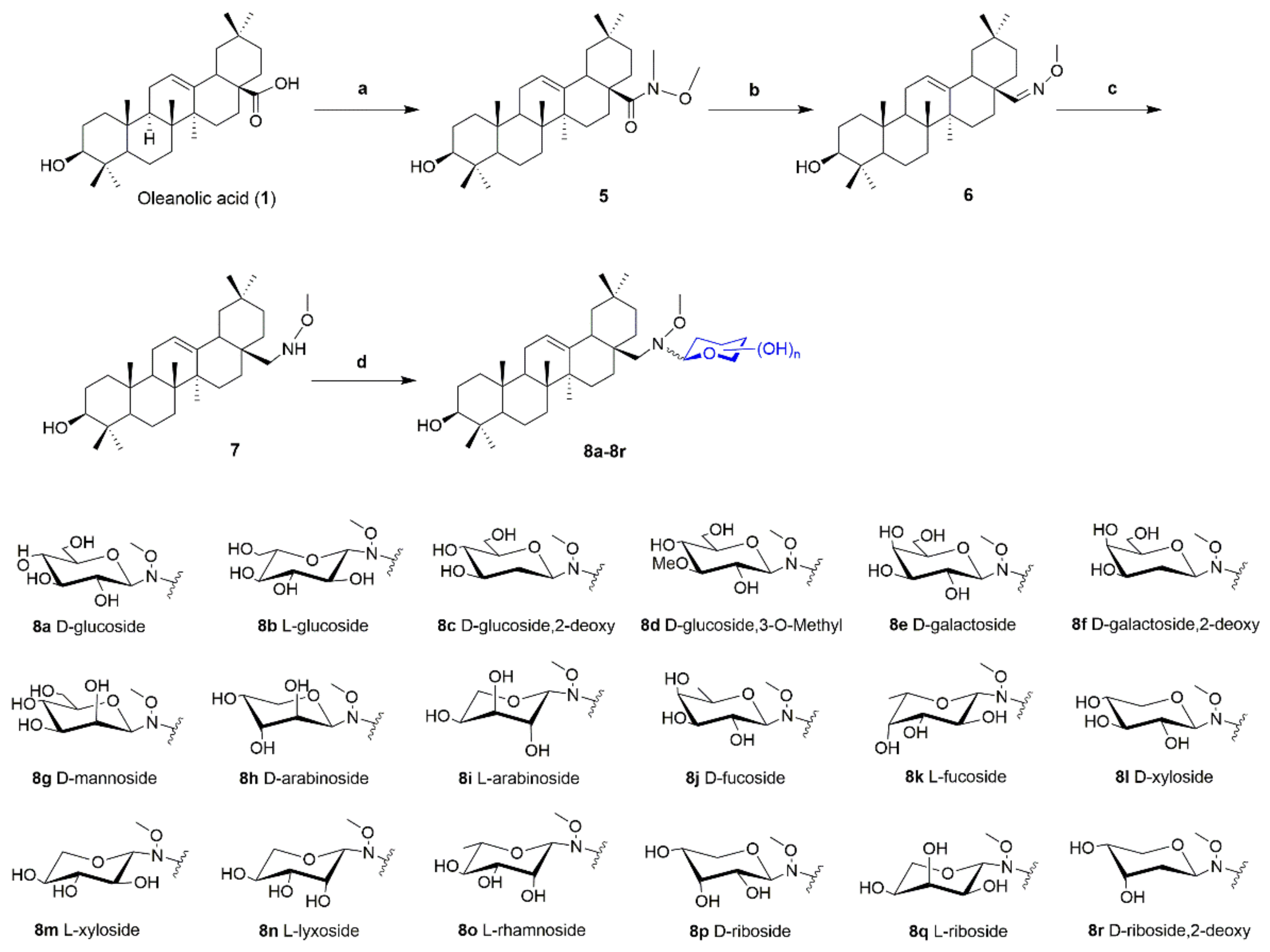
3.2.6. General Procedure for the Synthesis of Oleanolic Acid Neoglycosides 4a–4r and 8a–8r
To a solution of neoaglycone 3 (typically 0.1 mmol, 54.2 mg) and reducing sugar (2 eq.) were dissolved in MeOH/CHCl3 (6:1, 5 mL). Neoaglycon 7 (0.1 mmol, 47.2 mg) and reducing sugar (2 eq.) was dissolved in MeOH/CHCl3 (4:1, 5 mL), external proton source AcOH (10 eq.) were added, and then reaction at 40 °C for 48 h on a rotary shaker at 250 rpm. The target neoglycosides was purified with MeOH/CH2Cl2 by silica gel column chromatography. The configuration of the glycosidic bond of all the neoglycosides was identified by the J value of JH1′−H2′.
(3S)-O-(N-Methoxy-N-d-glucosylglycyl) oleanolic acid (4a)
White solid (15.2 mg, 21%). 1H-NMR (500 MHz, C5D5N) δ 5.50 (t, J = 3.3 Hz, 1H), 4.86–4.77 (m, 2H), 4.58 (dd, J = 11.9, 2.2 Hz, 1H), 4.43–4.33 (m, 2H), 4.32–4.19 (m, 3H), 4.15 (d, J = 16.9 Hz, 1H), 4.05 (s, 3H), 4.00–3.94 (m, 1H), 3.33 (dd, J = 13.8, 4.0 Hz, 1H), 1.31 (s, 3H), 1.04 (s, 3H), 1.01 (s, 3H), 0.98 (s, 3H), 0.89 (s, 3H), 0.84 (s, 3H), 0.82 (s, 3H). 13C-NMR (125 MHz, C5D5N) δ 180.5, 170.7, 145.2, 122.8, 95.6, 81.4, 80.8, 80.2, 72.1, 71.9, 63.3, 62.8, 55.9, 55.7, 48.3, 47.0, 46.8, 42.5, 42.4, 40.1, 38.6, 38.3, 37.5, 34.6, 33.6, 33.6, 33.4, 31.3, 28.7, 28.5, 26.6, 24.3, 24.1, 24.1, 24.1, 18.8, 17.7, 17.3, 15.8. HRMS (ESI) m/z for C39H64NO10 ([M+H]+) 706.4525, calc. 706.4525.
(3S)-O-(N-Methoxy-N-β-l-glucosylglycyl) oleanolic acid (4b)
White solid (11.0 mg, 16%). 1H-NMR (500 MHz, C5D5N) δ 5.50 (t, J = 3.1 Hz, 1H), 4.84 (d, J = 8.7 Hz, 1H), 4.81 (dd, J = 11.2, 5.4 Hz, 1H), 4.57 (dd, J = 11.8, 2.1 Hz, 1H), 4.40 (dd, J = 11.9, 5.4 Hz, 1H), 4.35–4.12 (m, 5H), 4.05 (s, 3H), 4.00–3.93 (m, 1H), 3.33 (dd, J = 13.7, 3.9 Hz, 1H), 1.30 (s, 3H), 1.04 (s, 3H), 1.01 (s, 3H), 0.98 (s, 3H), 0.92 (s, 3H), 0.84 (s, 3H), 0.83 (s, 3H). 13C-NMR (125 MHz, C5D5N) δ 180.0, 170.2, 144.7, 122.2, 94.9, 80.8, 80.2, 79.6, 71.6, 71.3, 62.7, 62.1, 55.3, 54.9, 47.7, 46.5, 46.3, 42.0, 41.8, 39.5, 38.0, 37.8, 37.0, 33.8, 33.1, 33.0, 32.8, 31.3, 28.1, 27.9, 26.0, 23.7, 23.6, 23.5, 22.7, 18.3, 17.2, 16.6, 15.2. HRMS (ESI) m/z for C39H64NO10 ([M+H]+) 706.4533, calc. 706.4525.
