N-Phenyl Cinnamamide Derivatives Protect Hepatocytes against Oxidative Stress by Inducing Cellular Glutathione Synthesis via Nuclear Factor (Erythroid-Derived 2)-Like 2 Activation
Abstract
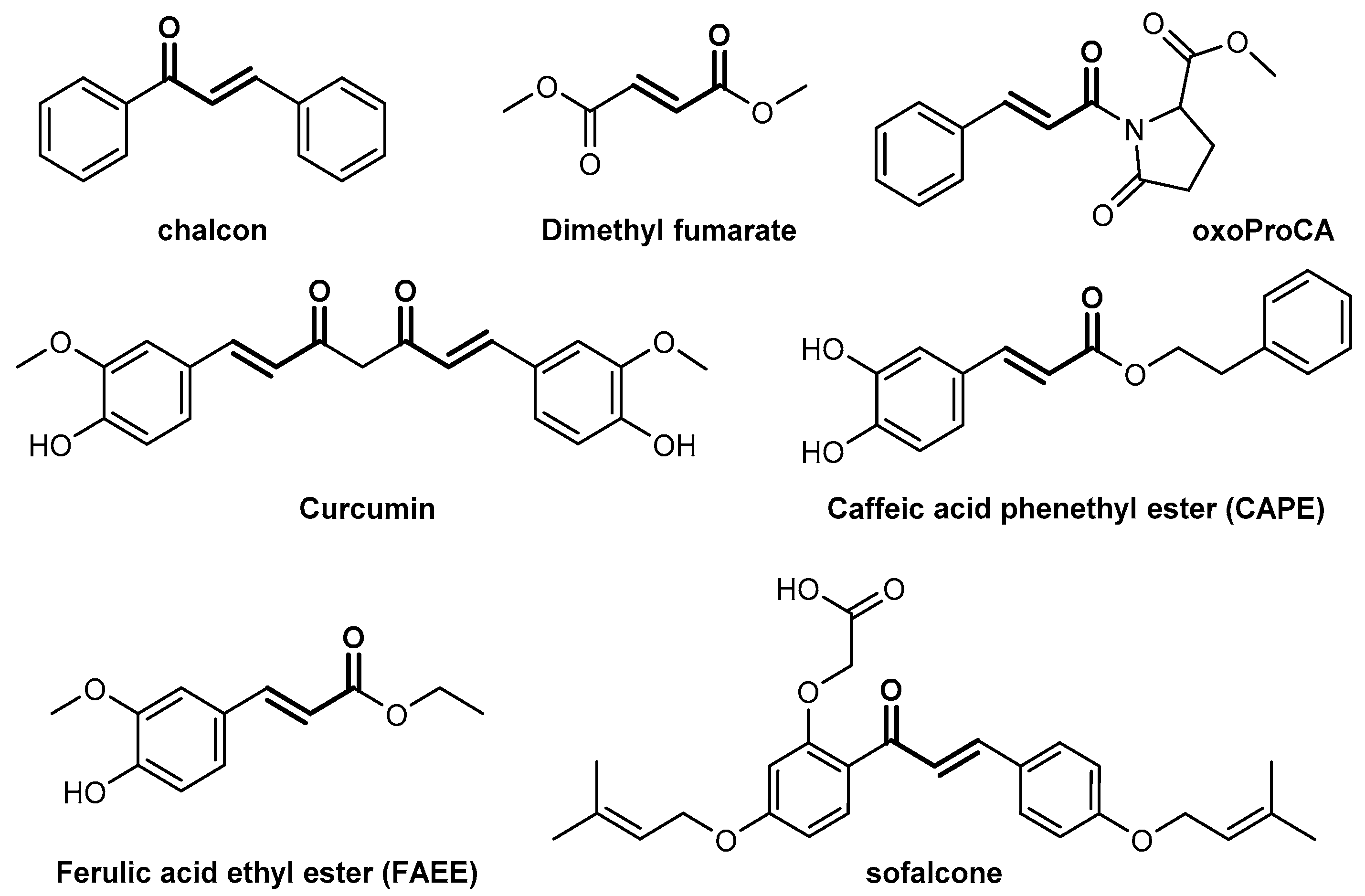
1. Introduction
2. Results and Discussion
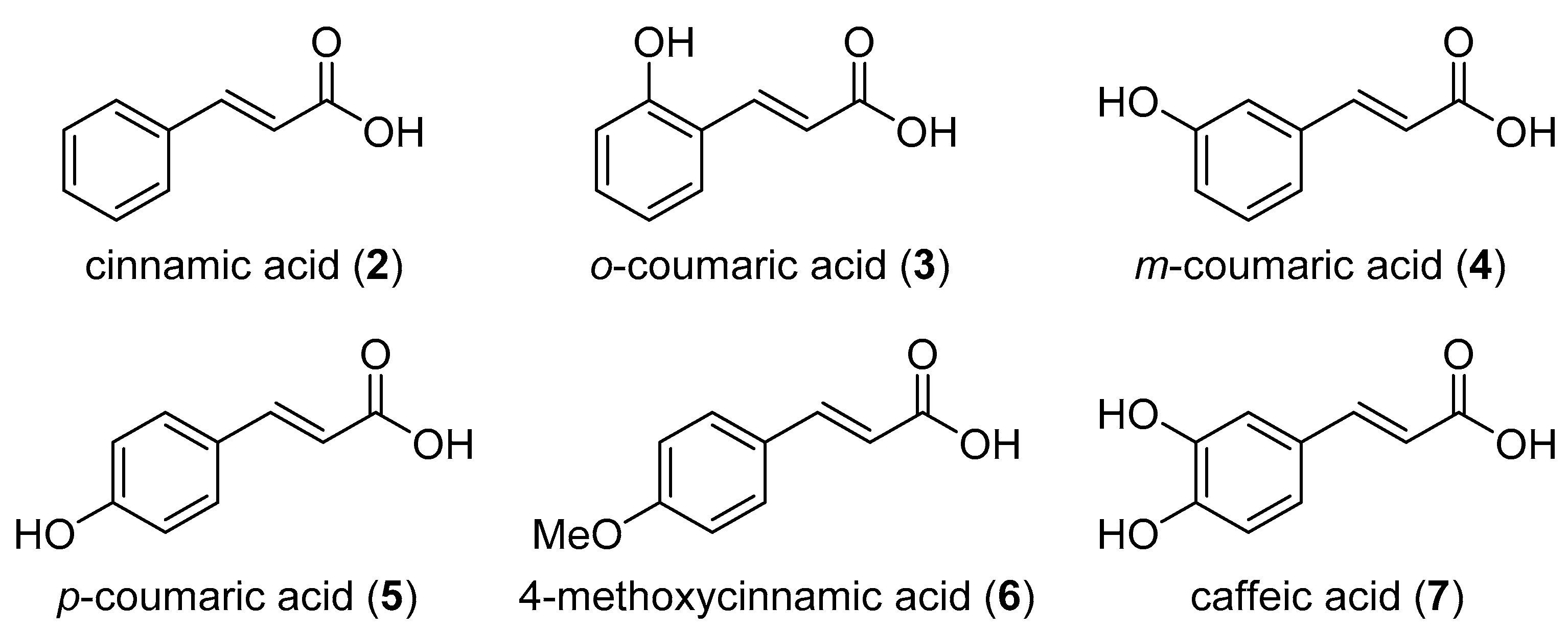
2.1. Nrf2/ARE Agonistic Activity of Aromatic Carboxylic Acids (C6–C3)
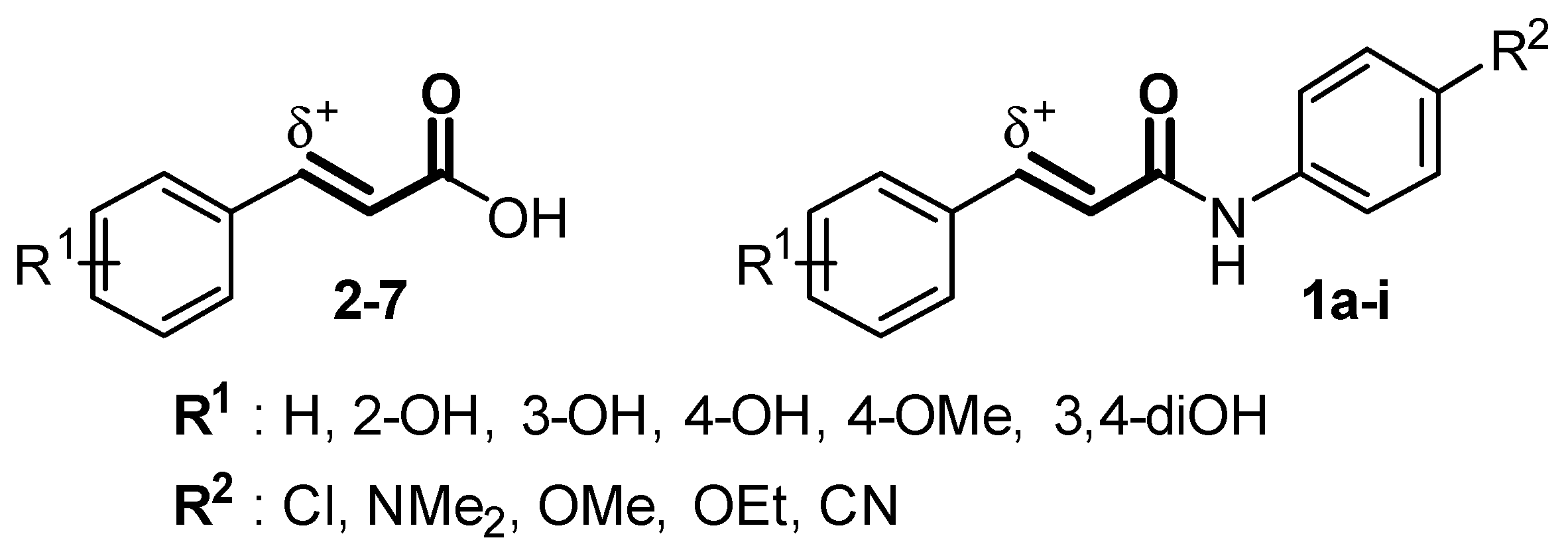
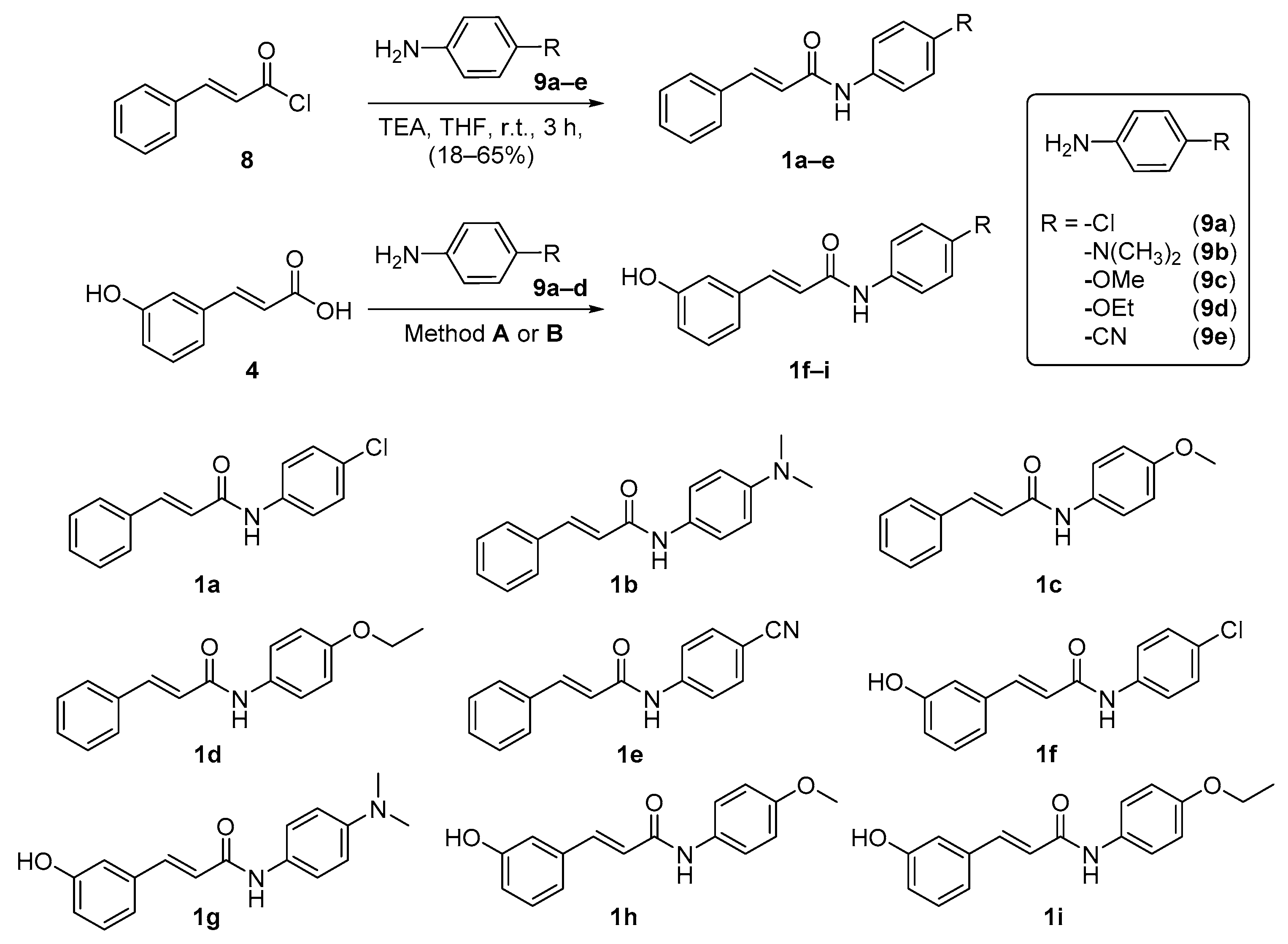
2.2. Synthesis of Cinnamamide Derivatives
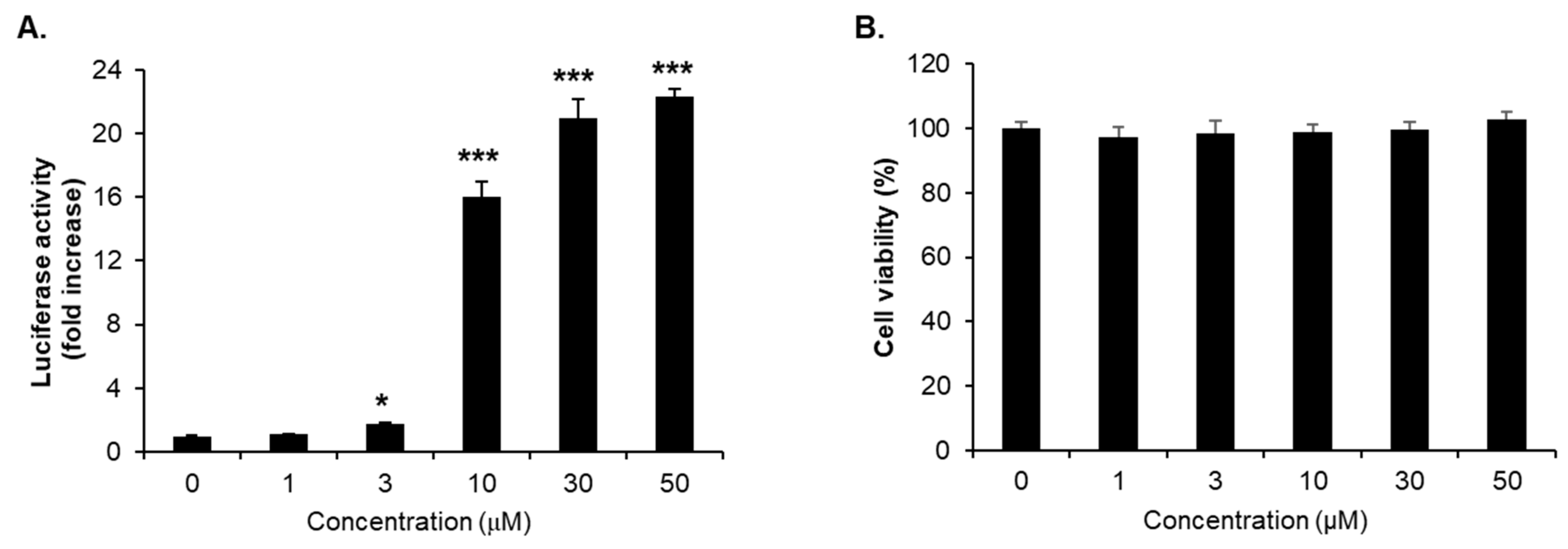
2.3. Nrf2/ARE Agonistic Activity of Cinnamamide Derivatives
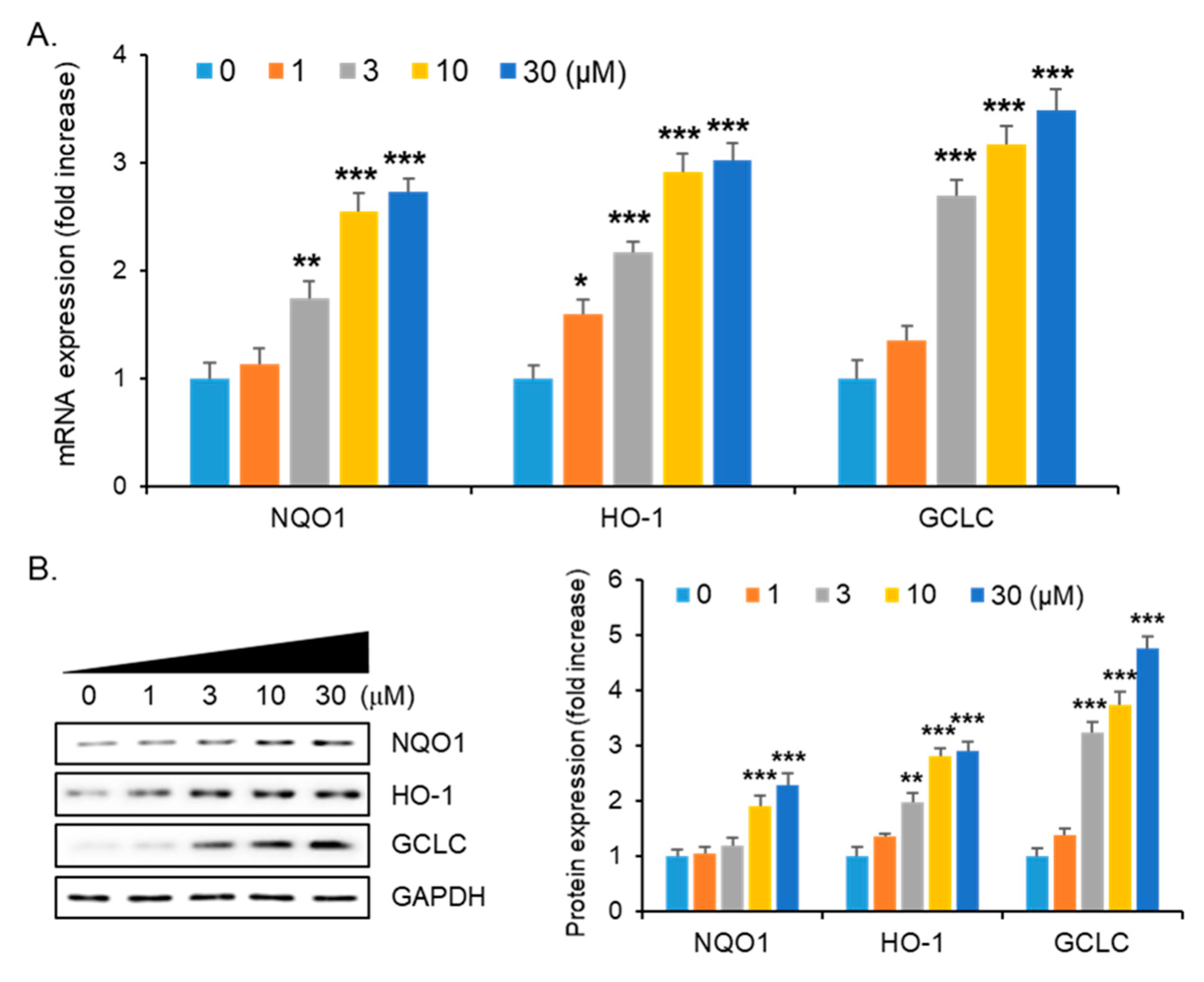
2.4. Enhancement of Nrf2/ARE-Dependent Gene Expression by Compound 1g Treatment
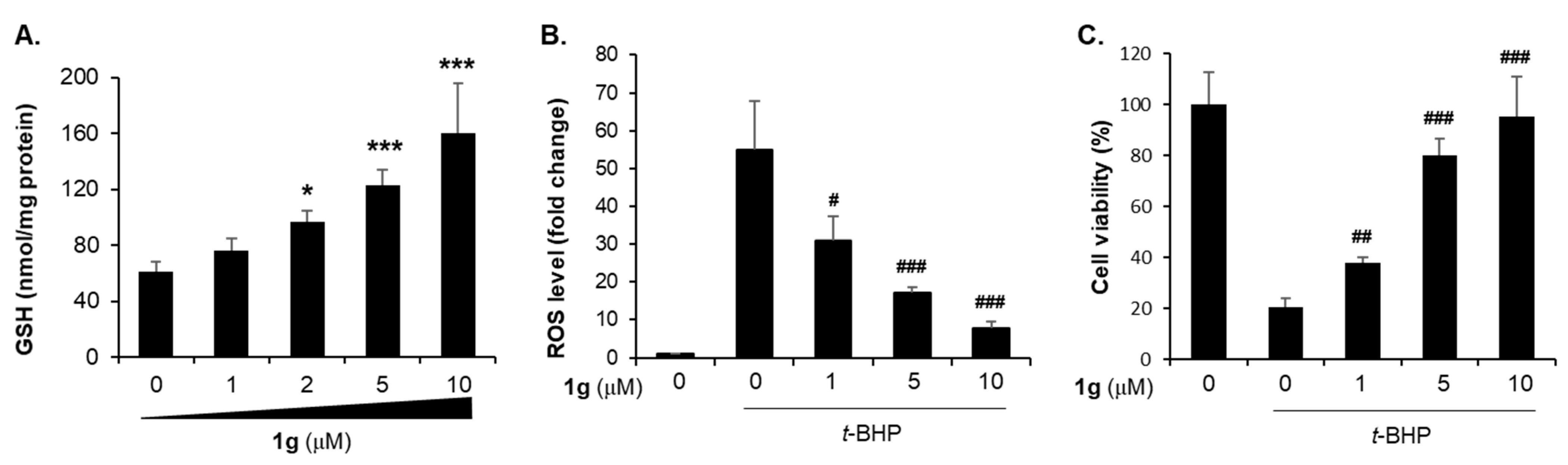
2.5. Protective Effect of Compound 1g Against Oxidative Cell Death via Increasing GSH Level in HepG2 Cell
3. Materials and Methods
3.1. Synthesis Procedure for N-Phenyl Cinnamamide Derivatives
3.1.1. General Procedure for 4-Substituted N-Phenyl Cinnamamide Derivatives (1a–e)
N-(4-chlorophenyl)cinnamamide (1a)
N-(4-(dimethylamino)phenyl)cinnamamide (1b)
N-(4-methoxyphenyl)cinnamamide (1c)
N-(4-ethoxyphenyl)cinnamamide (1d)
N-(4-cyanophenyl)cinnamamide (1e)
3.1.2. Procedure for (E)-N-(4-chlorophenyl)-3-(3-hydroxyphenyl)acrylamide (Method A; 1f)
