New Acetophenones and Chromenes from the Leaves of Melicope barbigera A. Gray
Abstract
1. Introduction
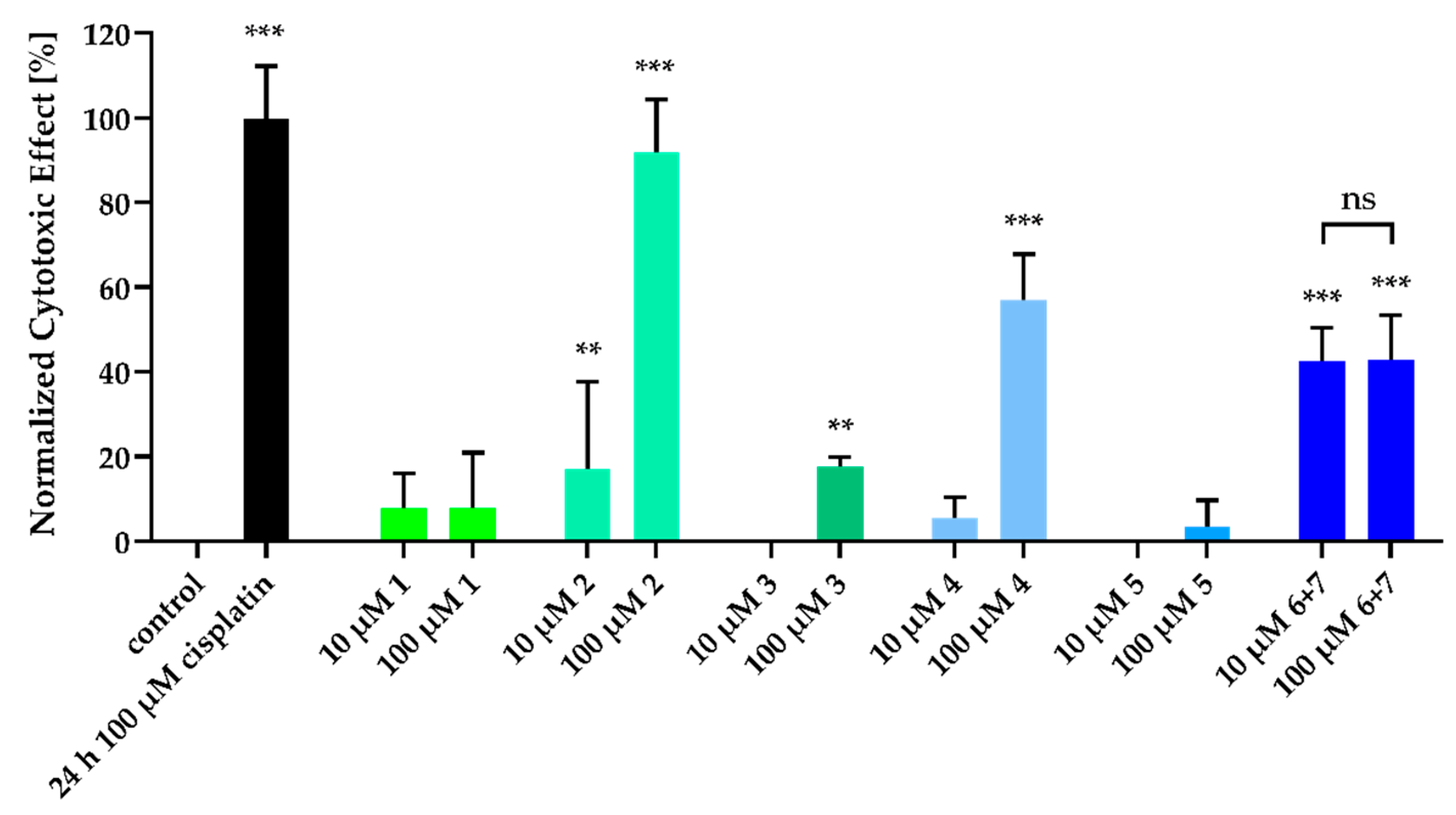
2. Results and Discussion
3. Materials and Methods
3.1. General Experimental Procedures
3.2. Plant Material
3.3. Extraction and Isolation
3.4. Preparation of (R)-and (S)-MTPA Esters
3.5. Cell Lines and Cell Culture
3.6. Cytotoxicity Assay (Nuclear Shrinkage)
3.7. Caspase 3/7-Activation Assay
3.8. MTT-Assay
3.9. Data Analysis
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Appelhans, M.S.; Wen, J.; Duretto, M.; Crayn, D.; Wagner, W.L. Historical biogeography of Melicope (Rutaceae) and its close relatives with a special emphasis on Pacific dispersals. J. Syst. Evol. 2018, 56, 576–599. [Google Scholar] [CrossRef]
- Hartley, T.G. On the Taxonomy and Biogeography of Euodia and Melicope (Rutaceae). Allertonia 2000, 8, 1–319. [Google Scholar]
- Paetzold, C.; Wood, K.R.; Eaton, D.; Wagner, W.L.; Appelhans, M.S. Phylogeny of Hawaiian Melicope (Rutaceae): RAD-seq resolves species relationships and reveals ancient introgression of Hawaiian Melicope. Front. Plant. Sci. 2019, 10, 1074. [Google Scholar] [CrossRef]
- Wagner, W.L.; Herbst, D.R.; Sohmer, S.H. Manual of the Flowering Plants of Hawai’i, Vols. 1 and 2; University of Hawai’i and Bishop Museum Press: Honolulu, HI, USA, 1999. [Google Scholar]
- Wood, K.R. Possible Extinctions, Rediscoveries, and New Plant Records within the Hawaiian Islands. Bish. Mus. Occas. Pap. 2012, 113, 91–102. [Google Scholar]
- Wood, K.R.; Appelhans, M.S.; Wagner, W.L. Melicope oppenheimeri, section Pelea (Rutaceae), a new species from West Maui, Hawaiian Islands: With notes on its ecology, conservation, and phylogenetic placement. PhytoKeys 2016, 69, 51–64. [Google Scholar] [CrossRef]
- Wood, K.R.; Appelhans, M.S.; Wagner, W.L. Melicope stonei, section Pelea (Rutaceae), a new species from Kaua‘i, Hawaiian Islands: With notes on its distribution, ecology, conservation status, and phylogenetic placement. PhytoKeys 2017, 83, 119–132. [Google Scholar] [CrossRef]
- Wood, K.R. Rediscovery, conservation status and taxonomic assessment of Melicope degeneri (Rutaceae), Kaua’i, Hawai’i. Endanger. Species Res. 2011, 14, 61–68. [Google Scholar] [CrossRef]
- Xu, J.; Sun, X.; Liu, X.; Peng, M.; Li, S.; Jin, D.-Q.; Lee, D.; Bartlam, M.; Guo, Y. Phytochemical constituents from Melicope pteleifolia that promote neurite outgrowth in PC12 cells. J. Funct. Foods 2016, 23, 565–572. [Google Scholar] [CrossRef]
- Nakashima, K.-i.; Oyama, M.; Ito, T.; Akao, Y.; Witono, J.R.; Darnaedi, D.; Tanaka, T.; Murata, J.; Iinuma, M. Novel quinolinone alkaloids bearing a lignoid moiety and related constituents in the leaves of Melicope denhamii. Tetrahedron 2012, 68, 2421–2428. [Google Scholar] [CrossRef]
- Saputri, R.D.; Tjahjandarie, T.S.; Tanjung, M. Two novel coumarins bearing an acetophenone derivative from the leaves of Melicope Quercifolia. Nat. Prod. Res. 2019, 1–6. [Google Scholar] [CrossRef]
- Tjahjandarie, T.S.; Saputri, R.D.; Hasanah, U.; Rachmadiarti, F.; Tanjung, M. 5,7-Dihydroxy-3,6-Dimethoxy-3′,4′-Methylendioxyflavone. Molbank 2018, 2018, M1007. [Google Scholar] [CrossRef]