(3S)-O-(N-Methoxy-N-(2-deoxy-d-glucosyl)glycyl) oleanolic acid (4c)
White solid (38.5 mg, 56%). 1H-NMR (500 MHz, C5D5N) δ 5.49 (s, 1H), 4.83–4.77 (m, 2H), 4.55 (dd, J = 11.8, 2.3 Hz, 1H), 4.45–3.99 (m, 5H), 3.88 (s, 3H), 3.86–3.82 (m, 1H), 3.32 (dd, J = 13.7, 3.8 Hz, 1H), 2.71–2.54 (m, 1H), 1.30 (s, 3H), 1.03 (s, 3H), 1.01 (s, 3H), 0.98 (s, 3H), 0.92 (s, 3H), 0.85 (d, J = 4.0 Hz, 6H). 13C-NMR (125 MHz, C5D5N) δ 179.9, 170.0, 144.7, 122.2, 91.0, 80.9, 80.3, 73.2, 73.0, 62.8, 62.4, 55.5, 55.3, 47.7, 46.5, 46.3, 42.0, 41.8, 39.5, 38.0, 37.8, 37.5, 37.0, 34.0, 33.1, 33.0, 32.8, 30.8, 28.1, 27.9, 26.0, 23.7, 23.6, 23.6, 23.5, 18.3, 17.2, 16.8, 15.2. HRMS (ESI) m/z for C39H64NO9 ([M+H]+) 690.4575, calc. 690.4576.
(3S)-O-(N-Methoxy-N-(3-O-methyl-d-glucosyl)glycyl) oleanolic acid (4d)
White solid (10.7 mg, 15%). 1H-NMR (500 MHz, C5D5N) δ 5.48 (s, 1H), 4.89–4.70 (m, 2H), 4.56–4.29 (m, 5H), 4.26–4.05 (m, 2H), 4.02 (s, 3H), 3.96 (s, 3H), 3.93–3.84 (m, 1H), 3.32 (d, J = 13.2 Hz, 1H), 1.30 (s, 3H), 1.02 (d, J = 9.6 Hz, 3H), 1.00 (d, J = 4.8 Hz, 3H), 0.98 (d, J = 2.4 Hz, 3H), 0.88 (s, 3H), 0.84 (s, 3H), 0.82 (d, J = 3.8 Hz, 3H). 13C-NMR (125 MHz, C5D5N) δ 180.5, 168.2, 145.2, 122.7, 95.5, 82.0, 81.5, 80.6, 71.7, 71.1, 66.0, 63.0, 62.7, 55.9, 55.8, 48.3, 47.0, 46.8, 42.5, 42.3, 40.1, 38.5, 38.3, 37.5, 34.6, 33.6, 33.6, 33.4, 31.2, 28.7, 28.5, 26.5, 24.3, 24.1, 24.0, 23.3, 19.8, 17.7, 17.3, 14.1. HRMS (ESI) m/z for C40H66NO10 ([M+H]+) 720.4678, calc. 720.4681.
(3S)-O-(N-Methoxy-N-d-galactosylglycyl) oleanolic acid (4e)
White solid (16.1 mg, 23%). 1H-NMR (500 MHz, C5D5N) δ 5.50 (d, J = 11.1 Hz, 1H), 5.34 (d, J = 5.7 Hz, 0.67H, αH1′), 4.87–4.81 (m, 0.67H), 4.78 (dd, J = 10.4, 5.7 Hz, 1H), 4.63–4.43 (m, 2H), 4.42–4.13 (m, 4H), 3.99 (s, 3H), 3.95–3.89 (m, 1H), 3.44–3.25 (m, 1H), 1.30 (s, 3H), 1.03 (s, 3H), 1.01 (s, 3H), 0.98 (s, 3H), 0.83 (d, J = 2.0 Hz, 3H), 0.80 (s, 3H), 0.78 (d, J = 7.4 Hz, 3H). 13C-NMR (125 MHz, C5D5N) δ 179.9, 170.1, 144.6, 122.2, 99.8, 95.3, 84.0, 81.0, 80.8, 78.7, 78.6, 77.5, 76.3, 74.1, 72.6, 69.0, 65.3, 64.3, 55.3, 54.5, 47.7, 46.5, 46.3, 42.0, 41.8, 39.5, 38.0, 37.7, 37.0, 34.0, 33.1, 33.0, 32.8, 30.6, 28.1, 27.9, 26.0, 23.7, 23.6, 23.5, 23.5, 19.1, 17.2, 15.2, 13.5. HRMS (ESI) m/z for C39H64NO10 ([M+H]+) 706.4520, calc. 706.4525.
(3S)-O-(N-Methoxy-N-β-(2-deoxy-d-galactosyl)glycyl) oleanolic acid (4f)
White solid (29.8 mg, 43%). 1H-NMR (500 MHz, C5D5N) δ 5.49 (s, 1H), 4.79 (dd, J = 12.2, 5.2 Hz, 1H), 4.76 (d, J = 10.9 Hz, 1H), 4.52–4.01 (m, 6H), 3.99–3.90 (m, 1H), 3.85 (s, 3H), 3.33 (dd, J = 13.7, 3.7 Hz, 1H), 1.30 (s, 20H), 1.04 (s, 3H), 1.01 (s, 3H), 0.98 (s, 3H), 0.90 (s, 3H), 0.85 (s, 6H). 13C-NMR (125 MHz, C5D5N) δ 180.0, 170.1, 144.7, 122.2, 91.2, 80.9, 78.8, 70.2, 68.6, 62.5, 62.1, 55.3, 55.1, 47.7, 46.5, 46.3, 42.0, 41.8, 39.5, 38.0, 37.8, 37.0, 34.0, 33.1, 33.1, 33.0, 32.83, 30.18, 28.1, 27.9, 26.0, 23.7, 23.6, 23.5, 22.7, 18.3, 17.2, 16.8, 15.2. HRMS (ESI) m/z for C39H64NO9 ([M+H]+) 690.4581, calc. 690.4576.
(3S)-O-(N-Methoxy-N-d-mannosylglycyl) oleanolic acid (4g)
White solid (17.2 mg, 24%). 1H-NMR (500 MHz, C5D5N) δ 5.50 (d, J = 12.8 Hz, 1H), 5.13 (dd, J = 38.5, 21.1 Hz, 2H), 4.93 (d, J = 1.7 Hz, 0.5H, αH1′), 4.82–4.54 (m, 3H), 4.53–4.28 (m, 3H), 4.25–4.18 (m, 0.5H), 3.96 (s, 3H), 3.79 (s, 1H), 3.44–3.25 (m, 1H), 1.03 (s, 3H), 1.00 (d, J = 1.6 Hz, 3H), 0.98 (s, 3H), 0.84 (d, J = 7.0 Hz, 3H), 0.82 (d, J = 2.8 Hz, 3H), 0.78 (s, 3H). 13C-NMR (125 MHz, C5D5N) δ 180.0, 170.1, 144.6, 122.2, 93.7, 92.1, 81.0, 81.0, 80.5, 77.9, 76.2, 74.1, 73.2, 72.2, 68.8, 65.3, 64.3, 55.6, 55.3, 55.3, 47.7, 46.5, 46.3, 42.0, 41.8, 39.5, 38.0, 37.7, 34.0, 34.0, 33.1, 33.0, 32.8, 30.6, 28.1, 27.9, 26.0, 23.8, 23.6, 23.5, 23.5, 19.1, 17.2, 15.2, 13.5. HRMS (ESI) m/z for C39H64NO10 ([M+H]+) 706.4528, calc. 706.4525.
