; van Diepen, A.; Hokke, C.H.; Reichardt, N.-C. Synthesis and Microarray-Assisted Binding Studies of Core Xylose and Fucose Containing N-Glycans. ACS Chem. Biol. 2015, 10, 1290–1302. [Google Scholar] [CrossRef] [PubMed]
- Echeverria, B.; Serna, S.; Achilli, S.; Vivès, C.; Pham, J.; Thépaut, M.; Hokke, C.H.; Fieschi, F.; Reichardt, N.-C. Chemoenzymatic Synthesis of N-glycan Positional Isomers and Evidence for Branch Selective Binding by Monoclonal Antibodies and Human C-type Lectin Receptors. ACS Chem. Biol. 2018, 13, 2269–2279. [Google Scholar] [CrossRef]
- Wu, Z.; Liu, Y.; Ma, C.; Li, L.; Bai, J.; Byrd-Leotis, L.; Lasanajak, Y.; Guo, Y.; Wen, L.; Zhu, H.; et al. Identification of the binding roles of terminal and internal glycan epitopes using enzymatically synthesized N-glycans containing tandem epitopes. Org. Biomol. Chem. 2016, 14, 11106–11116. [Google Scholar] [CrossRef] [PubMed]
- Pawar, S.; Hsu, L.; Narendar Reddy, T.; Ravinder, M.; Ren, C.-T.; Lin, Y.-W.; Cheng, Y.-Y.; Lin, T.-W.; Hsu, T.-L.; Wang, S.-K.; et al. Synthesis of Asymmetric N-Glycans as Common Core Substrates for Structural Diversification through Selective Enzymatic Glycosylation. ACS Chem. Biol. 2020, 15, 2382–2394. [Google Scholar] [CrossRef]
- Wu, Z.; Liu, Y.; Li, L.; Wan, X.-F.; Zhu, H.; Guo, Y.; Wei, M.; Guan, W.; Wang, P.G. Decoding glycan protein interactions by a new class of asymmetric N-glycans. Org. Biomol. Chem. 2017, 15, 8946–8951. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Guan, W.; Zhang, G.; Wu, Z.; Yu, H.; Chen, X.; Wang, P.G. Microarray analyses of closely related glycoforms reveal different accessibilities of glycan determinants on N-glycan branches. Glycobiology 2019, 30, 334–345. [Google Scholar] [CrossRef] [PubMed]
- Ratner, D.M.; Adams, E.W.; Su, J.; O’Keefe, B.R.; Mrksich, M.; Seeberger, P.H. Probing Protein–Carbohydrate Interactions with Microarrays of Synthetic Oligosaccharides. ChemBioChem 2004, 5, 379–383. [Google Scholar] [CrossRef]
- Song, X.; Xia, B.; Stowell, S.R.; Lasanajak, Y.; Smith, D.F.; Cummings, R.D. Novel Fluorescent Glycan Microarray Strategy Reveals Ligands for Galectins. Chem. Biol. 2009, 16, 36–47. [Google Scholar] [CrossRef]
- Song, X.; Yu, H.; Chen, X.; Lasanajak, Y.; Tappert, M.M.; Air, G.M.; Tiwari, V.K.; Cao, H.; Chokhawala, H.A.; Zheng, H.; et al. A Sialylated Glycan Microarray Reveals Novel Interactions of Modified Sialic Acids with Proteins and Viruses. J. Biol. Chem. 2011, 286, 31610–31622. [Google Scholar] [CrossRef]
- Huang, W.; Wang, D.; Yamada, M.; Wang, L.-X. Chemoenzymatic Synthesis and Lectin Array Characterization of a Class of N-Glycan Clusters. J. Am. Chem. Soc. 2009, 131, 17963–17971. [Google Scholar] [CrossRef]
- Gao, C.; Hanes, M.S.; Byrd-Leotis, L.A.; Wei, M.; Jia, N.; Kardish, R.J.; McKitrick, T.R.; Steinhauer, D.A.; Cummings, R.D. Unique Binding Specificities of Proteins toward Isomeric Asparagine-Linked Glycans. Cell Chem. Biol. 2019, 26, 535–547.e4. [Google Scholar] [CrossRef]
- Peng, W.; de Vries, R.P.; Grant, O.C.; Thompson, A.J.; McBride, R.; Tsogtbaatar, B.; Lee, P.S.; Razi, N.; Wilson, I.A.; Woods, R.J.; et al. Recent H3N2 Viruses Have Evolved Specificity for Extended, Branched Human-type Receptors, Conferring Potential for Increased Avidity. Cell Host Microbe 2017, 21, 23–34. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.-R.; Anwar, M.T.; Fan, C.-Y.; Low, P.Y.; Angata, T.; Lin, C.-C. Expedient assembly of Oligo-LacNAcs by a sugar nucleotide regeneration system: Finding the role of tandem LacNAc and sialic acid position towards siglec binding. Eur. J. Med. Chem. 2019, 180, 627–636. [Google Scholar] [CrossRef]
- Iwaki, J.; Hirabayashi, J. Carbohydrate-Binding Specificity of Human Galectins: An Overview by Frontal Affinity Chromatography. Trends Glycosci. Glycotechnol. 2018, 30, SE137–SE153. [Google Scholar] [CrossRef]
- Stevens, J.; Blixt, O.; Paulson, J.C.; Wilson, I.A. Glycan microarray technologies: Tools to survey host specificity of influenza viruses. Nat. Rev. Microbiol. 2006, 4, 857–864. [Google Scholar] [CrossRef]
- Neu, U.; Bauer, J.; Stehle, T. Viruses and sialic acids: Rules of engagement. Curr. Opin. Struct. Biol. 2011, 21, 610–618. [Google Scholar] [CrossRef]
- Day, C.J.; Semchenko, E.A.; Korolik, V. Glycoconjugates play a key role in Campylobacter jejuni Infection: Interactions between host and pathogen. Front. Cell. Infect. Microbiol. 2012, 2, 9. [Google Scholar] [CrossRef]
- Gulati, S.; Lasanajak, Y.; Smith, D.F.; Cummings, R.D.; Air, G.M. Glycan array analysis of influenza H1N1 binding and release. Cancer Biomark 2014, 14, 43–53. [Google Scholar] [CrossRef] [PubMed]