- Xu, J.-F.; Han, C.; Xu, Q.-Q.; Wang, X.-B.; Zhao, H.-J.; Xue, G.-M.; Luo, J.-G.; Kong, L.-Y. Isolation, Chiral-Phase Resolution, and Determination of the Absolute Configurations of a Complete Series of Stereoisomers of a Rearranged Acetophenone with Three Stereocenters. J. Nat. Prod. 2019, 82, 1399–1404. [Google Scholar] [CrossRef] [PubMed]
- Vu, V.-T.; Nguyen, M.-T.; Khoi, N.-M.; Xu, X.-J.; Kong, L.-Y.; Luo, J.-G. New lignans and acetophenone derivatives with α-glucosidase inhibitory activity from the leaves of Melicope patulinervia. Fitoterapia 2021, 148, 104805. [Google Scholar] [CrossRef] [PubMed]
- Yao, Q.; Gao, Y.; Lai, C.; Wu, C.; Zhao, C.-L.; Wu, J.-L.; Tang, D.-X. The phytochemistry, pharmacology and applications of Melicope pteleifolia: A review. J. Ethnopharmacol. 2020, 251, 112546. [Google Scholar] [CrossRef]
- Johnson, A.J.; Kumar, A.; Rasheed, S.A.; Chandrika, S.P.; Chandrasekhar, A.; Baby, S.; Subramoniam, A. Antipyretic, analgesic, anti-inflammatory and antioxidant activities of two major chromenes from Melicope lunu-ankenda. J. Ethnopharmacol. 2010, 130, 267–271. [Google Scholar] [CrossRef]
- Simonsen, H.T.; Adsersen, A.; Bremner, P.; Heinrich, M.; Wagner Smitt, U.; Jaroszewski, J.W. Antifungal constituents of Melicope borbonica. Phytother. Res. 2004, 18, 542–545. [Google Scholar] [CrossRef]
- Passreiter, C.M.; Suckow-Schnitker, A.-K.; Kulawik, A.; Addae-Kyereme, J.; Wright, C.W.; Wätjen, W. Prenylated flavanone derivatives isolated from Erythrina addisoniae are potent inducers of apoptotic cell death. Phytochemistry 2015, 117, 237–244. [Google Scholar]
- Hauschild, W.; Mutiso, P.B.C.; Passreiter, C.M. Prenylated pterocarpanes from Erythrina melanacantha. Nat. Prod. Commun. 2010, 5, 721–725. [Google Scholar] [CrossRef]
- Higa, T.; Scheuer, P.J. Hawaiian plant studies. Part XVI. Coumarins and flavones from Pelea barbigera (Gray) Hillebrand (Rutaceae). J. Chem. Soc. Perkin Trans. 1974, 1, 1350–1352. [Google Scholar] [CrossRef]
- Kamperdick, C.; Van, N.H.; van Sung, T.; Adam, G. Benzopyrans from Melicope ptelefolia leaves. Phytochemistry 1997, 45, 1049–1056. [Google Scholar] [CrossRef]
- Goh, S.H.; Chung, V.C.; Sha, C.K.; Mak, T.C.W. Monoterpenoid phloroacetophenones from Euodia latifolia. Phytochemistry 1990, 29, 1704–1706. [Google Scholar] [CrossRef]
- Ohtani, I.; Kusumi, T.; Kashman, Y.; Kakisawa, H. High-field FT NMR application of Mosher’s method. The absolute configurations of marine terpenoids. J. Am. Chem. Soc. 1991, 113, 4092–4096. [Google Scholar] [CrossRef]
- Hoye, T.R.; Jeffrey, C.S.; Shao, F. Mosher ester analysis for the determination of absolute configuration of stereogenic (chiral) carbinol carbons. Nat. Protoc. 2007, 2, 2451–2458. [Google Scholar] [PubMed]
- Su, C.-R.; Kuo, P.-C.; Wang, M.-L.; Liou, M.-J.; Damu, A.G.; Wu, T.-S. Acetophenone Derivatives from Acronychia pedunculata. J. Nat. Prod. 2003, 66, 990–993. [Google Scholar] [CrossRef] [PubMed]
- Appelhans, M.S.; Wen, J.; Wagner, W.L. A molecular phylogeny of Acronychia, Euodia, Melicope and relatives (Rutaceae) reveals polyphyletic genera and key innovations for species richness. Mol. Phylogenet. Evol. 2014, 79, 54–68. [Google Scholar] [CrossRef] [PubMed]