(3S)-O-(N-Methoxy-N-α-d-arabinosylglycyl) oleanolic acid (4h)
White solid (35.5 mg, 53%). 1H-NMR (500 MHz, C5D5N) δ 5.49 (s, 1H), 5.33 (d, J = 5.4 Hz, 1H), 4.81 (dd, J = 11.6, 4.7 Hz, 2H), 4.70–4.64 (m, 1H), 4.46–4.09 (m, 4H), 4.03–3.94 (m, 1H), 3.92 (s, 3H), 3.37–3.27 (m, 1H), 1.30 (d, J = 1.9 Hz, 3H), 1.03 (s, 3H), 1.01 (s, 3H), 0.98 (s, 3H), 0.93 (d, J = 4.0 Hz, 3H), 0.89 (s, 3H), 0.84 (s, 3H). 13C-NMR (125 MHz, C5D5N) δ 180.5, 170.5, 145.2, 122.8, 100.3, 85.7, 81.6, 79.4, 77.7, 63.1, 62.7, 55.9, 48.3, 47.0, 46.8, 42.5, 42.4, 40.1, 38.6, 38.4, 37.5, 34.6, 33.6, 33.6, 33.4, 31.9, 31.3, 30.7, 28.7, 28.5, 26.6, 24.3, 24.1, 24.1, 18.8, 17.7, 17.4, 15.7. HRMS (ESI) m/z for C38H62NO9 ([M+H]+) 676.4413, calc. 676.4419.
(3S)-O-(N-Methoxy-N-l-arabinosylglycyl) oleanolic acid (4i)
White solid (13.7 mg, 20%). 1H-NMR (500 MHz, C5D5N) δ 5.50 (s, 1H), 5.32 (d, J = 5.4 Hz, 0.5H, αH1′), 4.86–4.72 (m, 2H), 4.70–4.66 (m, 0.5H), 4.64 (d, J = 9.0 Hz, 0.5H, βH1′), 4.52 (t, J = 8.9 Hz, 0.5H), 4.42–4.26 (m, 2H), 4.19 (ddd, J = 29.1, 11.0, 5.8 Hz, 2H), 3.96 (d, J = 32.9 Hz, 3H), 3.33 (dd, J = 13.8, 3.7 Hz, 1H), 1.30 (s, 3H), 1.04 (s, 3H), 1.01 (s, 3H), 0.98 (s, 3H), 0.88 (s, 3H), 0.86 (d, J = 5.8 Hz, 3H), 0.83 (d, J = 4.6 Hz, 3H). 13C-NMR (125 MHz, C5D5N) δ 179.9, 170.2, 144.7, 122.2, 99.8, 95.5, 85.1, 81.0, 80.9, 78.9, 77.2, 75.6, 69.8, 69.1, 62.4, 62.2, 61.7, 55.6, 55.3, 47.7, 46.5, 46.3, 42.0, 41.8, 39.5, 37.8, 37.7, 37.0, 34.0, 33.1, 33.0, 32.8, 30.8, 28.1, 27.9, 26.0, 23.7, 23.6, 23.5, 18.3, 17.2, 16.8, 15.2. HRMS (ESI) m/z for C38H62NO9 ([M+H]+) 676.4411, calc. 676.4419.
(3S)-O-(N-Methoxy-N-d-fucosylglycyl) oleanolic acid (4j)
White solid (19.4 mg, 28%). 1H-NMR (500 MHz, C5D5N) δ 5.49 (s, 1H), 5.29 (d, J = 5.3 Hz, 0.67H, αH1′), 4.70 (d, J = 9.1 Hz, 0.33H, βH1′), 4.53–4.09 (m, 3H), 3.99 (s, 3H), 3.93 (s, 3H), 3.85 (dd, J = 12.7, 6.1 Hz, 0.5H), 3.70–3.60 (m, 0.5H), 3.32 (d, J = 13.1 Hz, 1H), 1.30 (s, 3H), 1.03 (s, 3H), 1.01 (s, 3H), 0.98 (s, 3H), 0.92 (d, J = 9.7 Hz, 3H), 0.87 (s, 3H), 0.84 (s, 3H). 13C-NMR (125 MHz, C5D5N) δ 180.5, 170.5, 145.2, 122.7, 100.2, 95.6, 88.0, 81.6, 81.5, 79.6, 78.4, 76.9, 73.6, 68.0, 62.8, 62.2, 56.0, 55.9, 52.3, 48.3, 47.0, 46.8, 42.5, 42.3, 40.1, 38.6, 38.3, 37.5, 34.6, 33.6, 33.6, 33.4, 31.3, 28.7, 28.5, 26.6, 24.3, 24.1, 24.1, 18.8, 17.7, 17.4, 17.3, 15.7, 14.6. HRMS (ESI) m/z for C39H64NO9 ([M+H]+) 690.4585, calc. 690.4576.
(3S)-O-(N-Methoxy-N-l-fucosylglycyl) oleanolic acid (4k)
White solid (17.3 mg, 25%). 1H-NMR (500 MHz, C5D5N) δ 5.49 (s, 1H), 5.31 (d, J = 5.3 Hz, 0.67H, αH1′), 4.72 (d, J = 9.0 Hz, 0.33H, βH1′), 4.49–4.04 (m, 3H), 3.99 (s, 3H), 3.93 (s, 3H), 3.86 (q, J = 6.4 Hz, 0.5H), 3.64 (dd, J = 9.3, 5.1 Hz, 0.5H), 3.32 (d, J = 13.6 Hz, 1H), 1.30 (s, 3H), 1.03 (s, 3H), 1.01 (s, 3H), 0.98 (s, 3H), 0.94–0.91 (m, 3H), 0.88 (s, 3H), 0.84 (s, 3H). 13C-NMR (125 MHz, C5D5N) δ 180.0, 170.0, 144.7, 122.2, 99.6, 95.0, 87.5, 81.0, 80.8, 79.0, 77.8, 76.3, 73.0, 67.4, 62.2, 61.6, 56.1, 55.3, 47.7, 46.5, 46.3, 42.0, 41.8, 39.5, 38.0, 37.8, 37.0, 34.0, 33.1, 33.0, 32.8, 30.8, 28.1, 27.9, 26.0, 23.7, 23.6, 23.5, 22.7, 18.3, 17.2, 16.8, 16.7, 15.2, 14.1. HRMS (ESI) m/z for C39H62NO9 ([M+H]−) 688.4434, calc. 688.4430.
(3S)-O-(N-Methoxy-N-β-d-xylosylglycyl) oleanolic acid (4l)
White solid (15.0 mg, 22%). 1H-NMR (500 MHz, C5D5N) δ 5.49 (s, 1H), 4.70 (d, J = 8.4 Hz, 1H), 4.53–4.29 (m, 2H), 4.27–4.12 (m, 2H), 4.04 (s, 3H), 3.77–3.58 (m, 1H), 3.32 (d, J = 13.3 Hz, 1H), 1.30 (s, 3H), 1.03 (s, 3H), 1.00 (s, 3H), 0.98 (s, 3H), 0.91 (s, 3H), 0.89 (s, 3H), 0.83 (s, 3H). 13C-NMR (125 MHz, C5D5N) δ 180.7, 170.7, 145.2, 123.4, 96.2, 81.6, 80.3, 72.0, 71.3, 69.6, 62.7, 55.9, 55.6, 48.3, 47.0, 46.9, 42.5, 42.4, 40.1, 38.6, 38.4, 37.5, 34.6, 33.7, 33.6, 33.4, 30.8, 28.7, 28.5, 26.6, 24.3, 24.2, 24.1, 23.3, 18.8, 17.7, 17.3, 15.7. HRMS (ESI) m/z for C38H62NO9 ([M+H]+) 676.4426, calc. 676.4419.
(3S)-O-(N-Methoxy-N-β-l-xylosylglycyl) oleanolic acid (4m)
White solid (8.6 mg, 13%). 1H-NMR (500 MHz, C5D5N) δ 5.49 (s, 1H), 4.72 (d, J = 8.2 Hz, 1H), 4.58–4.36 (m, 2H), 4.34–4.13 (m, 3H), 4.04 (s, 3H), 3.32 (d, J = 13.6 Hz, 1H), 1.30 (s, 3H), 1.03 (s, 3H), 1.00 (s, 3H), 0.98 (s, 3H), 0.94 (s, 3H), 0.90 (s, 3H), 0.84 (s, 3H). 13C-NMR (125 MHz, C5D5N) δ 179.8, 170.2, 144.5, 122.9, 95.6, 80.9, 79.7, 71.4, 70.7, 69.0, 62.0, 55.3, 54.8, 47.7, 46.5, 46.3, 42.0, 41.8, 39.5, 38.0, 37.8, 37.0, 34.0, 33.1, 33.0, 32.8, 30.2, 28.1, 27.9, 26.0, 23.7, 23.6, 23.5, 22.7, 18.3, 17.2, 16.8, 15.2. HRMS (ESI) m/z for C38H62NO9 ([M+H]+) 676.4430, calc. 676.4419.