3.1.3. General Procedure for 4-Substituted N-Phenyl m-Coumaric Amide Derivatives (Method B; 1g–i)
(E)-N-(4-(Dimethylamino)phenyl)-3-(3-hydroxyphenyl)acrylamide (1g)
(E)-3-(3-Hydroxyphenyl)-N-(4-methoxyphenyl)acrylamide (1h)
(E)-N-(4-Ethoxyphenyl)-3-(3-hydroxyphenyl)acrylamide (1i)
3.2. Evaluation of Biological Activity
3.2.1. Cell Culture
3.2.2. Nrf2/ARE Luciferase Assay
3.2.3. Cell Viability Assay
3.2.4. Real-Time Reverse Transcription-Polymerase Chain Reaction (RT-PCR)
3.2.5. Western Blot Analysis
3.2.6. GSH Level in HepG2 Cell Line
3.2.7. Evaluation of ROS Level
3.2.8. Statistical Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Sample Availability
References
- Abramovič, H. Chapter 93-Antioxidant Properties of Hydroxycinnamic Acid Derivatives: A Focus on Biochemistry, Physicochemical Parameters, Reactive Species, and Biomolecular Interactions. In Coffee in Health and Disease Prevention; Preedy, V.R., Ed.; Academic Press: San Diego, CA, USA, 2015; pp. 843–852. [Google Scholar] [CrossRef]
- Muriel, P. Role of free radicals in liver diseases. Hepatol. Int. 2009, 3, 526–536. [Google Scholar] [CrossRef]
- Surh, Y.J. Cancer chemoprevention with dietary phytochemicals. Nat. Rev. Cancer 2003, 3, 768–780. [Google Scholar] [CrossRef]