- Adsersen, A.; Smitt, U.W.; Simonsen, H.T.; Christensen, S.B.; Jaroszewski, J.W. Prenylated acetophenones from Melicope obscura and Melicope obtusifolia ssp. obtusifolia var. arborea and their distribution in Rutaceae. Biochem. Syst. Ecol. 2007, 35, 447–453. [Google Scholar] [CrossRef]
- Yang, L.-J.; Jiang, K.; Tan, J.-J.; Qu, S.-J.; Luo, H.-F.; Tan, C.-H.; Zhu, D.-Y. Prenylated Benzene Metabolites from Melicope pteleifolia. Helv. Chim. Acta 2013, 96, 119–123. [Google Scholar] [CrossRef]
- Lee, Y.R.; Li, X.; Lee, S.W.; Yong, C.S.; Hwang, M.; Lyoo, W.S. Concise total synthesis of biologically interesting prenylated chalcone natural products: 4′-O-methylxanthohumol, xanthohumol E, and sericone. Bull. Korean Chem. Soc. 2008, 29, 1205–1210. [Google Scholar]
- Lee, Y.; Xia, L. Concise Total Synthesis of Biologically Interesting Pyranochalcone Natural Products: Citrunobin, Boesenbergin A, Boesenbergin B, Xanthohumol, C, and Glabrachromene. Synthesis 2007, 20, 3240–3246. [Google Scholar] [CrossRef]
- Schmidt, S.; Jürgenliemk, G.; Schmidt, T.J.; Skaltsa, H.; Heilmann, J. Bi-, tri-, and Polycyclic Acylphloroglucinols from Hypericum empetrifolium. J. Nat. Prod. 2012, 75, 1697–1705. [Google Scholar] [CrossRef]
- Su, B.-N.; Park, E.J.; Mbwambo, Z.H.; Santarsiero, B.D.; Mesecar, A.D.; Fong, H.H.S.; Pezzuto, J.M.; Kinghorn, A.D. New Chemical Constituents of Euphorbia quinquecostata and Absolute Configuration Assignment by a Convenient Mosher Ester Procedure Carried Out in NMR Tubes. J. Nat. Prod. 2002, 65, 1278–1282. [Google Scholar] [CrossRef] [PubMed]
- Miyake, K.; Suzuki, A.; Morita, C.; Goto, M.; Newman, D.J.; O’Keefe, B.R.; Morris-Natschke, S.L.; Lee, K.-H.; Nakagawa-Goto, K. Acetophenone Monomers from Acronychia trifoliolata. J. Nat. Prod. 2016, 79, 2883–2889. [Google Scholar] [CrossRef] [PubMed]
- Hemphill, C.F.P.; Sureechatchaiyan, P.; Kassack, M.U.; Orfali, R.S.; Lin, W.; Daletos, G.; Proksch, P. OSMAC approach leads to new fusarielin metabolites from Fusarium tricinctum. J. Antibiot. 2017, 70, 726–732. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Dai, H.; Makhloufi, G.; Heering, C.; Janiak, C.; Hartmann, R.; Mándi, A.; Kurtán, T.; Müller, W.E.G.; Kassack, M.U. Cytotoxic 14-membered macrolides from a mangrove-derived endophytic fungus, Pestalotiopsis microspora. J. Nat. Prod. 2016, 79, 2332–2340. [Google Scholar] [CrossRef]
- Wang, C.; Engelke, L.; Bickel, D.; Hamacher, A.; Frank, M.; Proksch, P.; Gohlke, H.; Kassack, M.U. The tetrahydroxanthone-dimer phomoxanthone A is a strong inducer of apoptosis in cisplatin-resistant solid cancer cells. Bioorg. Med. Chem. 2019, 27, 115044. [Google Scholar] [CrossRef]
- Ishikawa, C.; Senba, M.; Mori, N. Anti-adult T-cell leukemia/lymphoma activity of cerdulatinib, a dual SYK/JAK kinase inhibitor. Int. J. Oncol. 2018, 53, 1681–1690. [Google Scholar] [CrossRef]
- Tayeh, M.; Nilwarangkoon, S.; Tanunyutthawongse, C.; Mahabusarakum, W.; Watanapokasin, R. Apoptosis and antimigration induction in human skin cancer cells by rhodomyrtone. Exp. Ther. Med. 2018, 15, 5035–5040. [Google Scholar] [CrossRef]