(3S)-O-(N-Methoxy-N-β-l-lyxosylglycyl) oleanolic acid (4n)
White solid (40.2 mg, 59%). 1H-NMR (500 MHz, C5D5N) δ 5.49 (t, J = 3.3 Hz, 1H), 5.18 (d, J = 8.2 Hz, 1H), 4.87–4.72 (m, 3H), 4.44–4.08 (m, 4H), 3.98 (s, 3H), 3.32 (dd, J = 13.9, 3.7 Hz, 1H), 1.30 (s, 3H), 1.03 (s, 3H), 1.01 (s, 3H), 0.98 (s, 3H), 0.91 (s, 3H), 0.88 (s, 3H), 0.84 (s, 3H). 13C-NMR (125 MHz, C5D5N) δ 179.9, 170.2, 144.6, 122.2, 92.2, 81.0, 72.9, 70.5, 67.6, 66.9, 61.8, 55.3, 55.1, 47.7, 46.5, 46.3, 42.0, 41.8, 39.5, 38.0, 37.8, 37.0, 34.0, 33.1, 33.0, 32.8, 30.8, 28.1, 27.9, 26.0, 23.7, 23.6, 23.6, 23.5, 18.3, 17.2, 16.8, 15.2. HRMS (ESI) m/z for C38H62NO9 ([M+H]+) 676.4417, calc. 676.4419.
(3S)-O-(N-Methoxy-N-l-rhamnosylglycyl) oleanolic acid (4o)
White solid (16.4 mg, 24%). 1H-NMR (500 MHz, C5D5N) δ 5.49 (s, 1H), 5.14-5.09 (m, 0.8H), 4.99 (s, 0.2H), 4.88 (s, 0.5H), 4.85–4.76 (m, 0.8H), 4.67 (d, J = 2.0 Hz, 0.2H, αH1′), 4.61 (dd, J = 8.8, 3.3 Hz, 0.5H), 4.47–4.38 (m, 0.6H), 4.38–4.24 (m, 1H), 4.20–4.08 (m, 0.4H), 3.97–3.77 (m, 2H), 3.74 (s, 3H), 3.31 (d, J = 13.2 Hz, 1H), 1.30 (s, 3H), 1.03 (s, 3H), 1.00 (d, J = 3.7 Hz, 3H), 0.97 (s, 3H), 0.95 (d, J = 6.6 Hz, 3H), 0.93 (s, 3H), 0.87 (dd, J = 10.8, 5.1 Hz, 3H), 0.82 (d, J = 10.4 Hz, 3H). 13C-NMR (125 MHz, C5D5N) δ 180.5, 170.4, 145.2, 122.7, 94.4, 92.4, 81.7, 81.1, 74.3, 74.0, 73.5, 72.9, 72.7, 70.4, 62.4, 61.8, 56.7, 55.9, 48.2, 47.0, 46.8, 42.5, 42.3, 40.1, 38.5, 38.3, 37.5, 34.6, 33.6, 33.6, 33.4, 31.3, 28.7, 28.6, 26.6, 24.3, 24.1, 24.1, 24.1, 19.2, 17.7, 17.4, 17.4, 15.7, 14.6. HRMS (ESI) m/z for C39H64NO9 ([M+H]+) 690.4578, calc. 690.4576.
(3S)-O-(N-Methoxy-N-d-ribosylglycyl) oleanolic acid (4p)
White solid (42.1 mg, 62%). 1H-NMR (500 MHz, C5D5N) δ 5.49 (s, 1H), 5.41 (d, J = 2.9 Hz, 0.33H, αH1′), 5.08 (d, J = 8.2 Hz, 0.67H, βH1′), 4.86–4.72 (m, 2H), 4.67 (d, J = 4.9 Hz, 1H), 4.41–4.06 (m, 4H), 4.05 (s, 3H), 3.91 (d, J = 15.8 Hz, 1H), 3.32 (d, J = 12.9 Hz, 1H), 1.30 (s, 3H), 1.03 (s, 3H), 1.01 (s, 3H), 0.98 (s, 3H), 0.94 (s, 3H), 0.91 (s, 3H), 0.83 (d, J = 4.4 Hz, 3H). 13C-NMR (125 MHz, C5D5N) δ 180.3, 170.5, 145.0, 122.6, 100.7, 92.0, 85.6, 81.3, 73.2, 72.7, 72.5, 69.2, 68.8, 66.4, 63.9, 62.6, 62.5, 55.7, 48.1, 46.8, 46.6, 42.3, 42.2, 39.9, 38.4, 38.1, 37.3, 34.4, 33.4, 33.4, 33.2, 31.1, 28.5, 28.3, 26.4, 24.1, 23.9, 23.9, 23.9, 18.6, 17.5, 17.1, 15.5. HRMS (ESI) m/z for C38H62NO9 ([M+H]+) 676.4423, calc. 676.4419.
(3S)-O-(N-Methoxy-N-l-ribosylglycyl) oleanolic acid (4q)
White solid (40.8 mg, 60%). 1H-NMR (500 MHz, C5D5N) δ 5.49 (s, 1H), 5.42 (d, J = 3.7 Hz, 0.33H, αH1′), 5.10 (d, J = 8.6 Hz, 0.67H, βH1′), 4.86–4.70 (m, 2H), 4.67 (dd, J = 9.3, 4.7 Hz, 1H), 4.35–4.16 (m, 4H), 4.04 (s, 3H), 3.95 (s, 0.33H), 3.89 (s, 0.67H), 3.32 (d, J = 13.6 Hz, 1H), 1.30 (d, J = 2.8 Hz, 3H), 1.03 (s, 3H), 1.01 (s, 3H), 0.98 (s, 3H), 0.94 (s, 3H), 0.91 (d, J = 6.1 Hz, 3H), 0.83 (d, J = 4.6 Hz, 3H). 13C-NMR (125 MHz, C5D5N) δ 180.5, 170.8, 145.2, 122.8, 100.9, 92.2, 85.8, 81.5, 73.4, 72.9, 72.7, 69.5, 69.0, 66.6, 64.0, 62.8, 62.7, 55.9, 48.3, 47.0, 46.8, 42.5, 42.4, 40.1, 38.6, 38.4, 37.5, 34.6, 33.6, 33.6, 33.4, 31.3, 28.7, 28.5, 26.6, 24.3, 24.1, 24.1, 24.1, 18.8, 17.7, 17.3, 15.7. HRMS (ESI) m/z for C38H62NO9 ([M+H]+) 676.4413, calc. 676.4419.
(3S)-O-(N-Methoxy-N-(2-deoxy-d-ribosyl)glycyl) oleanolic acid (4r)
White solid (32.5 mg, 49%). 1H-NMR (500 MHz, C5D5N) δ 5.49 (s, 1H), 5.32 (d, J = 2.9 Hz, 0.8H, αH1′), 4.73–4.46 (m, 2H), 4.24–4.05 (m, 4H), 3.86 (s, 3H), 3.50 (d, J = 10.1 Hz, 1H), 3.35–3.29 (m, 1H), 1.30 (s, 3H), 1.03 (s, 3H), 1.01 (s, 3H), 0.98 (s, 3H), 0.93 (d, J = 7.5 Hz, 3H), 0.88 (d, J = 10.6 Hz, 3H), 0.84 (s, 3H). 13C-NMR (125 MHz, C5D5N) δ 180.5, 168.2, 145.2, 122.8, 106.2, 91.8, 82.3, 81.5, 72.5, 71.1, 70.8, 68.8, 68.6, 66.0, 63.3, 62.8, 62.5, 55.9, 48.3, 47.0, 46.8, 42.5, 42.4, 40.1, 38.6, 38.4, 37.5, 34.6, 33.6, 33.6, 33.4, 31.2, 28.7, 28.5, 26.6, 24.3, 24.2, 24.2, 24.1, 18.9, 17.7, 17.4, 15.8. HRMS (ESI) m/z for C38H60NO8 ([M+H]−) 658.4326, calc. 658.4324.