- Kim, J.; Cha, Y.N.; Surh, Y.J. A protective role of nuclear factor-erythroid 2-related factor-2 (Nrf2) in inflammatory disorders. Mutat. Res. 2010, 690, 12–23. [Google Scholar] [CrossRef]
- Itoh, K.; Chiba, T.; Takahashi, S.; Ishii, T.; Igarashi, K.; Katoh, Y.; Oyake, T.; Hayashi, N.; Satoh, K.; Hatayama, I.; et al. An Nrf2/small Maf heterodimer mediates the induction of phase II detoxifying enzyme genes through antioxidant response elements. Biochem. Biophys. Res. Commun. 1997, 236, 313–322. [Google Scholar] [CrossRef]
- Hwang, Y.P.; Choi, J.H.; Han, E.H.; Kim, H.K.; Kang, S.K.; Chung, Y.C.; Jeong, H.G. Protective mechanisms of Aralia continentalis extract against tert-butyl hydroperoxide-induced hepatotoxicity: In vivo and in vitro studies. Food Chem. Toxicol. 2008, 46, 3512–3521. [Google Scholar] [CrossRef]
- Subramaniam, S.R.; Ellis, E.M. Esculetin-induced protection of human hepatoma HepG2 cells against hydrogen peroxide is associated with the Nrf2-dependent induction of the NAD(P)H: Quinone oxidoreductase 1 gene. Toxicol. Appl. Pharmacol. 2011, 250, 130–136. [Google Scholar] [CrossRef] [PubMed]
- Senthil Kumar, K.J.; Liao, J.W.; Xiao, J.H.; Gokila Vani, M.; Wang, S.Y. Hepatoprotective effect of lucidone against alcohol-induced oxidative stress in human hepatic HepG2 cells through the up-regulation of HO-1/Nrf-2 antioxidant genes. Toxicol. In Vitro 2012, 26, 700–708. [Google Scholar] [CrossRef]