- Kozaki, S.; Takenaka, Y.; Mizushina, Y.; Yamaura, T.; Tanahashi, T. Three acetophenones from Acronychia pedunculata. J. Nat. Med. 2014, 68, 421–426. [Google Scholar] [CrossRef]
- Huang, P.-L.; Won, S.-J.; Day, S.-H.; Lin, C.-N. A Cytotoxic Acetophenone with a Novel Skeleton, Isolated from Cynanchum taiwanianum. Helv. Chim. Acta 1999, 82, 1716–1720. [Google Scholar] [CrossRef]
- El Gaafary, M.; Ezzat, S.M.; El Sayed, A.M.; Sabry, O.M.; Hafner, S.; Lang, S.; Schmiech, M.; Syrovets, T.; Simmet, T. Acovenoside A Induces Mitotic Catastrophe Followed by Apoptosis in Non-Small-Cell Lung Cancer Cells. J. Nat. Prod. 2017, 80, 3203–3210. [Google Scholar] [CrossRef]
- Jitsuno, M.; Yokosuka, A.; Hashimoto, K.; Amano, O.; Sakagami, H.; Mimaki, Y. Chemical Constituents of Lycoris albiflora and their Cytotoxic Activities. Nat. Prod. Commun. 2011, 6, 187–192. [Google Scholar] [CrossRef] [PubMed]
- Sriphana, U.; Yenjai, C.; Koatthada, M. Cytotoxicity of chemical constituents from the roots of Knema globularia. Phytochem. Lett. 2016, 16, 129–133. [Google Scholar] [CrossRef]
- Giap, T.H.; Thoa, H.T.; Oanh, V.T.K.; Hang, N.T.M.; Dang, N.H.; Thuc, D.N.; van Hung, N.; Le Thanh, N. New Acetophenone and Cardanol Derivatives from Knema pachycarpa. Nat. Prod. Commun. 2019, 14. [Google Scholar] [CrossRef]
- Kouloura, E.; Halabalaki, M.; Lallemand, M.-C.; Nam, S.; Jove, R.; Litaudon, M.; Awang, K.; Hadi, H.A.; Skaltsounis, A.-L. Cytotoxic Prenylated Acetophenone Dimers from Acronychia pedunculata. J. Nat. Prod. 2012, 75, 1270–1276. [Google Scholar] [CrossRef] [PubMed]
- Ito, C.; Hosono, M.; Tokuda, H.; Wu, T.-S.; Itoigawa, M. Acetophenones from Acronychia pedunculata and their Cancer Chemopreventive Activity. Nat. Prod. Commun. 2016, 11, 1299–1302. [Google Scholar] [CrossRef] [PubMed]
- Oyama, M.; Bastow, K.F.; Tachibana, Y.; Shirataki, Y.; Yamaguchi, S.; Cragg, G.M.; Wu, T.-S.; Lee, K.-H. Antitumor agents 225. Acrofoliones A and B, two novel cytotoxic acetophenone dimers from Acronychia trifoliolata. Chin. Pharm. J. 2003, 55, 239–245. [Google Scholar]
- Wätjen, W.; Kulawik, A.; Suckow-Schnitker, A.K.; Chovolou, Y.; Rohrig, R.; Ruhl, S.; Kampkötter, A.; Addae-Kyereme, J.; Wright, C.W.; Passreiter, C.M. Pterocarpans phaseollin and neorautenol isolated from Erythrina addisoniae induce apoptotic cell death accompanied by inhibition of ERK phosphorylation. Toxicology 2007, 242, 71–79. [Google Scholar] [CrossRef]
- Koch, K.; Schulz, G.; Döring, W.; Büchter, C.; Havermann, S.; Mutiso, P.C.; Passreiter, C.; Wätjen, W. Abyssinone V, a prenylated flavonoid isolated from the stem bark of Erythrina melanacantha increases oxidative stress and decreases stress resistance in Caenorhabditis elegans. J. Pharm. Pharmacol. 2019, 71, 1007–1016. [Google Scholar] [CrossRef]
- Wätjen, W.; Suckow-Schnitker, A.K.; Rohrig, R.; Kulawik, A.; Addae-Kyereme, J.; Wright, C.W.; Passreiter, C.M. Prenylated Flavonoid Derivatives from the Bark of Erythrina addisoniae. J. Nat. Prod. 2008, 71, 735–738. [Google Scholar] [CrossRef]