- Stencel-Baerenwald, J.E.; Reiss, K.; Reiter, D.M.; Stehle, T.; Dermody, T.S. The sweet spot: Defining virus–sialic acid interactions. Nat. Rev. Microbiol. 2014, 12, 739–749. [Google Scholar] [CrossRef] [PubMed]
- Ramani, S.; Hu, L.; Venkataram Prasad, B.V.; Estes, M.K. Diversity in Rotavirus–Host Glycan Interactions: A “Sweet” Spectrum. Cell. Mol. Gastroenterol. Hepatol. 2016, 2, 263–273. [Google Scholar] [CrossRef]
- Tzarum, N.; McBride, R.; Nycholat, C.M.; Peng, W.; Paulson, J.C.; Wilson, I.A. Unique Structural Features of Influenza Virus H15 Hemagglutinin. J. Virol. 2017, 91, e00046-17. [Google Scholar] [CrossRef]
- Zhu, X.; Turner, H.L.; Lang, S.; McBride, R.; Bangaru, S.; Gilchuk, I.M.; Yu, W.; Paulson, J.C.; Crowe, J.E.; Ward, A.B.; et al. Structural Basis of Protection against H7N9 Influenza Virus by Human Anti-N9 Neuraminidase Antibodies. Cell Host Microbe 2019, 26, 729–738.e4. [Google Scholar] [CrossRef]
- Byrd-Leotis, L.; Gao, C.; Jia, N.; Mehta, A.Y.; Trost, J.; Cummings, S.F.; Heimburg-Molinaro, J.; Cummings, R.D.; Steinhauer, D.A. Antigenic Pressure on H3N2 Influenza Virus Drift Strains Imposes Constraints on Binding to Sialylated Receptors but Not Phosphorylated Glycans. J. Virol. 2019, 93, e01178-19. [Google Scholar] [CrossRef]
- Blixt, O.; Han, S.; Liao, L.; Zeng, Y.; Hoffmann, J.; Futakawa, S.; Paulson, J.C. Sialoside Analogue Arrays for Rapid Identification of High Affinity Siglec Ligands. J. Am. Chem. Soc. 2008, 130, 6680–6681. [Google Scholar] [CrossRef] [PubMed]
- Rillahan, C.D.; Schwartz, E.; McBride, R.; Fokin, V.V.; Paulson, J.C. Click and Pick: Identification of Sialoside Analogues for Siglec-Based Cell Targeting. Angew. Chem. Int. Ed. 2012, 51, 11014–11018. [Google Scholar] [CrossRef]
- Medve, L.; Achilli, S.; Serna, S.; Zuccotto, F.; Varga, N.; Thépaut, M.; Civera, M.; Vivès, C.; Fieschi, F.; Reichardt, N.; et al. On-Chip Screening of a Glycomimetic Library with C-Type Lectins Reveals Structural Features Responsible for Preferential Binding of Dectin-2 over DC-SIGN/R and Langerin. Chemistry 2018, 24, 14448–14460. [Google Scholar] [CrossRef]
- Ardá, A.; Blasco, P.; Varón Silva, D.; Schubert, V.; André, S.; Bruix, M.; Cañada, F.J.; Gabius, H.-J.; Unverzagt, C.; Jiménez-Barbero, J. Molecular Recognition of Complex-Type Biantennary N-Glycans by Protein Receptors: A Three-Dimensional View on Epitope Selection by NMR. J. Am. Chem. Soc. 2013, 135, 2667–2675. [Google Scholar] [CrossRef]
- Blaum, B.S.; Hannan, J.P.; Herbert, A.P.; Kavanagh, D.; Uhrín, D.; Stehle, T. Structural basis for sialic acid–mediated self-recognition by complement factor H. Nat. Chem. Biol. 2015, 11, 77–82. [Google Scholar] [CrossRef] [PubMed]
- Macchi, E.; Rudd, T.R.; Raman, R.; Sasisekharan, R.; Yates, E.A.; Naggi, A.; Guerrini, M.; Elli, S. Nuclear Magnetic Resonance and Molecular Dynamics Simulation of the Interaction between Recognition Protein H7 of the Novel Influenza Virus H7N9 and Glycan Cell Surface Receptors. Biochemistry 2016, 55, 6605–6616. [Google Scholar] [CrossRef] [PubMed]
- Di Carluccio, C.; Crisman, E.; Manabe, Y.; Forgione, R.E.; Lacetera, A.; Amato, J.; Pagano, B.; Randazzo, A.; Zampella, A.; Lanzetta, R.; et al. Characterisation of the Dynamic Interactions between Complex N-Glycans and Human CD22. ChemBioChem 2020, 21, 129–140. [Google Scholar] [CrossRef] [PubMed]
- Forgione, R.E.; Di Carluccio, C.; Guzmán-Caldentey, J.; Gaglione, R.; Battista, F.; Chiodo, F.; Manabe, Y.; Arciello, A.; Del Vecchio, P.; Fukase, K.; et al. Unveiling Molecular Recognition of Sialoglycans by Human Siglec-10. iScience 2020, 23, 101231. [Google Scholar] [CrossRef]
- Gimeno, A.; Reichardt, N.-C.; Cañada, F.J.; Perkams, L.; Unverzagt, C.; Jiménez-Barbero, J.; Ardá, A. NMR and Molecular Recognition of N-Glycans: Remote Modifications of the Saccharide Chain Modulate Binding Features. ACS Chem. Biol. 2017, 12, 1104–1112. [Google Scholar] [CrossRef]
- Diercks, T.; Infantino, A.S.; Unione, L.; Jiménez-Barbero, J.; Oscarson, S.; Gabius, H.-J. Fluorinated Carbohydrates as Lectin Ligands: Synthesis of OH/F-Substituted N-Glycan Core Trimannoside and Epitope Mapping by 2D STD-TOCSYreFNMR spectroscopy. Chem. Eur. J. 2018, 24, 15761–15765. [Google Scholar] [CrossRef] [PubMed]
- Manabe, Y.; Marchetti, R.; Takakura, Y.; Nagasaki, M.; Nihei, W.; Takebe, T.; Tanaka, K.; Kabayama, K.; Chiodo, F.; Hanashima, S.; et al. The Core Fucose on an IgG Antibody is an Endogenous Ligand of Dectin-1. Angew. Chem. Int. Ed. 2019, 58, 18697–18702. [Google Scholar] [CrossRef] [PubMed]
- Kötzler, M.P.; Blank, S.; Bantleon, F.I.; Wienke, M.; Spillner, E.; Meyer, B. Donor Assists Acceptor Binding and Catalysis of Human α1,6-Fucosyltransferase. ACS Chem. Biol. 2013, 8, 1830–1840. [Google Scholar] [CrossRef]