- Truong, V.L.; Bak, M.J.; Jun, M.; Kong, A.N.; Ho, C.T.; Jeong, W.S. Antioxidant defense and hepatoprotection by procyanidins from almond (Prunus amygdalus) skins. J. Agric. Food Chem. 2014, 62, 8668–8678. [Google Scholar] [CrossRef]
- Liu, L.; Hudgins, W.R.; Shack, S.; Yin, M.Q.; Samid, D. Cinnamic acid: A natural product with potential use in cancer intervention. Int. J. Cancer 1995, 62, 345–350. [Google Scholar] [CrossRef]
- Yilmaz, S.; Sova, M.; Ergun, S. Antimicrobial activity of trans-cinnamic acid and commonly used antibiotics against important fish pathogens and nonpathogenic isolates. J. Appl. Microbiol. 2018. [Google Scholar] [CrossRef] [PubMed]
- Taofiq, O.; Gonzalez-Paramas, A.M.; Barreiro, M.F.; Ferreira, I.C. Hydroxycinnamic Acids and Their Derivatives: Cosmeceutical Significance, Challenges and Future Perspectives, a Review. Molecules 2017, 22, 281. [Google Scholar] [CrossRef]
- Perluigi, M.; Joshi, G.; Sultana, R.; Calabrese, V.; De Marco, C.; Coccia, R.; Cini, C.; Butterfield, D.A. In vivo protective effects of ferulic acid ethyl ester against amyloid-beta peptide 1-42-induced oxidative stress. J. Neurosci. Res. 2006, 84, 418–426. [Google Scholar] [CrossRef]