- Xu, J.-F.; Han, C.; Xue, G.-M.; Wang, X.-B.; Luo, J.; Yang, M.-H.; Luo, J.-G.; Kong, L.-Y. Novel rearranged acetophenone derivatives possessing diverse architectures from the leaves of Melicope ptelefolia. Tetrahedron 2019, 75, 130784. [Google Scholar] [CrossRef]
- Li, W.; Rao, L.; Liu, Y.; He, Q.; Fan, Y.; You, Y.-X.; Su, Y.; Hu, F.; Xu, Y.-K.; Lin, B. (±)-Meliviticines A and B: Rearranged prenylated acetophenone derivatives from Melicope viticina and their antimicrobial activity. Bioorganic Chem. 2019, 90, 103099. [Google Scholar] [CrossRef] [PubMed]
- Xu, Q.-Q.; Chen, X.-L.; Xu, J.-F.; Wang, S.-B.; Luo, J.-G.; Kong, L.-Y. Acetophenone derivatives from the roots of Melicope ptelefolia. Fitoterapia 2019, 132, 40–45. [Google Scholar] [CrossRef]
- Simonsen, H.T. Four novel geminally dialkylated, non-aromatic acetophenone derivatives from Melicope coodeana. Phytochem. Lett. 2012, 5, 371–375. [Google Scholar] [CrossRef]
- Muyard, F.; Bissoue, A.N.; Bevalot, F.; Tillequin, F.; Cabalion, P.; Vaquette, J. Acetophenones and other constituents from the roots of Melicope erromangensis. Phytochemistry 1996, 42, 1175–1179. [Google Scholar] [CrossRef]
- Parsons, I.C.; Gray, A.I.; Hartley, T.G.; Waterman, P.G. Acetophenones and coumarins from stem bark and leaves of Melicope Stipitata. Phytochemistry 1994, 37, 565–570. [Google Scholar] [CrossRef]
- Chen, J.-J.; Cho, J.-Y.; Hwang, T.-L.; Chen, I.-S. Benzoic Acid Derivatives, Acetophenones, and Anti-inflammatory Constituents from Melicope semecarpifolia. J. Nat. Prod. 2008, 71, 71–75. [Google Scholar] [CrossRef]
- Nakashima, K.-i.; Oyama, M.; Ito, T.; Witono, J.R.; Darnaedi, D.; Tanaka, T.; Murata, J.; Iinuma, M. Melicodenines A and B, novel Diels–Alder type adducts isolated from Melicope denhamii. Tetrahedron Lett. 2011, 52, 4694–4696. [Google Scholar] [CrossRef]
- Niu, Q.-W.; Chen, N.-H.; Wu, Z.-N.; Luo, D.; Li, Y.-Y.; Zhang, Y.-B.; Li, Q.-G.; Li, Y.-L.; Wang, G.-C. Isolation and identification of new prenylated acetophenone derivatives from Acronychia oligophlebia. Nat. Prod. Res. 2019, 33, 2230–2235. [Google Scholar] [CrossRef]
- Robertson, L.P.; Lucantoni, L.; Duffy, S.; Avery, V.M.; Carroll, A.R. Acrotrione: An Oxidized Xanthene from the Roots of Acronychia pubescens. J. Nat. Prod. 2019, 82, 1019–1023. [Google Scholar] [CrossRef]
- Ito, C.; Matsui, T.; Ban, Y.; Wu, T.-S.; Itoigawa, M. Acetophenones Isolated from Acronychia pedunculata and their Anti-proliferative Activities. Nat. Prod. Commun. 2016, 11, 83–86. [Google Scholar] [CrossRef]
- Miyake, K.; Morita, C.; Suzuki, A.; Matsushita, N.; Saito, Y.; Goto, M.; Newman, D.J.; O’Keefe, B.R.; Lee, K.-H.; Nakagawa-Goto, K. Prenylated Acetophloroglucinol Dimers from Acronychia trifoliolata: Structure Elucidation and Total Synthesis. J. Nat. Prod. 2019, 82, 2852–2858. [Google Scholar] [CrossRef] [PubMed]
- Tran, T.D.; Olsson, M.A.; McMillan, D.J.; Cullen, J.K.; Parsons, P.G.; Reddell, P.W.; Ogbourne, S.M. Potent antibacterial prenylated acetophenones from the australian endemic plant Acronychia crassipetala. Antibiotics 2020, 9, 487. [Google Scholar] [CrossRef] [PubMed]