- Claasen, B.; Axmann, M.; Meinecke, R.; Meyer, B. Direct Observation of Ligand Binding to Membrane Proteins in Living Cells by a Saturation Transfer Double Difference (STDD) NMR Spectroscopy Method Shows a Significantly Higher Affinity of Integrin αIIbβ3 in Native Platelets than in Liposomes. J. Am. Chem. Soc. 2005, 127, 916–919. [Google Scholar] [CrossRef]
- Xia, Y.; Zhu, Q.; Jun, K.-Y.; Wang, J.; Gao, X. Clean STD-NMR spectrum for improved detection of ligand-protein interactions at low concentration of protein. Magn. Reson. Chem. 2010, 48, 918–924. [Google Scholar] [CrossRef] [PubMed]
- Vogtherr, M.; Peters, T. Application of NMR Based Binding Assays to Identify Key Hydroxy Groups for Intermolecular Recognition. J. Am. Chem. Soc. 2000, 122, 6093–6099. [Google Scholar] [CrossRef]
- Nagae, M.; Kanagawa, M.; Morita-Matsumoto, K.; Hanashima, S.; Kizuka, Y.; Taniguchi, N.; Yamaguchi, Y. Atomic visualization of a flipped-back conformation of bisected glycans bound to specific lectins. Sci. Rep. 2016, 6, 22973. [Google Scholar] [CrossRef]
- Re, S.; Miyashita, N.; Yamaguchi, Y.; Sugita, Y. Structural Diversity and Changes in Conformational Equilibria of Biantennary Complex-Type N-Glycans in Water Revealed by Replica-Exchange Molecular Dynamics Simulation. Biophys. J. 2011, 101, L44–L46. [Google Scholar] [CrossRef]
- Nishima, W.; Miyashita, N.; Yamaguchi, Y.; Sugita, Y.; Re, S. Effect of Bisecting GlcNAc and Core Fucosylation on Conformational Properties of Biantennary Complex-Type N-Glycans in Solution. J. Phys. Chem. B 2012, 116, 8504–8512. [Google Scholar] [CrossRef] [PubMed]
- Davis, B.G. Synthesis of Glycoproteins. Chem. Rev. 2002, 102, 579–602. [Google Scholar] [CrossRef]
- Unverzagt, C.; Kajihara, Y. Chemical assembly of N-glycoproteins: A refined toolbox to address a ubiquitous posttranslational modification. Chem. Soc. Rev. 2013, 42, 4408–4420. [Google Scholar] [CrossRef]
- Wang, L.-X.; Amin, M.N. Chemical and Chemoenzymatic Synthesis of Glycoproteins for Deciphering Functions. Chem. Biol. 2014, 21, 51–66. [Google Scholar] [CrossRef] [PubMed]
- Carlo, U.; Yasuhiro, K. Recent advances in the chemical synthesis of N-linked glycoproteins. Curr. Opin. Chem. Biol. 2018, 46, 130–137. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Tran, A.H.; Danishefsky, S.J.; Tan, Z. Chapter Twelve—Chemical biology of glycoproteins: From chemical synthesis to biological impact. In Methods Enzymol; Shukla, A.K., Ed.; Academic Press: Cambridge, MA, USA, 2019; Volume 621, pp. 213–229. [Google Scholar]
- Fairbanks, A.J. The ENGases: Versatile biocatalysts for the production of homogeneous N-linked glycopeptides and glycoproteins. Chem. Soc. Rev. 2017, 46, 5128–5146. [Google Scholar] [CrossRef]
- Takeda, Y.; Totani, K.; Matsuo, I.; Ito, Y. Chemical approaches toward understanding glycan-mediated protein quality control. Curr. Opin. Chem. Biol. 2009, 13, 582–591. [Google Scholar] [CrossRef] [PubMed]
- Ito, Y.; Takeda, Y.; Seko, A.; Izumi, M.; Kajihara, Y. Functional analysis of endoplasmic reticulum glucosyltransferase (UGGT): Synthetic chemistry’s initiative in glycobiology. Semin. Cell Dev. Biol. 2015, 41, 90–98. [Google Scholar] [CrossRef] [PubMed]
- Jaeken, J.; Matthijs, G. Congenital Disorders of Glycosylation: A Rapidly Expanding Disease Family. Annu. Rev. Genomics Hum. Genet. 2007, 8, 261–278. [Google Scholar] [CrossRef] [PubMed]
- Mori, K. The unfolded protein response: The dawn of a new field. Proc. Jpn. Acad. Ser. B 2015, 91, 469–480. [Google Scholar] [CrossRef]
- Totani, K.; Matsuo, I.; Ito, Y. Tight binding ligand approach to oligosaccharide-grafted protein. Bioorg. Med. Chem. Lett. 2004, 14, 2285–2289. [Google Scholar] [CrossRef]
- Totani, K.; Matsuo, I.; Ihara, Y.; Ito, Y. High-mannose-type glycan modifications of dihydrofolate reductase using glycan–methotrexate conjugates. Biorg. Med. Chem. 2006, 14, 5220–5229. [Google Scholar] [CrossRef]
- Totani, K.; Ihara, Y.; Matsuo, I.; Koshino, H.; Ito, Y. Synthetic Substrates for an Endoplasmic Reticulum Protein-Folding Sensor, UDP-Glucose: Glycoprotein Glucosyltransferase. Angew. Chem. Int. Ed. 2005, 44, 7950–7954. [Google Scholar] [CrossRef]
- Totani, K.; Ihara, Y.; Tsujimoto, T.; Matsuo, I.; Ito, Y. The Recognition Motif of the Glycoprotein-Folding Sensor Enzyme UDP-Glc:Glycoprotein Glucosyltransferase. Biochemistry 2009, 48, 2933–2940. [Google Scholar] [CrossRef]
- Sakono, M.; Seko, A.; Takeda, Y.; Hachisu, M.; Ito, Y. Biophysical properties of UDP-glucose: Glycoprotein glucosyltransferase, a folding sensor enzyme in the ER, delineated by synthetic probes. Biochem. Biophys. Res. Commun. 2012, 426, 504–510. [Google Scholar] [CrossRef]