28-N-Methoxyaminooleanane-β-d-glucoside (8a)
White solid (36.2 mg, 60%). 1H-NMR (500 MHz, C5D5N) δ 5.20 (d, J = 3.0 Hz, 1H), 4.61 (d, J = 7.9 Hz, 1H), 4.48 (dd, J = 11.8, 2.6 Hz, 1H), 4.36 (dd, J = 11.8, 4.9 Hz, 1H), 4.27–4.18 (m, 3H), 3.93–3.88 (m, 1H), 3.80 (s, 3H), 3.46–3.36 (m, 2H), 3.10 (d, J = 15.3 Hz, 1H), 2.27–2.09 (m, 1H), 1.21 (s, 3H), 1.20 (s, 3H), 1.05 (s, 3H), 1.02 (s, 3H), 0.89 (d, J = 1.3 Hz, 6H), 0.88 (s, 3H). 13C-NMR (125 MHz, C5D5N) δ 145.7, 123.7, 97.6, 81.1, 80.8, 78.8, 72.6, 72.5, 63.7, 56.5, 48.8, 48.0, 45.6, 42.6, 41.0, 40.2, 39.9, 38.0, 37.1, 35.6, 34.2, 33.7, 33.6, 32.3, 31.8, 31.1, 30.7, 29.5, 28.9, 27.2, 26.9, 24.8, 24.7, 19.6, 18.1, 17.3, 16.5. HRMS (ESI) m/z for C37H64NO7 ([M+H]+) 634.4676, calc. 634.4677.
28-N-Methoxyaminooleanane-β-l-glucoside (8b)
White solid (32.2 mg, 51%). 1H-NMR (500 MHz, C5D5N) δ 5.19 (d, J = 3.0 Hz, 1H), 4.60 (d, J = 8.0 Hz, 1H), 4.48 (dd, J = 11.8, 2.5 Hz, 1H), 4.37 (dd, J = 11.8, 4.9 Hz, 1H), 4.30–4.22 (m, 3H), 3.86 (s, 1H), 3.81 (s, 3H), 3.44–3.38 (m, 2H), 3.22 (d, J = 14.9 Hz, 1H), 2.26–2.18 (m, 1H), 1.23 (s, 3H), 1.20 (s, 3H), 1.09 (s, 3H), 1.02 (s, 3H), 0.95 (s, 3H), 0.90 (s, 3H), 0.86 (s, 3H). 13C-NMR (125 MHz, C5D5N) δ 145.1, 123.0, 97.2, 80.5, 80.3, 78.2, 72.2, 71.9, 63.0, 55.9, 48.1, 47.4, 44.5, 42.0, 40.4, 39.5, 39.2, 37.3, 36.7, 35.0, 33.5, 33.2, 33.1, 31.6, 31.2, 30.5, 30.1, 28.9, 28.2, 26.4, 26.1, 24.0, 23.9, 18.9, 17.7, 16.7, 15.9. HRMS (ESI) m/z for C37H64NO7 ([M+H]+) 634.4673, calc. 634.4677.
28-N-Methoxyaminooleanane-β-2-deoxy-d-glucoside (8c)
White solid (33.9 mg, 53%). 1H-NMR (500 MHz, C5D5N) δ 5.21 (t, J = 3.2 Hz, 1H), 4.72 (d, J = 10.5 Hz, 1H), 4.50 (dd, J = 11.6, 2.8 Hz, 1H), 4.39 (dd, J = 11.6, 4.9 Hz, 1H), 4.30–4.23 (m, 1H), 4.06 (t, J = 9.0 Hz, 1H), 3.86–3.80 (m, 1H), 3.66 (s, 3H), 3.44 (dd, J = 10.9, 4.8 Hz, 1H), 3.34 (d, J = 15.1 Hz, 1H), 2.96 (d, J = 15.2 Hz, 1H), 2.60–2.47 (m, 1H), 1.26 (s, 3H), 1.24 (s, 3H), 1.08 (s, 3H), 1.06 (s, 3H), 0.95 (s, 3H), 0.94 (s, 3H), 0.91 (s, 3H). 13C-NMR (125 MHz, C5D5N) δ 145.0, 123.2, 92.8, 80.5, 78.2, 73.9, 73.6, 63.3, 55.8, 48.2, 47.3, 45.0, 41.9, 40.4, 39.5, 39.2, 37.7, 37.3, 36.3, 34.9, 33.5, 33.0, 31.6, 31.1, 30.5, 30.1, 28.9, 28.2, 26.5, 26.1, 24.1, 24.0, 23.9, 18.9, 17.4, 16.6, 15.9. HRMS (ESI) m/z for C37H64NO6 ([M+H]+) 618.4730, calc. 618.4728.
28-N-Methoxyaminooleanane-β-3-O-methyl-d-glucoside (8d)
White solid (25.2 mg, 39%). 1H-NMR (500 MHz, C5D5N) δ 5.27 (s, 1H), 4.63 (d, J = 8.9 Hz, 1H), 4.50 (dd, J = 11.8, 2.5 Hz, 1H), 4.39 (dd, J = 11.8, 4.8 Hz, 1H), 4.23 (q, J = 9.1 Hz, 2H), 3.95 (s, 3H), 3.85–3.77 (m, 4H), 3.50–3.41 (m, 2H), 3.13 (d, J = 15.3 Hz, 1H), 2.32–2.18 (m, 1H), 1.27 (s, 3H), 1.27 (s, 3H), 1.11 (s, 3H), 1.09 (s, 3H), 0.96 (s, 3H), 0.95 (s, 3H), 0.95 (s, 3H). 13C-NMR (125 MHz, C5D5N) δ 144.9, 122.9, 96.6, 90.0, 80.1, 78.1, 71.2, 70.8, 62.6, 60.9, 55.7, 48.0, 47.2, 44.7, 41.8, 40.2, 39.4, 39.1, 37.2, 36.3, 34.8, 33.4, 32.8, 32.8, 31.5, 31.0, 30.3, 28.7, 28.1, 26.4, 26.1, 24.1, 23.9, 23.8, 18.8, 17.3, 16.5, 15.7. HRMS (ESI) m/z for C38H66NO7 ([M+H]+) 648.4825, calc. 648.4834.
28-N-Methoxyaminooleanane-d-galactoside (8e)
White solid (35.8 mg, 56%). 1H-NMR (500 MHz, C5D5N) δ 5.23 (t, J = 5.4 Hz, 1H), 5.11 (d, J = 6.2 Hz, 0.2H, βH1′), 4.67 (d, J = 2.8 Hz, 0.8H, αH1′), 4.62 (s, 1H), 4.56–4.46 (m, 1H), 4.43–4.33 (m, 2H), 4.24 (dt, J = 9.5, 4.7 Hz, 1H), 4.10 (t, J = 6.3 Hz, 1H), 3.83–3.78 (m, 3H), 3.70–3.60 (m, 1H), 3.45 (dd, J = 10.9, 5.1 Hz, 1H), 3.11 (dd, J = 15.2, 8.0 Hz, 1H), 1.28 (s, 3H), 1.25 (s, 3H), 1.14 (d, J = 6.5 Hz, 3H), 1.06 (s, 3H), 0.94 (s, 3H), 0.92 (s, 3H), 0.86 (s, 3H). 13C-NMR (125 MHz, C5D5N) δ 145.0, 122.8, 101.7, 97.1, 83.8, 78.7, 78.3, 78.2, 78.0, 76.9, 73.0, 70.2, 69.6, 64.9, 62.2, 61.6, 55.8, 48.2, 47.3, 45.4, 42.0, 40.4, 39.5, 39.2, 37.3, 36.5, 34.8, 33.5, 33.3, 33.0, 31.1, 30.5, 30.0, 28.8, 28.2, 26.5, 26.5, 26.2, 24.0, 18.9, 17.4, 16.6, 15.8. HRMS (ESI) m/z for C37H64NO7 ([M+H]+) 634.4676, calc. 634.4677.