- Brophy, J.J.; Goldsack, R.J.; Forster, P.I. The Leaf Oils of the Australian species of Medicosma (Rutaceae). J. Essent. Oil Res. 2004, 16, 161–166. [Google Scholar] [CrossRef]
- Nguyen, N.H.; Ha, T.K.Q.; Choi, S.; Eum, S.; Lee, C.H.; Bach, T.T.; Chinh, V.T.; Oh, W.K. Chemical constituents from Melicope pteleifolia leaves. Phytochemistry 2016, 130, 291–300. [Google Scholar] [CrossRef] [PubMed]
- Chan, J.A.; Shultis, E.A.; Carr, S.A.; DeBrosse, C.W.; Eggleston, D.S.; Francis, T.A.; Hyland, L.J.; Johnson, W.P.; Killmer, L.B. Novel phloroglucinols from the plant Melicope sessiliflora (Rutaceae). J. Org. Chem. 1989, 54, 2098–2103. [Google Scholar] [CrossRef]
- Matsui, T.; Ito, C.; Kato, A.; Wu, T.-S.; Itoigawa, M. Acrofolione A and B, acetophenone dimers from Acronychia pendunculata, induce an apoptotic effect on human NALM-6 pre-B cell leukaemia cells. J. Pharm. Pharmacol 2019, 71, 348–361. [Google Scholar] [CrossRef]
- Wu, T.S.; Wang, M.L.; Jong, T.T.; McPhail, A.T.; McPhail, D.R.; Lee, K.H. X-ray Crystal Structure of Acrovestone, a Cytotoxic Principle from Acronychia pedunculata. J. Nat. Prod. 1989, 52, 1284–1289. [Google Scholar] [CrossRef]
- Shaari, K.; Safri, S.; Abas, F.; Lajis, N.H.; Israf, D.A. A geranylacetophenone from the leaves of Melicope ptelefolia. Nat. Prod. Res. 2006, 20, 415–419. [Google Scholar] [CrossRef]
- Xu, J.-F.; Zhao, H.-J.; Wang, X.-B.; Li, Z.-R.; Luo, J.; Yang, M.-H.; Yang, L.; Yu, W.-Y.; Yao, H.-Q.; Luo, J.-G.; et al. (±)-Melicolones A and B, Rearranged Prenylated Acetophenone Stereoisomers with an Unusual 9-oxatricyclo[3.2.1.1(3,8)]nonane core from the Leaves of Melicope ptelefolia. Org. Lett. 2015, 17, 146–149. [Google Scholar] [CrossRef] [PubMed]
- Sabulal, B.; George, V.; Shiburaj, S. Volatile Constituents and Antibacterial Activity of the Flower Oil of Evodia lunu-ankenda (Gaertn) Merr. J. Essent. Oil Res. 2006, 18, 462–464. [Google Scholar] [CrossRef]
- Cambie, R.C.; Pan, Y.J.; Bowden, B.F. Flavonoids of the barks of Melicope simplex and Melicope ternata. Biochem. Syst. Ecol. 1996, 5, 461–462. [Google Scholar] [CrossRef]
- Bandolik, J.J.; Hamacher, A.; Schrenk, C.; Weishaupt, R.; Kassack, M.U. Class i-histone deacetylase (hdac) inhibition is superior to pan-hdac inhibition in modulating cisplatin potency in high grade serous ovarian cancer cell lines. Int. J. Mol. Sci. 2019, 20, 3052. [Google Scholar] [CrossRef]
- Engelke, L.H.; Hamacher, A.; Proksch, P.; Kassack, M.U. Ellagic acid and resveratrol prevent the development of cisplatin resistance in the epithelial ovarian cancer cell line A2780. J. Cancer 2016, 7, 353. [Google Scholar] [CrossRef] [PubMed]
- Marek, L.; Hamacher, A.; Hansen, F.K.; Kuna, K.; Gohlke, H.; Kassack, M.U.; Kurz, T. Histone deacetylase (HDAC) inhibitors with a novel connecting unit linker region reveal a selectivity profile for HDAC4 and HDAC5 with improved activity against chemoresistant cancer cells. J. Med. Chem. 2013, 56, 427–436. [Google Scholar] [CrossRef]
- Hemmerich, W. StatistikGuru: Normalverteilung Online Prüfen. Available online: https://statistikguru.de/rechner/normalverteilung-rechner.html (accessed on 18 November 2020).