- Scapagnini, G.; Butterfield, D.A.; Colombrita, C.; Sultana, R.; Pascale, A.; Calabrese, V. Ethyl ferulate, a lipophilic polyphenol, induces HO-1 and protects rat neurons against oxidative stress. Antioxid. Redox Signal. 2004, 6, 811–818. [Google Scholar] [CrossRef]
- Wondrak, G.T.; Cabello, C.M.; Villeneuve, N.F.; Zhang, S.; Ley, S.; Li, Y.; Sun, Z.; Zhang, D.D. Cinnamoyl-based Nrf2-activators targeting human skin cell photo-oxidative stress. Free Radic. Biol. Med. 2008, 45, 385–395. [Google Scholar] [CrossRef]
- de Freitas Silva, M.; Pruccoli, L.; Morroni, F.; Sita, G.; Seghetti, F.; Viegas, C.; Tarozzi, A. The Keap1/Nrf2-ARE Pathway as a Pharmacological Target for Chalcones. Molecules 2018, 23, 1803. [Google Scholar] [CrossRef]
- Robledinos-Antón, N.; Fernández-Ginés, R.; Manda, G.; Cuadrado, A. Activators and Inhibitors of NRF2: A Review of Their Potential for Clinical Development. Oxidative Med. Cell. Longev. 2019, 2019, 9372182. [Google Scholar] [CrossRef]
- Magesh, S.; Chen, Y.; Hu, L. Small molecule modulators of Keap1-Nrf2-ARE pathway as potential preventive and therapeutic agents. Med. Res. Rev. 2012, 32, 687–726. [Google Scholar] [CrossRef]