- Izumi, M.; Makimura, Y.; Dedola, S.; Seko, A.; Kanamori, A.; Sakono, M.; Ito, Y.; Kajihara, Y. Chemical Synthesis of Intentionally Misfolded Homogeneous Glycoprotein: A Unique Approach for the Study of Glycoprotein Quality Control. J. Am. Chem. Soc. 2012, 134, 7238–7241. [Google Scholar] [CrossRef]
- Izumi, M.; Kuruma, R.; Okamoto, R.; Seko, A.; Ito, Y.; Kajihara, Y. Substrate Recognition of Glycoprotein Folding Sensor UGGT Analyzed by Site-Specifically 15N-Labeled Glycopeptide and Small Glycopeptide Library Prepared by Parallel Native Chemical Ligation. J. Am. Chem. Soc. 2017, 139, 11421–11426. [Google Scholar] [CrossRef]
- Kiuchi, T.; Izumi, M.; Mukogawa, Y.; Shimada, A.; Okamoto, R.; Seko, A.; Sakono, M.; Takeda, Y.; Ito, Y.; Kajihara, Y. Monitoring of Glycoprotein Quality Control System with a Series of Chemically Synthesized Homogeneous Native and Misfolded Glycoproteins. J. Am. Chem. Soc. 2018, 140, 17499–17507. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.; Harada, Y.; Hosomi, A.; Masahara-Negishi, Y.; Seino, J.; Fujihira, H.; Funakoshi, Y.; Suzuki, T.; Dohmae, N.; Suzuki, T. Endo-β-N-acetylglucosaminidase forms N-GlcNAc protein aggregates during ER-associated degradation in Ngly1-defective cells. Proc. Natl. Acad. Sci. USA 2015, 112, 1398–1403. [Google Scholar] [CrossRef]
- Maynard, J.C.; Fujihira, H.; Dolgonos, G.E.; Suzuki, T.; Burlingame, A.L. Cytosolic N-GlcNAc proteins are formed by the action of endo-β-N-acetylglucosaminidase. Biochem. Biophys. Res. Commun. 2020, 530, 719–724. [Google Scholar] [CrossRef]
- Bi, Y.; Might, M.; Vankayalapati, H.; Kuberan, B. Repurposing of Proton Pump Inhibitors as first identified small molecule inhibitors of endo-β-N-acetylglucosaminidase (ENGase) for the treatment of NGLY1 deficiency, a rare genetic disease. Bioorg. Med. Chem. Lett. 2017, 27, 2962–2966. [Google Scholar] [CrossRef]
- Fujihira, H.; Masahara-Negishi, Y.; Tamura, M.; Huang, C.; Harada, Y.; Wakana, S.; Takakura, D.; Kawasaki, N.; Taniguchi, N.; Kondoh, G.; et al. Lethality of mice bearing a knockout of the Ngly1-gene is partially rescued by the additional deletion of the Engase gene. PLoS Genet. 2017, 13, e1006696. [Google Scholar] [CrossRef]
- Wang, P.; Dong, S.; Shieh, J.-H.; Peguero, E.; Hendrickson, R.; Moore, M.A.S.; Danishefsky, S.J. Erythropoietin Derived by Chemical Synthesis. Science 2013, 342, 1357–1360. [Google Scholar] [CrossRef] [PubMed]
- Okamoto, R.; Mandal, K.; Ling, M.; Luster, A.D.; Kajihara, Y.; Kent, S.B.H. Total Chemical Synthesis and Biological Activities of Glycosylated and Non-Glycosylated Forms of the Chemokines CCL1 and Ser-CCL1. Angew. Chem. Int. Ed. 2014, 53, 5188–5193. [Google Scholar]
- Reif, A.; Siebenhaar, S.; Tröster, A.; Schmälzlein, M.; Lechner, C.; Velisetty, P.; Gottwald, K.; Pöhner, C.; Boos, I.; Schubert, V.; et al. Semisynthesis of Biologically Active Glycoforms of the Human Cytokine Interleukin 6. Angew. Chem. Int. Ed. 2014, 53, 12125–12131. [Google Scholar] [CrossRef]
- Murakami, M.; Kiuchi, T.; Nishihara, M.; Tezuka, K.; Okamoto, R.; Izumi, M.; Kajihara, Y. Chemical synthesis of erythropoietin glycoforms for insights into the relationship between glycosylation pattern and bioactivity. Sci. Adv. 2016, 2, e1500678. [Google Scholar] [CrossRef] [PubMed]
- Streichert, K.; Seitz, C.; Hoffmann, E.; Boos, I.; Jelkmann, W.; Brunner, T.; Unverzagt, C.; Rubini, M. Synthesis of Erythropoietins Site-Specifically Conjugated with Complex-Type N-Glycans. ChemBioChem 2019, 20, 1914–1918. [Google Scholar] [CrossRef]
- Sakamoto, I.; Tezuka, K.; Fukae, K.; Ishii, K.; Taduru, K.; Maeda, M.; Ouchi, M.; Yoshida, K.; Nambu, Y.; Igarashi, J.; et al. Chemical Synthesis of Homogeneous Human Glycosyl-interferon-β That Exhibits Potent Antitumor Activity In Vivo. J. Am. Chem. Soc. 2012, 134, 5428–5431. [Google Scholar] [CrossRef]
- Murakami, M.; Okamoto, R.; Izumi, M.; Kajihara, Y. Chemical Synthesis of an Erythropoietin Glycoform Containing a Complex-type Disialyloligosaccharide. Angew. Chem. Int. Ed. 2012, 51, 3567–3572. [Google Scholar] [CrossRef]
- Yang, Q.; An, Y.; Zhu, S.; Zhang, R.; Loke, C.M.; Cipollo, J.F.; Wang, L.-X. Glycan Remodeling of Human Erythropoietin (EPO) Through Combined Mammalian Cell Engineering and Chemoenzymatic Transglycosylation. ACS Chem. Biol. 2017, 12, 1665–1673. [Google Scholar] [CrossRef]
- Maki, Y.; Okamoto, R.; Izumi, M.; Kajihara, Y. Chemical Synthesis of an Erythropoietin Glycoform Having a Triantennary N-Glycan: Significant Change of Biological Activity of Glycoprotein by Addition of a Small Molecular Weight Trisaccharide. J. Am. Chem. Soc. 2020, 142, 20671–20679. [Google Scholar] [CrossRef]