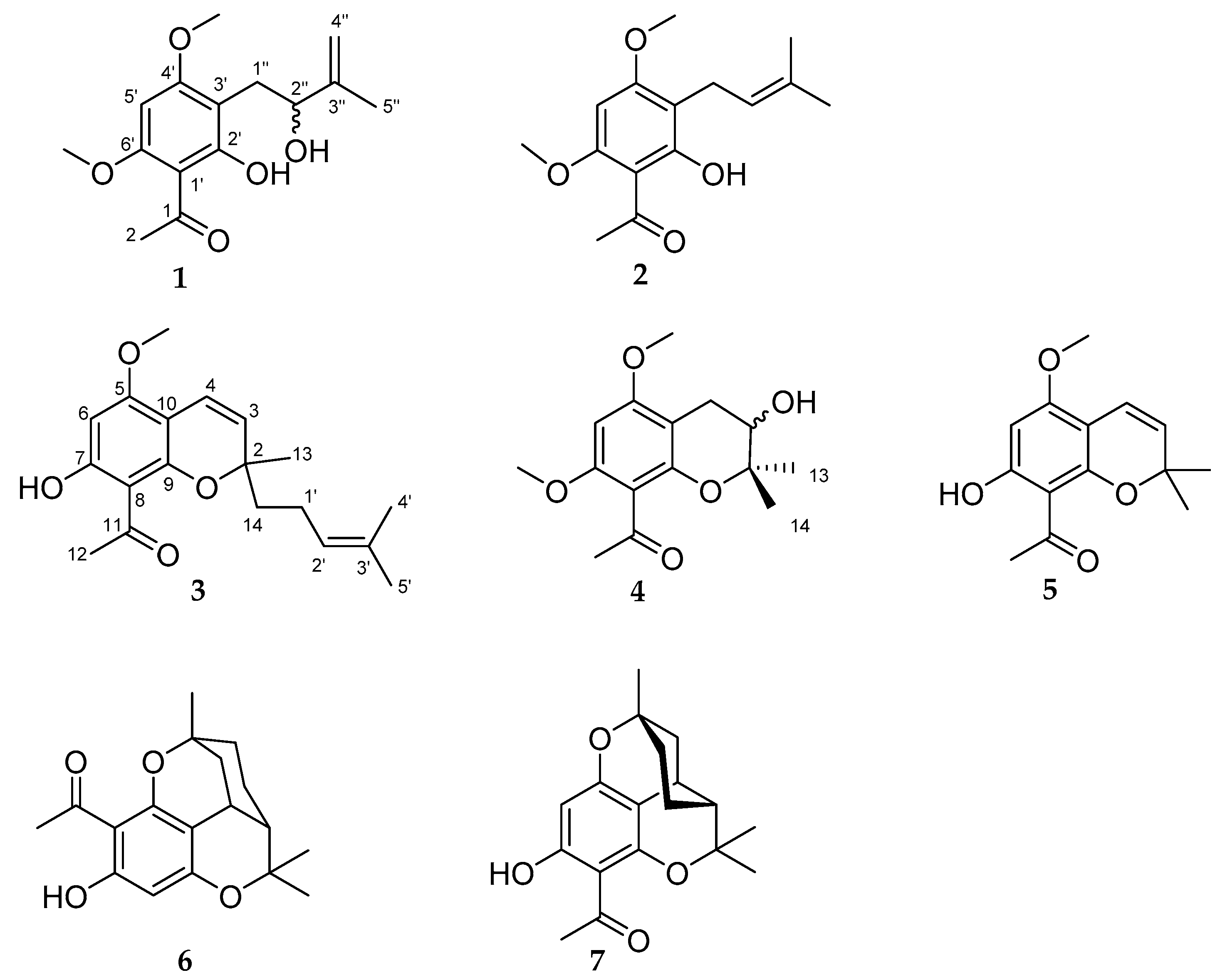
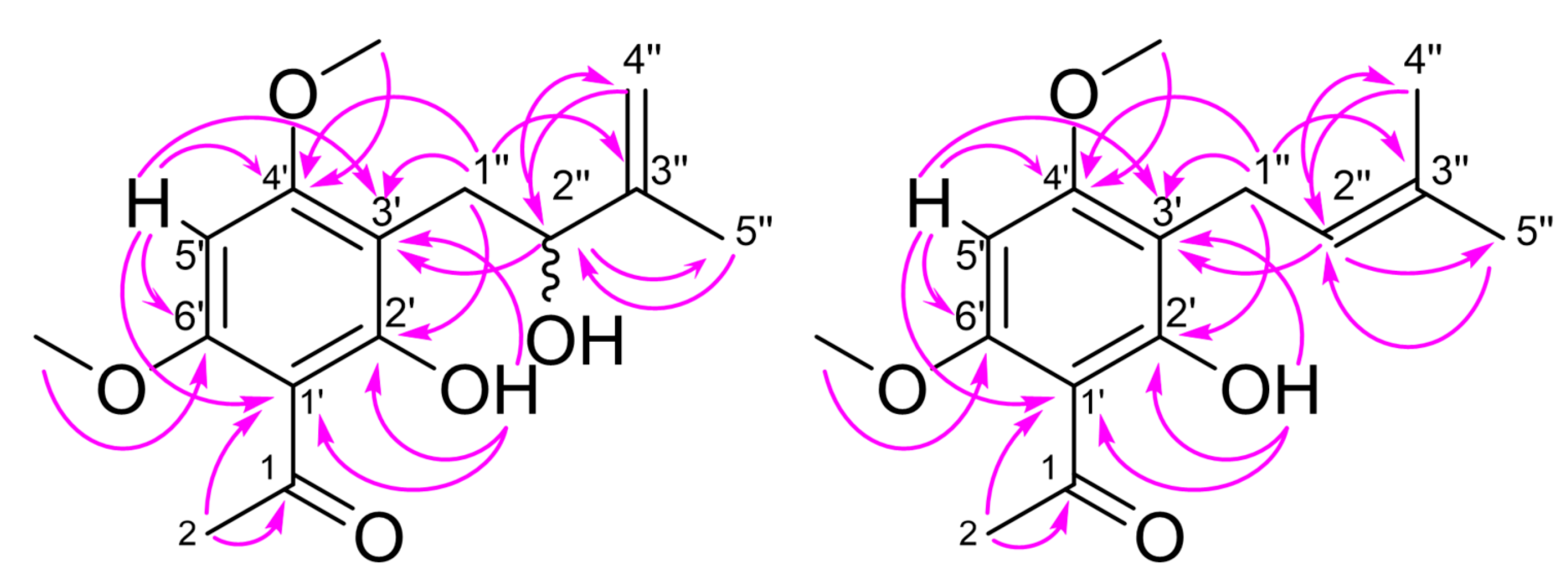