- Rucker, H.; Al-Rifai, N.; Rascle, A.; Gottfried, E.; Brodziak-Jarosz, L.; Gerhauser, C.; Dick, T.P.; Amslinger, S. Enhancing the anti-inflammatory activity of chalcones by tuning the Michael acceptor site. Org. Biomol. Chem. 2015, 13, 3040–3047. [Google Scholar] [CrossRef]
- Kim, M.; Baek, I.-h.; Kwak, J.-H. Synthesis of Phenyl m-coumarylamide Analogs and Their Anti-platelet Aggregation Activity. Yakhak Hoeji 2019, 63, 70–74. [Google Scholar] [CrossRef]
- Jung, Y.S. Metabolism of Sulfur-Containing Amino Acids in the Liver: A Link between Hepatic Injury and Recovery. Biol. Pharm. Bull. 2015, 38, 971–974. [Google Scholar] [CrossRef] [PubMed]
- Meister, A.; Anderson, M.E. Glutathione. Annu. Rev. Biochem. 1983, 52, 711–760. [Google Scholar] [CrossRef]
- Lee, Y.H.; Kim, S.H.; Lee, S.; Kim, K.M.; Jung, J.C.; Son, T.G.; Ki, S.H.; Seo, W.D.; Kwak, J.H.; Hong, J.T.; et al. Antioxidant Effect of Barley Sprout Extract via Enhancement of Nuclear Factor-Erythroid 2 Related Factor 2 Activity and Glutathione Synthesis. Nutrients 2017, 9, 1252. [Google Scholar] [CrossRef]
| Entry | Concentration (µM) | Relative Luciferase Activity (Fold) |
|---|---|---|
| Con | 0 | 1.00 ± 0.056 |
| Cinnamic acid (2) | 10 | 1.22 ± 0.036 |
| o-Coumaric acid (3) | 10 | 1.16 ± 0.061 |
| m-Coumaric acid (4) | 10 | 1.45 ± 0.033 |
| p-Coumaric acid (5) | 10 | 1.12 ± 0.031 |