- Macmillan, D.; Bill, R.M.; Sage, K.A.; Fern, D.; Flitsch, S.L. Selective in vitro glycosylation of recombinant proteins: Semi-synthesis of novel homogeneous glycoforms of human erythropoietin. Chem. Biol. 2001, 8, 133–145. [Google Scholar] [CrossRef]
- Kochendoerfer, G.G.; Chen, S.-Y.; Mao, F.; Cressman, S.; Traviglia, S.; Shao, H.; Hunter, C.L.; Low, D.W.; Cagle, E.N.; Carnevali, M.; et al. Design and Chemical Synthesis of a Homogeneous Polymer-Modified Erythropoiesis Protein. Science 2003, 299, 884–887. [Google Scholar] [CrossRef]
- Hirano, K.; Macmillan, D.; Tezuka, K.; Tsuji, T.; Kajihara, Y. Design and Synthesis of a Homogeneous Erythropoietin Analogue with Two Human Complex-Type Sialyloligosaccharides: Combined Use of Chemical and Bacterial Protein Expression Methods. Angew. Chem. Int. Ed. 2009, 48, 9557–9560. [Google Scholar] [CrossRef] [PubMed]
- Lee, D.J.; Cameron, A.J.; Wright, T.H.; Harris, P.W.R.; Brimble, M.A. A synthetic approach to ‘click’ neoglycoprotein analogues of EPO employing one-pot native chemical ligation and CuAAC chemistry. Chem. Sci. 2019, 10, 815–828. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, K.; Siwu, E.R.O.; Minami, K.; Hasegawa, K.; Nozaki, S.; Kanayama, Y.; Koyama, K.; Chen, W.C.; Paulson, J.C.; Watanabe, Y.; et al. Noninvasive Imaging of Dendrimer-Type N-Glycan Clusters: In Vivo Dynamics Dependence on Oligosaccharide Structure. Angew. Chem. Int. Ed. 2010, 49, 8195–8200. [Google Scholar] [CrossRef]
- Vong, K.; Yamamoto, T.; Tanaka, K. Artificial Glycoproteins as a Scaffold for Targeted Drug Therapy. Small 2020, 16, 1906890. [Google Scholar] [CrossRef] [PubMed]
- Valverde, P.; Ardá, A.; Reichardt, N.-C.; Jiménez-Barbero, J.; Gimeno, A. Glycans in drug discovery. MedChemComm 2019, 10, 1678–1691. [Google Scholar] [CrossRef]
- Eon-Duval, A.; Broly, H.; Gleixner, R. Quality attributes of recombinant therapeutic proteins: An assessment of impact on safety and efficacy as part of a quality by design development approach. Biotechnol. Progr. 2012, 28, 608–622. [Google Scholar] [CrossRef]
- Beck, A.; Wagner-Rousset, E.; Ayoub, D.; Van Dorsselaer, A.; Sanglier-Cianférani, S. Characterization of Therapeutic Antibodies and Related Products. Anal. Chem. 2013, 85, 715–736. [Google Scholar] [CrossRef]
- Shinkawa, T.; Nakamura, K.; Yamane, N.; Shoji-Hosaka, E.; Kanda, Y.; Sakurada, M.; Uchida, K.; Anazawa, H.; Satoh, M.; Yamasaki, M.; et al. The Absence of Fucose but Not the Presence of Galactose or Bisecting N-Acetylglucosamine of Human IgG1 Complex-type Oligosaccharides Shows the Critical Role of Enhancing Antibody-dependent Cellular Cytotoxicity. J. Biol. Chem. 2003, 278, 3466–3473. [Google Scholar] [CrossRef]
- Shields, R.L.; Lai, J.; Keck, R.; O’Connell, L.Y.; Hong, K.; Meng, Y.G.; Weikert, S.H.A.; Presta, L.G. Lack of Fucose on Human IgG1 N-Linked Oligosaccharide Improves Binding to Human FcγRIII and Antibody-dependent Cellular Toxicity. J. Biol. Chem. 2002, 277, 26733–26740. [Google Scholar] [CrossRef] [PubMed]
- Satoh, M.; Shitara, K.; Hanai, N. The Current Stream and Prospect of Glycoscience Application Therapeutic Antibodies. Trends Glycosci. Glycotechnol. 2006, 18, 129–136. [Google Scholar] [CrossRef]
- Niwa, R.; Shoji-Hosaka, E.; Sakurada, M.; Shinkawa, T.; Uchida, K.; Nakamura, K.; Matsushima, K.; Ueda, R.; Hanai, N.; Shitara, K. Defucosylated Chimeric Anti-CC Chemokine Receptor 4 IgG1 with Enhanced Antibody-Dependent Cellular Cytotoxicity Shows Potent Therapeutic Activity to T-Cell Leukemia and Lymphoma. Cancer Res. 2004, 64, 2127–2133. [Google Scholar] [CrossRef] [PubMed]
- Davies, J.; Jiang, L.; Pan, L.-Z.; LaBarre, M.J.; Anderson, D.; Reff, M. Expression of GnTIII in a recombinant anti-CD20 CHO production cell line: Expression of antibodies with altered glycoforms leads to an increase in ADCC through higher affinity for FCγRIII. Biotechnol. Bioeng. 2001, 74, 288–294. [Google Scholar] [CrossRef]
- Hodoniczky, J.; Zheng, Y.Z.; James, D.C. Control of Recombinant Monoclonal Antibody Effector Functions by Fc N-Glycan Remodeling In Vitro. Biotechnol. Progr. 2005, 21, 1644–1652. [Google Scholar] [CrossRef] [PubMed]
- Ferrara, C.; Brünker, P.; Suter, T.; Moser, S.; Püntener, U.; Umaña, P. Modulation of therapeutic antibody effector functions by glycosylation engineering: Influence of Golgi enzyme localization domain and co-expression of heterologous β1, 4-N-acetylglucosaminyltransferase III and Golgi α-mannosidase II. Biotechnol. Bioeng. 2006, 93, 851–861. [Google Scholar] [CrossRef] [PubMed]
- Huang, W.; Giddens, J.; Fan, S.-Q.; Toonstra, C.; Wang, L.-X. Chemoenzymatic Glycoengineering of Intact IgG Antibodies for Gain of Functions. J. Am. Chem. Soc. 2012, 134, 12308–12318. [Google Scholar] [CrossRef]