28-N-Methoxyaminooleanane-2-deoxy-d-galactoside (8f)
White solid (33.1 mg, 54%). 1H-NMR (500 MHz, C5D5N) δ 5.36 (d, J = 6.5 Hz, 0.33H, αH1′), 5.15 (t, J = 3.3 Hz, 1H), 4.69–4.62 (m, 1H), 4.52–4.35 (m, 3H), 4.32–4.28 (m, 0.67 H), 3.95 (t, J = 6.1 Hz, 1H), 3.67 (d, J = 8.1 Hz, 3H), 3.52–3.42 (m, 3H), 2.95 (d, J = 15.3 Hz, 1H), 1.28–1.25 (m, 6H), 1.12 (s, 3H), 1.07 (s, 3H), 0.95 (s, 6H), 0.88 (s, 3H). 13C-NMR (125 MHz, C5D5N) δ 144.8, 122.7, 98.0, 97.0, 92.8, 86.8, 86.3, 78.8, 78.0, 73.7, 73.1, 72.1, 71.8, 70.7, 68.6, 65.0, 64.7, 62.3, 55.7, 48.0, 48.0, 47.2, 45.1, 41.8, 41.8, 40.2, 39.3, 39.0, 37.2, 36.2, 34.7, 30.9, 28.7, 28.0, 26.3, 25.9, 23.8, 23.7, 23.6, 18.7, 17.3, 16.4, 15.7. HRMS (ESI) m/z for C37H64NO6 ([M+H]+) 618.4739, calc. 618.4728.
28-N-Methoxyaminooleanane-d-mannoside (8g)
White solid (33.5 mg, 52%). 1H-NMR (500 MHz, C5D5N) δ 5.24 (t, J = 5.4 Hz, 1H), 5.00 (d, J = 2.5 Hz, 0.5H, αH1′), 4.77–4.70 (m, 0.5H), 4.67 (dt, J = 8.3, 4.0 Hz, 1H), 4.59–4.43 (m, 2H), 4.38–4.17 (m, 1H), 3.99–3.57 (m, 5H), 3.54–3.32 (m, 2H), 3.03 (dd, J = 20.5, 14.9 Hz, 1H), 1.28 (d, J = 3.1 Hz, 3H), 1.27 (d, J = 2.8 Hz, 3H), 1.07 (t, J = 3.1 Hz, 3H), 0.98 (t, J = 3.5 Hz, 3H), 0.95 (s, 3H), 0.93 (d, J = 2.8 Hz, 3H), 0.89 (d, J = 11.2 Hz, 3H). 13C-NMR (125 MHz, C5D5N) δ 145.0, 122.9, 102.6, 96.1, 93.8, 82.0, 81.3, 78.2, 77.1, 73.4, 72.5, 71.5, 69.8, 65.2, 63.5, 62.3, 60.6, 55.9, 48.2, 47.3, 45.2, 42.0, 40.3, 39.5, 39.2, 37.3, 36.6, 35.0, 33.5, 31.6, 31.1, 30.5, 28.9, 28.2, 26.6, 26.4, 24.1, 24.0, 23.9, 18.9, 17.5, 16.6, 15.9. HRMS (ESI) m/z for C37H64NO7 ([M+H]+) 634.4672, calc. 634.4677.
28-N-Methoxyaminooleanane-d-arabinoside (8h)
White solid (35.2 mg, 58%). 1H-NMR (500 MHz, C5D5N) δ 5.25 (t, J = 3.1 Hz, 1H), 5.18 (d, J = 5.5 Hz, 0.67H, αH1′), 4.67–4.63 (m, 1H), 4.61–4.50 (m, 1H), 4.46–4.34 (m, 2H), 4.29–4.21 (m, 1H), 4.16 (dd, J = 8.9, 3.4 Hz, 0.33H), 3.89–3.78 (m, 3H), 3.49–3.42 (m, 2H), 3.34 (dd, J = 50.9, 15.5 Hz, 1H), 2.28 (dd, J = 13.6, 3.8 Hz, 0.33H), 1.30 (s, 3H), 1.26 (d, J = 1.8 Hz, 3H), 1.08 (d, J = 2.8 Hz, 3H), 1.06 (s, 3H), 0.95 (d, J = 3.4 Hz, 3H), 0.93 (s, 3H), 0.90 (d, J = 5.7 Hz, 3H). 13C-NMR (125 MHz, C5D5N) δ 144.8, 122.7, 102.0, 97.6, 84.3, 78.0, 77.7, 76.3, 70.2, 69.5, 69.4, 62.7, 61.9, 55.7, 48.0, 47.2, 45.1, 41.9, 40.2, 39.3, 39.1, 37.2, 36.5, 34.8, 33.3, 33.1, 33.0, 31.4, 30.9, 30.3, 29.9, 28.7, 28.1, 26.3, 26.0, 23.9, 23.7, 18.7, 17.5, 16.5, 15.7. HRMS (ESI) m/z for C36H62NO6 ([M+H]+) 604.4581, calc. 604.4572.
28-N-Methoxyaminooleanane-l-arabinoside (8i)
White solid (37.3 mg, 62%). 1H-NMR (500 MHz, C5D5N) δ 5.18 (d, J = 5.9 Hz, 0.5H, αH1′), 5.14 (t, J = 3.2 Hz, 2H), 4.63–4.59 (m, 0.5H), 4.53–4.41 (m, 2H), 4.31–4.11 (m, 3H), 3.73 (d, J = 6.3 Hz, 3H), 3.42 (dd, J = 10.9, 4.9 Hz, 1H), 3.01 (d, J = 15.5 Hz, 1H), 2.17 (d, J = 10.6 Hz, 3H), 1.25 (s, 3H), 1.21 (s, 3H), 1.11 (s, 3H), 1.02 (s, 3H), 0.94 (s, 3H), 0.88 (s, 3H), 0.78 (s, 3H). 13C-NMR (125 MHz, C5D5N) δ 144.9, 122.6, 101.4, 97.0, 84.6, 78.1, 76.2, 70.2, 69.6, 69.5, 55.8, 55.7, 48.1, 48.1, 47.1, 45.6, 41.9, 41.9, 40.3, 40.2, 39.4, 39.1, 37.3, 36.4, 33.4, 33.0, 31.5, 31.0, 30.4, 30.0, 28.8, 28.1, 26.4, 26.1, 23.9, 23.9, 23.5, 18.8, 17.4, 16.5, 15.8. HRMS (ESI) m/z for C36H62NO6 ([M+H]+) 604.4576, calc. 604.4572.
28-N-Methoxyaminooleanane-d-fucoside (8j)
White solid (35.2 mg, 57%). 1H-NMR (500 MHz, C5D5N) δ 5.15 (s, 1H), 4.51 (dt, J = 17.5, 8.7 Hz, 2H), 4.18 (dd, J = 8.7, 3.4 Hz, 1H), 4.09 (d, J = 2.8 Hz, 1H), 3.88 (q, J = 6.0 Hz, 1H), 3.78 (s, 3H), 3.66 (t, J = 9.8 Hz, 1H), 3.47 (dd, J = 11.0, 4.9 Hz, 1H), 3.10–3.04 (m, 1H), 1.29 (s, 3H), 1.27 (s, 3H), 1.19 (s, 3H), 1.08 (s, 3H), 0.99 (s, 3H), 0.93 (s, 3H), 0.86 (s, 3H). 13C-NMR (125 MHz, C5D5N) δ 145.5, 123.3, 97.2, 87.6, 78.7, 77.4, 74.0, 73.5, 69.7, 56.4, 48.7, 47.8, 46.1, 42.5, 40.9, 40.0, 39.7, 37.9, 37.0, 35.3, 34.0, 33.9, 33.6, 32.1, 31.6, 31.0, 30.6, 29.4, 28.7, 27.0, 26.7, 24.5, 19.4, 18.1, 18.0, 17.1, 16.4. HRMS (ESI) m/z for C37H64NO6 ([M+H]+)618.4733, calc. 618.4728.