| 4-Methoxycinnamic acid (6) | 10 | 1.22 ± 0.058 |
| Caffeic acid (7) | 10 | 1.01 ± 0.047 |
| m-Coumaric acid (4) | 1 | 1.21 ± 0.001 |
| 3 | 1.32 ± 0.150 | |
| 5 | 1.63 ± 0.207 | |
| 10 | 1.45 ± 0.033 |
| Entry | Concentration (µM) | Relative Luciferase Activity (Fold) |
|---|---|---|
| Con. | 0 | 1.00 ± 0.056 |
| t-BHQ | 5 | 5.74 ± 0.439 |
| 10 | 15.6 ± 0.334 | |
| 1a | 10 | 15.3 ± 0.903 |
| 1b | 10 | 10.3 ± 0.665 |
| 1c | 10 | 7.28 ± 0.437 |
| 1d | 10 | 6.55 ± 0.356 |
| 1e | 10 | 3.57 ± 0.543 |
| 1f | 1 | 0.95 ± 0.038 |
| 3 | 1.18 ± 0.029 | |
| 5 | 3.36 ± 0.653 | |
| 10 | 8.85 ± 1.081 | |
| 1g | 1 | 0.84 ± 0.024 |
| 3 | 1.50 ± 0.018 | |
| 5 | 5.00 ± 0.488 | |
| 10 | 15.6 ± 1.783 | |
| 1h | 1 | 0.81 ± 0.008 |
| 3 | 0.90 ± 0.020 | |
| 5 | 1.51 ± 0.154 | |
| 10 | 4.10 ± 0.306 | |
| 1i | 1 | 0.90 ± 0.030 |
| 3 | 0.91 ± 0.003 | |
| 5 | 1.78 ± 0.217 | |
| 10 | 3.34 ± 0.303 |
| Genes | Primer Sequences | |
|---|---|---|
| NQO1 | F: AGGCTGGTTTGAGCGAGT | R: ATTGAATTCGGGCGTCTGCTG |
| HO-1 | F: CGGGCCAGCAACAAAGTG | R: AGTGTAAGGACCCATCGGAGAA |
| GCLC | F: TGGCAATGCAGTGGTGGAT | R: AACACACCTTCCTTCCCATTGA |