- Qasba, P.K. Glycans of Antibodies as a Specific Site for Drug Conjugation Using Glycosyltransferases. Bioconjugate Chem. 2015, 26, 2170–2175. [Google Scholar] [CrossRef]
- Manabe, S. Attempts to synthesize homogeneous glycan-conjugated antibody-drug conjugates. Transl. Regul. Sci. 2020, 2, 84–89. [Google Scholar]
- Wang, L.-X.; Tong, X.; Li, C.; Giddens, J.P.; Li, T. Glycoengineering of Antibodies for Modulating Functions. Annu. Rev. Biochem 2019, 88, 433–459. [Google Scholar] [CrossRef]
- Manabe, S.; Yamaguchi, Y.; Matsumoto, K.; Fuchigami, H.; Kawase, T.; Hirose, K.; Mitani, A.; Sumiyoshi, W.; Kinoshita, T.; Abe, J.; et al. Characterization of Antibody Products Obtained through Enzymatic and Nonenzymatic Glycosylation Reactions with a Glycan Oxazoline and Preparation of a Homogeneous Antibody–Drug Conjugate via Fc N-Glycan. Bioconjugate Chem. 2019, 30, 1343–1355. [Google Scholar] [CrossRef]
- Parsons, T.B.; Struwe, W.B.; Gault, J.; Yamamoto, K.; Taylor, T.A.; Raj, R.; Wals, K.; Mohammed, S.; Robinson, C.V.; Benesch, J.L.P.; et al. Optimal Synthetic Glycosylation of a Therapeutic Antibody. Angew. Chem. Int. Ed. 2016, 55, 2361–2367. [Google Scholar] [CrossRef]
- Boeggeman, E.; Ramakrishnan, B.; Pasek, M.; Manzoni, M.; Puri, A.; Loomis, K.H.; Waybright, T.J.; Qasba, P.K. Site Specific Conjugation of Fluoroprobes to the Remodeled Fc N-Glycans of Monoclonal Antibodies Using Mutant Glycosyltransferases: Application for Cell Surface Antigen Detection. Bioconjugate Chem. 2009, 20, 1228–1236. [Google Scholar] [CrossRef]
- Tang, F.; Yang, Y.; Tang, Y.; Tang, S.; Yang, L.; Sun, B.; Jiang, B.; Dong, J.; Liu, H.; Huang, M.; et al. One-pot N-glycosylation remodeling of IgG with non-natural sialylglycopeptides enables glycosite-specific and dual-payload antibody–drug conjugates. Org. Biomol. Chem. 2016, 14, 9501–9518. [Google Scholar] [CrossRef]
- Zeglis, B.M.; Davis, C.B.; Aggeler, R.; Kang, H.C.; Chen, A.; Agnew, B.J.; Lewis, J.S. Enzyme-Mediated Methodology for the Site-Specific Radiolabeling of Antibodies Based on Catalyst-Free Click Chemistry. Bioconjugate Chem. 2013, 24, 1057–1067. [Google Scholar] [CrossRef]
- van Geel, R.; Wijdeven, M.A.; Heesbeen, R.; Verkade, J.M.M.; Wasiel, A.A.; van Berkel, S.S.; van Delft, F.L. Chemoenzymatic Conjugation of Toxic Payloads to the Globally Conserved N-Glycan of Native mAbs Provides Homogeneous and Highly Efficacious Antibody–Drug Conjugates. Bioconjugate Chem. 2015, 26, 2233–2242. [Google Scholar] [CrossRef]
- Lo, H.-J.; Krasnova, L.; Dey, S.; Cheng, T.; Liu, H.; Tsai, T.-I.; Wu, K.B.; Wu, C.-Y.; Wong, C.-H. Synthesis of Sialidase-Resistant Oligosaccharide and Antibody Glycoform Containing α2,6-Linked 3Fax-Neu5Ac. J. Am. Chem. Soc. 2019, 141, 6484–6488. [Google Scholar] [CrossRef]
- Hossain, M.A.; Okamoto, R.; Karas, J.A.; Praveen, P.; Liu, M.; Forbes, B.E.; Wade, J.D.; Kajihara, Y. Total Chemical Synthesis of a Nonfibrillating Human Glycoinsulin. J. Am. Chem. Soc. 2020, 142, 1164–1169. [Google Scholar] [CrossRef]
- Wang, L.-X.; Ni, J.; Singh, S.; Li, H. Binding of High-Mannose-Type Oligosaccharides and Synthetic Oligomannose Clusters to Human Antibody 2G12: Implications for HIV-1 Vaccine Design. Chem. Biol. 2004, 11, 127–134. [Google Scholar] [PubMed]
- Amin, M.N.; McLellan, J.S.; Huang, W.; Orwenyo, J.; Burton, D.R.; Koff, W.C.; Kwong, P.D.; Wang, L.-X. Synthetic glycopeptides reveal the glycan specificity of HIV-neutralizing antibodies. Nat. Chem. Biol. 2013, 9, 521–526. [Google Scholar] [CrossRef] [PubMed]
- Aussedat, B.; Vohra, Y.; Park, P.K.; Fernández-Tejada, A.; Alam, S.M.; Dennison, S.M.; Jaeger, F.H.; Anasti, K.; Stewart, S.; Blinn, J.H.; et al. Chemical Synthesis of Highly Congested gp120 V1V2 N-Glycopeptide Antigens for Potential HIV-1-Directed Vaccines. J. Am. Chem. Soc. 2013, 135, 13113–13120. [Google Scholar] [CrossRef] [PubMed]
- Cai, H.; Orwenyo, J.; Giddens, J.P.; Yang, Q.; Zhang, R.; LaBranche, C.C.; Montefiori, D.C.; Wang, L.-X. Synthetic Three-Component HIV-1 V3 Glycopeptide Immunogens Induce Glycan-Dependent Antibody Responses. Cell Chem. Biol. 2017, 24, 1513–1522.e4. [Google Scholar] [CrossRef]
- Cai, H.; Zhang, R.-S.; Orwenyo, J.; Giddens, J.; Yang, Q.; LaBranche, C.C.; Montefiori, D.C.; Wang, L.-X. Synthetic HIV V3 Glycopeptide Immunogen Carrying a N334 N-Glycan Induces Glycan-Dependent Antibodies with Promiscuous Site Recognition. J. Med. Chem. 2018, 61, 10116–10125. [Google Scholar] [CrossRef]