28-N-Methoxyaminooleanane-β-l-fucoside (8k)
White solid (38.9 mg, 63%). 1H-NMR (500 MHz, C5D5N) δ 5.19 (d, J = 3.2 Hz, 1H), 4.49 (t, J = 8.8 Hz, 1H), 4.44 (d, J = 8.7 Hz, 1H), 4.10 (dd, J = 8.8, 3.4 Hz, 1H), 4.04 (d, J = 2.9 Hz, 1H), 3.80 (d, J = 6.9 Hz, 1H), 3.74 (s, 3H), 3.42 (dd, J = 10.9, 4.9 Hz, 1H), 3.35 (dd, J = 14.9, 3.9 Hz, 1H), 3.26 (d, J = 15.0 Hz, 1H), 2.26 (dd, J = 13.4, 3.9 Hz, 1H), 1.25 (s, 3H), 1.22 (s, 3H), 1.14 (s, 3H), 1.02 (d, J = 2.3 Hz, 3H), 0.97 (s, 3H), 0.91 (s, 6H). 13C-NMR (125 MHz, C5D5N) δ 145.0, 122.7, 96.9, 86.6, 78.0, 76.9, 73.3, 72.8, 68.9, 55.7, 48.0, 47.3, 44.3, 41.9, 40.2, 39.3, 39.1, 37.2, 37.2, 36.4, 34.8, 33.3, 33.0, 32.8, 31.0, 28.7, 28.0, 26.2, 25.9, 23.9, 23.8, 20.5, 18.7, 17.4, 17.4, 16.5, 15.7. HRMS (ESI) m/z for C37H64NO6 ([M+H]+) 618.4732, calc. 618.4728.
28-N-Methoxyaminooleanane-β-d-xyloside (8l)
White solid (38.2 mg, 63%). 1H-NMR (500 MHz, C5D5N) δ 5.26 (t, J = 3.3 Hz, 1H), 4.57 (d, J = 6.7 Hz, 1H), 4.40 (dd, J = 10.8, 4.5 Hz, 1H), 4.19 (d, J = 13.9 Hz, 3H), 3.80 (s, 3H), 3.69 (t, J = 10.4 Hz, 1H), 3.52 (d, J = 15.3 Hz, 1H), 3.41 (t, J = 14.6 Hz, 1H), 3.03 (d, J = 15.4 Hz, 1H), 2.25–2.14 (m, 1H), 1.26 (s, 3H), 1.23 (s, 3H), 1.10 (s, 3H), 1.04 (s, 3H), 0.92 (s, 3H), 0.92 (s, 3H), 0.89 (s, 3H). 13C-NMR (125 MHz, C5D5N) δ 144.9, 122.7, 97.1, 80.1, 78.0, 71.7, 70.9, 69.2, 55.7, 48.0, 47.0, 45.3, 41.8, 40.2, 39.3, 39.0, 37.2, 36.2, 34.7, 33.3, 33.1, 32.8, 31.4, 31.0, 30.3, 28.7, 28.0, 26.3, 26.1, 23.8, 23.8, 23.6, 18.7, 17.2, 16.4, 15.7. HRMS (ESI) m/z for C36H62NO6 ([M+H]+) 604.4572, calc. 604.4572.
28-N-Methoxyaminooleanane-β-l-xyloside (8m)
White solid (37.2 mg, 62%). 1H-NMR (500 MHz, C5D5N) δ 5.30 (t, J = 3.2 Hz, 1H), 4.55 (d, J = 8.7 Hz, 1H), 4.43 (dd, J = 11.1, 5.2 Hz, 1H), 4.28–4.18 (m, 3H), 3.85 (s, 3H), 3.70 (t, J = 10.7 Hz, 1H), 3.49–3.43 (m, 1H), 3.34 (d, J = 5.8 Hz, 2H), 2.29 (dd, J = 13.3, 3.8 Hz, 1H), 1.29 (s, 3H), 1.25 (s, 3H), 1.17 (s, 3H), 1.06 (s, 3H), 0.99 (s, 3H), 0.95 (s, 3H), 0.92 (s, 3H). 13C-NMR (125 MHz, C5D5N) δ 145.0, 122.8, 97.8, 80.2, 78.1, 71.7, 71.1, 69.3, 55.7, 49.6, 48.0, 47.2, 44.7, 42.0, 40.3, 39.4, 39.1, 37.2, 36.5, 34.8, 33.4, 33.1, 33.0, 31.5, 31.0, 30.3, 28.7, 28.1, 26.3, 26.0, 23.9, 23.8, 18.8, 17.5, 16.5, 15.7. HRMS (ESI) m/z for C36H62NO6 ([M+H]+) 604.4578, calc. 604.4572.
28-N-Methoxyaminooleanane-l-lyxoside (8n)
White solid (34.2 mg, 57%). 1H-NMR (500 MHz, C5D5N) δ 5.88 (d, J = 3.5 Hz, 0.2H, αH1′), 5.22 (d, J = 3.2 Hz, 1H), 4.81 (d, J = 3.3 Hz, 1H), 4.78–4.72 (m, 1H), 4.72–4.65 (m, 0.8H), 4.49–4.42 (m, 1H), 4.35 (dd, J = 11.2, 4.2 Hz, 1H), 4.23 (dd, J = 18.4, 8.8 Hz, 1H), 3.97–3.78 (m, 3H), 3.60–3.43 (m, 2H), 3.09 (dd, J = 16.5, 15.1 Hz, 1H), 1.29–1.27 (m, 3H), 1.25 (d, J = 11.2 Hz, 3H), 1.16 (d, J = 5.4 Hz, 3H), 1.09–1.06 (m, 3H), 0.98 (d, J = 7.0 Hz, 3H), 0.95 (d, J = 3.8 Hz, 3H), 0.91 (d, J = 3.2 Hz, 3H). 13C-NMR (125 MHz, C5D5N) δ 145.56, 123.23, 102.50, 96.98, 83.26, 78.68, 73.85, 73.61, 73.41, 71.79, 69.99, 68.58, 67.66, 56.35, 50.22, 48.67, 47.71, 46.17, 42.49, 40.87, 39.97, 39.69, 37.83, 36.97, 35.39, 34.02, 33.98, 33.79, 33.52, 31.59, 30.94, 29.33, 28.70, 26.97, 26.73, 24.49, 24.43, 19.39, 17.92, 17.11, 16.34. HRMS (ESI) m/z for C36H62NO6 ([M+H]+) 604.4575, calc. 604.4572.
28-N-Methoxyaminooleanane-l-rhamnoside (8o)
White solid (34.7 mg, 56%). 1H-NMR (500 MHz, C5D5N) δ 5.33 (t, J = 3.3 Hz, 1H), 5.28 (d, J = 6.1 Hz, 0.2H, βH1′), 4.69 (d, J = 2.9 Hz, 0.8H, αH1′), 4.34–4.29 (m, 1H), 4.19 (t, J = 9.1 Hz, 1H), 4.07 (dd, J = 9.2, 3.0 Hz, 1H), 3.84 (d, J = 7.8 Hz, 3H), 3.76 (dq, J = 9.3, 6.1 Hz, 1H), 3.51–3.39 (m, 4H), 3.12 (d, J = 14.6 Hz, 1H), 1.18 (s, 3H), 1.08 (d, J = 3.1 Hz, 3H), 1.07 (s, 3H), 1.01 (s, 3H), 0.99 (s, 3H), 0.98 (s, 6H). 13C-NMR (125 MHz, C5D5N) δ 145.8, 123.2, 102.2, 95.3, 86.4, 78.7, 77.3, 76.9, 74.6, 72.5, 63.8, 62.3, 61.6, 56.4, 48.7, 48.0, 45.2, 42.5, 40.9, 40.0, 40.0, 39.7, 37.9, 37.1, 34.0, 32.1, 31.6, 31.6, 31.0, 30.6, 29.4, 29.3, 28.7, 27.0, 26.6, 24.5, 24.4, 19.4, 19.3, 18.3, 17.1, 17.1, 16.4. HRMS (ESI) m/z for C37H64NO6 ([M+H]+) 618.4733, calc. 618.4728.