| 18s | F: CAGCCACCCGAGATTGAGCA | R: TAGTAGCGACGGGCGGTGTG |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, S.H.; Kim, M.; Kwon, D.; Pyo, J.S.; Kim, J.H.; Kwak, J.-H.; Jung, Y.-S. N-Phenyl Cinnamamide Derivatives Protect Hepatocytes against Oxidative Stress by Inducing Cellular Glutathione Synthesis via Nuclear Factor (Erythroid-Derived 2)-Like 2 Activation. Molecules 2021, 26, 1027. https://doi.org/10.3390/molecules26041027
Kim SH, Kim M, Kwon D, Pyo JS, Kim JH, Kwak J-H, Jung Y-S. N-Phenyl Cinnamamide Derivatives Protect Hepatocytes against Oxidative Stress by Inducing Cellular Glutathione Synthesis via Nuclear Factor (Erythroid-Derived 2)-Like 2 Activation. Molecules. 2021; 26(4):1027. https://doi.org/10.3390/molecules26041027
Chicago/Turabian StyleKim, Sou Hyun, Minwoo Kim, Doyoung Kwon, Jae Sung Pyo, Joo Hyun Kim, Jae-Hwan Kwak, and Young-Suk Jung. 2021. "N-Phenyl Cinnamamide Derivatives Protect Hepatocytes against Oxidative Stress by Inducing Cellular Glutathione Synthesis via Nuclear Factor (Erythroid-Derived 2)-Like 2 Activation" Molecules 26, no. 4: 1027. https://doi.org/10.3390/molecules26041027
APA StyleKim, S. H., Kim, M., Kwon, D., Pyo, J. S., Kim, J. H., Kwak, J.-H., & Jung, Y.-S. (2021). N-Phenyl Cinnamamide Derivatives Protect Hepatocytes against Oxidative Stress by Inducing Cellular Glutathione Synthesis via Nuclear Factor (Erythroid-Derived 2)-Like 2 Activation. Molecules, 26(4), 1027. https://doi.org/10.3390/molecules26041027