- Wang, C.-C.; Chen, J.-R.; Tseng, Y.-C.; Hsu, C.-H.; Hung, Y.-F.; Chen, S.-W.; Chen, C.-M.; Khoo, K.-H.; Cheng, T.-J.; Cheng, Y.-S.E.; et al. Glycans on influenza hemagglutinin affect receptor binding and immune response. Proc. Natl. Acad. Sci. USA 2009, 106, 18137–18142. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.-R.; Yu, Y.-H.; Tseng, Y.-C.; Chiang, W.-L.; Chiang, M.-F.; Ko, Y.-A.; Chiu, Y.-K.; Ma, H.-H.; Wu, C.-Y.; Jan, J.-T.; et al. Vaccination of monoglycosylated hemagglutinin induces cross-strain protection against influenza virus infections. Proc. Natl. Acad. Sci. USA 2014, 111, 2476–2481. [Google Scholar] [CrossRef]
- Watanabe, Y.; Allen, J.D.; Wrapp, D.; McLellan, J.S.; Crispin, M. Site-specific glycan analysis of the SARS-CoV-2 spike. Science 2020, 369, 330–333. [Google Scholar] [PubMed]
- Brun, J.; Vasiljevic, S.; Gangadharan, B.; Hensen, M.; Chandran, A.V.; Hill, M.L.; Kiappes, J.L.; Dwek, R.A.; Alonzi, D.S.; Struwe, W.B.; et al. Analysis of SARS-CoV-2 spike glycosylation reveals shedding of a vaccine candidate. bioRxiv 2020. [Google Scholar] [CrossRef]
- Yamazaki, N.; Kojima, S.; Bovin, N.V.; André, S.; Gabius, S.; Gabius, H.J. Endogenous lectins as targets for drug delivery. Adv. Drug Del. Rev. 2000, 43, 225–244. [Google Scholar] [CrossRef]
- Zhang, H.; Ma, Y.; Sun, X.-L. Recent developments in carbohydrate-decorated targeted drug/gene delivery. Med. Res. Revs. 2010, 30, 270–289. [Google Scholar] [CrossRef]
- Cecioni, S.; Imberty, A.; Vidal, S. Glycomimetics versus Multivalent Glycoconjugates for the Design of High Affinity Lectin Ligands. Chem. Rev. 2015, 115, 525–561. [Google Scholar] [CrossRef]
- Ogura, A.; Tahara, T.; Nozaki, S.; Morimoto, K.; Kizuka, Y.; Kitazume, S.; Hara, M.; Kojima, S.; Onoe, H.; Kurbangalieva, A.; et al. Visualizing Trimming Dependence of Biodistribution and Kinetics with Homo- and Heterogeneous N-Glycoclusters on Fluorescent Albumin. Sci. Rep. 2016, 6, 21797. [Google Scholar] [CrossRef] [PubMed]
- Tsubokura, K.; Vong, K.K.H.; Pradipta, A.R.; Ogura, A.; Urano, S.; Tahara, T.; Nozaki, S.; Onoe, H.; Nakao, Y.; Sibgatullina, R.; et al. In Vivo Gold Complex Catalysis within Live Mice. Angew. Chem. Int. Ed. 2017, 56, 3579–3584. [Google Scholar] [CrossRef] [PubMed]
- Eda, S.; Nasibullin, I.; Vong, K.; Kudo, N.; Yoshida, M.; Kurbangalieva, A.; Tanaka, K. Biocompatibility and therapeutic potential of glycosylated albumin artificial metalloenzymes. Nat. Catal. 2019, 2, 780–792. [Google Scholar] [CrossRef]
- Chen, W.C.; Completo, G.C.; Sigal, D.S.; Crocker, P.R.; Saven, A.; Paulson, J.C. In Vivo targeting of B-cell lymphoma with glycan ligands of CD22. Blood 2010, 115, 4778–4786. [Google Scholar] [CrossRef]
- Macauley, M.S.; Pfrengle, F.; Rademacher, C.; Nycholat, C.M.; Gale, A.J.; von Drygalski, A.; Paulson, J.C. Antigenic liposomes displaying CD22 ligands induce antigen-specific B cell apoptosis. J. Clin. Investig. 2013, 123, 3074–3083. [Google Scholar] [CrossRef]
- Kawasaki, N.; Rillahan, C.D.; Cheng, T.-Y.; Van Rhijn, I.; Macauley, M.S.; Moody, D.B.; Paulson, J.C. Targeted Delivery of Mycobacterial Antigens to Human Dendritic Cells via Siglec-7 Induces Robust T Cell Activation. J. Immunol. 2014, 193, 1560–1566. [Google Scholar] [CrossRef]
- Angata, T.; Nycholat, C.M.; Macauley, M.S. Therapeutic Targeting of Siglecs using Antibody- and Glycan-Based Approaches. Trends Pharmacol. Sci. 2015, 36, 645–660. [Google Scholar] [CrossRef]
- Peng, W.; Paulson, J.C. CD22 Ligands on a Natural N-Glycan Scaffold Efficiently Deliver Toxins to B-Lymphoma Cells. J. Am. Chem. Soc. 2017, 139, 12450–12458. [Google Scholar] [CrossRef]
- Nycholat, C.M.; Duan, S.; Knuplez, E.; Worth, C.; Elich, M.; Yao, A.; O’Sullivan, J.; McBride, R.; Wei, Y.; Fernandes, S.M.; et al. A Sulfonamide Sialoside Analogue for Targeting Siglec-8 and -F on Immune Cells. J. Am. Chem. Soc. 2019, 141, 14032–14037. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Lang, S.; Tian, Y.; Zhang, J.; Yan, X.; Fang, Z.; Weng, J.; Lu, N.; Wu, X.; Li, T.; et al. Glycoengineering of Natural Killer Cells with CD22 Ligands for Enhanced Anticancer Immunotherapy. ACS Cent. Sci. 2020, 6, 382–389. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Huang, W.; Orwenyo, J.; Banerjee, A.; Vasta, G.R.; Wang, L.-X. Design and synthesis of glycoprotein-based multivalent glyco-ligands for influenza hemagglutinin and human galectin-3. Biorg. Med. Chem. 2013, 21, 2037–2044. [Google Scholar] [CrossRef] [PubMed]
- Cagnoni, A.J.; Pérez Sáez, J.M.; Rabinovich, G.A.; Mariño, K.V. Turning-Off Signaling by Siglecs, Selectins, and Galectins: Chemical Inhibition of Glycan-Dependent Interactions in Cancer. Front. Oncol. 2016, 6, 109. [Google Scholar] [CrossRef]
- Laaf, D.; Bojarová, P.; Elling, L.; Křen, V. Galectin–Carbohydrate Interactions in Biomedicine and Biotechnology. Trends Biotechnol. 2019, 37, 402–415. [Google Scholar] [CrossRef]