28-N-Methoxyaminooleanane-d-riboside (8p)
White solid (34.9 mg, 58%). 1H-NMR (500 MHz, C5D5N) δ 5.29 (t, J = 3.0 Hz, 1H), 5.23 (d, J = 3.4 Hz, 0.5H, αH1′), 4.80–4.62 (m, 2H), 4.35 (dd, J = 11.6, 3.7 Hz, 0.5H), 4.31–4.14 (m, 3H), 3.91–3.77 (m, 3H), 3.50–3.34 (m, 2H), 3.04 (dd, J = 38.0, 15.1 Hz, 1H), 1.28 (d, J = 2.7 Hz, 3H), 1.27 (s, 3H), 1.08 (d, J = 2.6 Hz, 3H), 0.97 (dd, J = 12.2, 3.2 Hz, 6H), 0.95 (d, J = 4.4 Hz, 3H), 0.93 (d, J = 3.9 Hz, 3H). 13C-NMR (125 MHz, C5D5N) δ 145.3, 122.9, 103.6, 93.4, 84.7, 78.3, 73.0, 72.6, 69.3, 68.8, 66.4, 64.1, 62.2, 56.0, 55.9, 48.2, 47.3, 45.6, 42.1, 40.5, 39.6, 39.3, 37.4, 36.6, 33.6, 31.7, 31.3, 30.6, 29.0, 28.9, 28.3, 26.6, 26.5, 26.4, 24.1, 24.1, 24.0, 19.0, 17.4, 16.7, 15.9. HRMS (ESI) m/z for C36H62NO6 ([M+H]+) 604.4580, calc. 604.4572.
28-N-Methoxyaminooleanane-l-riboside (8q)
White solid (33.2 mg, 55%). 1H-NMR (500 MHz, C5D5N) δ 5.24 (t, J = 3.2 Hz, 1H), 5.20 (d, J = 3.1 Hz, 0.5H, αH1′), 4.81–4.60 (m, 2H), 4.38 (dd, J = 11.7, 3.6 Hz, 0.5H), 4.31–4.18 (m, 3H), 3.84 (s, 3H), 3.46 (dd, J = 10.8, 5.1 Hz, 1H), 3.33 (dd, J = 14.9, 8.7 Hz, 1H), 2.98 (d, J = 14.8 Hz, 1H), 1.26 (s, 3H), 1.17 (s, 3H), 1.07 (s, 3H), 1.03 (s, 3H), 0.95 (s, 3H), 0.94 (d, J = 2.3 Hz, 3H), 0.93 (s, 3H). 13C-NMR (125 MHz, C5D5N) δ 145.0, 123.0, 104.3, 94.0, 84.3, 78.3, 73.1, 72.7, 69.2, 68.9, 64.0, 62.5, 55.9, 48.3, 47.3, 45.2, 44.9, 42.2, 42.1, 40.6, 40.4, 39.6, 39.3, 37.4, 37.4, 36.8, 36.7, 35.0, 33.6, 33.6, 31.2, 28.9, 28.3, 26.5, 26.5, 26.3, 24.1, 24.0, 23.8, 19.0, 17.6, 16.7, 15.9. HRMS (ESI) m/z for C36H62NO6 ([M+H]+) 604.4575, calc. 604.4572.
28-N-Methoxyaminooleanane-2-deoxy-d-riboside (8r)
White solid (33.9 mg, 58%). 1H-NMR (500 MHz, C5D5N) δ 5.30 (d, J = 3.3 Hz, 0.8H, αH1′), 5.25 (s, 1H), 4.70–4.52 (m, 1H), 4.47–4.34 (m, 1H), 4.28 (d, J = 10.5 Hz, 0.2H), 4.26–4.07 (m, 3H), 3.66 (d, J = 10.0 Hz, 3H), 3.50–3.44 (m, 2H), 1.30 (s, 3H), 1.28 (d, J = 3.5 Hz, 3H), 1.27 (s, 3H), 1.08 (s, 3H), 0.98 (d, J = 6.0 Hz, 3H), 0.97 (t, J = 3.0 Hz, 3H), 0.95 (s, 3H). 13C-NMR (125 MHz, C5D5N) δ 145.6, 123.4, 98.5, 93.5, 88.3, 78.7, 72.8, 72.1, 68.9, 68.9, 67.2, 62.6, 61.7, 60.3, 56.3, 48.6, 47.7, 47.2, 46.0, 45.6, 42.5, 40.9, 39.99, 39.72, 37.8, 36.8, 34.0, 31.6, 31.0, 29.3, 28.7, 27.0, 26.8, 26.7, 24.5, 24.4, 24.4, 19.4, 17.8, 17.1, 16.4. HRMS (ESI) m/z for C36H62NO5 ([M+H]+) 588.4616, calc. 588.4623.
Oleanolic acid-28-O-β-d-glucopyranoside (1a)
White solid (75.2 mg, 28%). 1H-NMR (500 MHz, C5D5N) δ 6.34 (d, J = 8.0 Hz, 1H), 5.46 (s, 1H), 4.48–4.03 (m, 6H), 3.44 (dd, J = 10.9, 5.1 Hz, 1H), 3.22 (dd, J = 14.1, 4.4 Hz, 1H), 1.25 (s, 3H), 1.23 (s, 3H), 1.14 (s, 3H), 1.03 (s, 3H), 0.93 (s, 3H), 0.90 (s, 3H), 0.88 (s, 3H). 13C-NMR (125 MHz, C5D5N) δ 176.5, 144.2, 123.0, 95.8, 79.4, 78.9, 78.1, 75.7, 74.2, 62.3, 55.8, 48.2, 47.1, 46.3, 42.2, 41.8, 39.9, 39.4, 39.0, 37.4, 34.0, 33.2, 33.2, 32.6, 31.0, 29.1, 28.8, 28.3, 28.1, 26.1, 23.9, 23.5, 18.9, 17.6, 16.6, 15.7 HRMS (ESI) m/z for C36H58O8Na ([M+Na]+) 641.4029, calc. 641.4024.
Erythrodiol-28-O-β-d-glucopyranoside (1b)
White solid (62.8 mg, 23%). 1H-NMR (500 MHz, C5D5N) δ 5.24 (s, 1H), 4.92 (d, J = 7.7 Hz, 1H), 4.62 (d, J = 11.7 Hz, 1H), 4.48 (dd, J = 11.2, 4.4 Hz, 1H), 4.35–4.28 (m, 2H), 4.10 (dd, J = 17.3, 8.2 Hz, 2H), 3.86 (s, 2H), 3.51–3.41 (m, 1H), 1.26 (s, 6H), 1.08 (s, 3H), 1.02 (s, 3H), 0.97 (s, 3H), 0.92 (s, 3H), 0.88 (s, 3H). 13C-NMR (125 MHz, C5D5N) δ 144.7, 122.8, 105.9, 78.8, 78.6, 78.1, 77.2, 75.4, 71.8, 63.0, 55.8, 48.1, 46.7, 43.2, 42.0, 40.2, 39.5, 39.2, 37.3, 37.2, 34.5, 33.5, 32.9, 32.7, 31.2, 28.8, 28.2, 28.2, 26.3, 23.9, 23.9, 22.1, 18.9, 17.1, 16.6, 15.9. HRMS (ESI) m/z for C36H60O7Na ([M+Na]+) 627.4231, calc. 627.4231.