- Griffin, M.E.; Hsieh-Wilson, L.C. Glycan Engineering for Cell and Developmental Biology. Cell Chem. Biol. 2016, 23, 108–121. [Google Scholar] [CrossRef]
- Prescher, J.A.; Dube, D.H.; Bertozzi, C.R. Chemical remodelling of cell surfaces in living animals. Nature 2004, 430, 873–877. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Mooney, D.J. Metabolic glycan labelling for cancer-targeted therapy. Nat. Chem. 2020, 12, 1102–1114. [Google Scholar] [CrossRef] [PubMed]
- Zeng, Y.; Ramya, T.N.C.; Dirksen, A.; Dawson, P.E.; Paulson, J.C. High-efficiency labeling of sialylated glycoproteins on living cells. Nat. Methods 2009, 6, 207–209. [Google Scholar] [CrossRef]
- Hui, J.; Bao, L.; Li, S.; Zhang, Y.; Feng, Y.; Ding, L.; Ju, H. Localized Chemical Remodeling for Live Cell Imaging of Protein-Specific Glycoform. Angew. Chem. Int. Ed. 2017, 56, 8139–8143. [Google Scholar] [CrossRef]
- Boyce, M.; Carrico, I.S.; Ganguli, A.S.; Yu, S.-H.; Hangauer, M.J.; Hubbard, S.C.; Kohler, J.J.; Bertozzi, C.R. Metabolic cross-talk allows labeling of O-linked β-N-acetylglucosamine-modified proteins via the N-acetylgalactosamine salvage pathway. Proc. Natl. Acad. Sci. USA 2011, 108, 3141–3146. [Google Scholar] [CrossRef]
- Zheng, T.; Jiang, H.; Gros, M.; Soriano del Amo, D.; Sundaram, S.; Lauvau, G.; Marlow, F.; Liu, Y.; Stanley, P.; Wu, P. Tracking N-Acetyllactosamine on Cell-Surface Glycans In Vivo. Angew. Chem. Int. Ed. 2011, 50, 4113–4118. [Google Scholar] [CrossRef]
- Mbua, N.E.; Li, X.; Flanagan-Steet, H.R.; Meng, L.; Aoki, K.; Moremen, K.W.; Wolfert, M.A.; Steet, R.; Boons, G.-J. Selective Exo-Enzymatic Labeling of N-Glycans on the Surface of Living Cells by Recombinant ST6Gal I. Angew. Chem. Int. Ed. 2013, 52, 13012–13015. [Google Scholar] [CrossRef]
- Briard, J.G.; Jiang, H.; Moremen, K.W.; Macauley, M.S.; Wu, P. Cell-based glycan arrays for probing glycan–glycan binding protein interactions. Nat. Commun. 2018, 9, 880. [Google Scholar] [CrossRef]
- Tang, F.; Zhou, M.; Qin, K.; Shi, W.; Yashinov, A.; Yang, Y.; Yang, L.; Guan, D.; Zhao, L.; Tang, Y.; et al. Selective N-glycan editing on living cell surfaces to probe glycoconjugate function. Nat. Chem. Biol. 2020, 16, 766–775. [Google Scholar] [CrossRef] [PubMed]
- Frame, T.; Carroll, T.; Korchagina, E.; Bovin, N.; Henry, S. Synthetic glycolipid modification of red blood cell membranes. Transfusion 2007, 47, 876–882. [Google Scholar] [CrossRef]
- Huang, M.L.; Smith, R.A.A.; Trieger, G.W.; Godula, K. Glycocalyx Remodeling with Proteoglycan Mimetics Promotes Neural Specification in Embryonic Stem Cells. J. Am. Chem. Soc. 2014, 136, 10565–10568. [Google Scholar] [CrossRef]
- Hudak, J.E.; Canham, S.M.; Bertozzi, C.R. Glycocalyx engineering reveals a Siglec-based mechanism for NK cell immunoevasion. Nat. Chem. Biol. 2014, 10, 69–75. [Google Scholar] [CrossRef]
- Paszek, M.J.; DuFort, C.C.; Rossier, O.; Bainer, R.; Mouw, J.K.; Godula, K.; Hudak, J.E.; Lakins, J.N.; Wijekoon, A.C.; Cassereau, L.; et al. The cancer glycocalyx mechanically primes integrin-mediated growth and survival. Nature 2014, 511, 319–325. [Google Scholar] [CrossRef]
- Pulsipher, A.; Griffin, M.E.; Stone, S.E.; Brown, J.M.; Hsieh-Wilson, L.C. Directing Neuronal Signaling through Cell-Surface Glycan Engineering. J. Am. Chem. Soc. 2014, 136, 6794–6797. [Google Scholar] [CrossRef] [PubMed]
- Pulsipher, A.; Griffin, M.E.; Stone, S.E.; Hsieh-Wilson, L.C. Long-Lived Engineering of Glycans to Direct Stem Cell Fate. Angew. Chem. Int. Ed. 2015, 54, 1466–1470. [Google Scholar] [CrossRef] [PubMed]
- Latypova, L.; Sibgatullina, R.; Ogura, A.; Fujiki, K.; Khabibrakhmanova, A.; Tahara, T.; Nozaki, S.; Urano, S.; Tsubokura, K.; Onoe, H.; et al. Sequential Double “Clicks” toward Structurally Well-Defined Heterogeneous N-Glycoclusters: The Importance of Cluster Heterogeneity on Pattern Recognition In Vivo. Adv. Sci. 2017, 4, 1600394. [Google Scholar] [CrossRef] [PubMed]








Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shirakawa, A.; Manabe, Y.; Fukase, K. Recent Advances in the Chemical Biology of N-Glycans. Molecules 2021, 26, 1040. https://doi.org/10.3390/molecules26041040
Shirakawa A, Manabe Y, Fukase K. Recent Advances in the Chemical Biology of N-Glycans. Molecules. 2021; 26(4):1040. https://doi.org/10.3390/molecules26041040
Chicago/Turabian StyleShirakawa, Asuka, Yoshiyuki Manabe, and Koichi Fukase. 2021. "Recent Advances in the Chemical Biology of N-Glycans" Molecules 26, no. 4: 1040. https://doi.org/10.3390/molecules26041040
APA StyleShirakawa, A., Manabe, Y., & Fukase, K. (2021). Recent Advances in the Chemical Biology of N-Glycans. Molecules, 26(4), 1040. https://doi.org/10.3390/molecules26041040