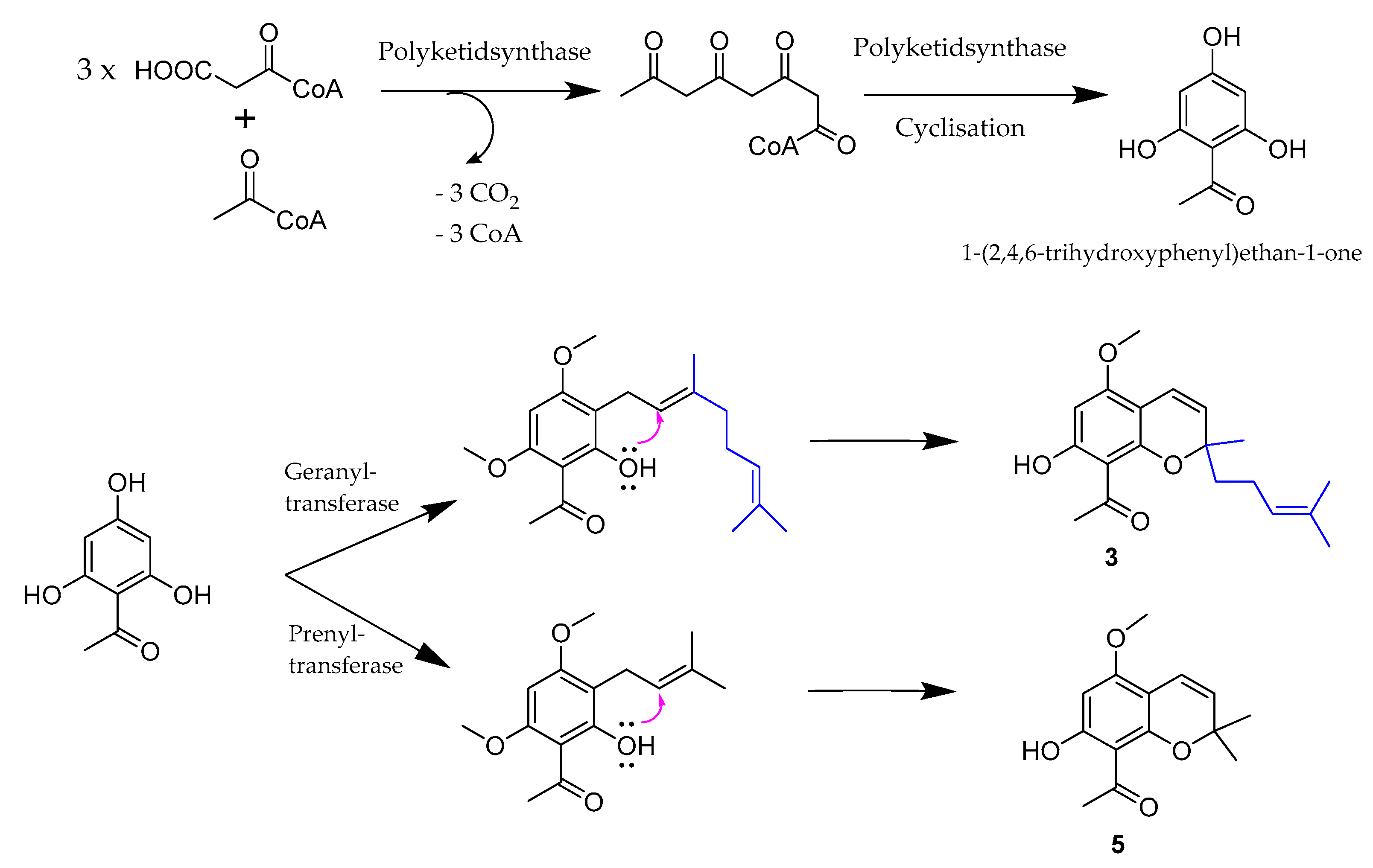
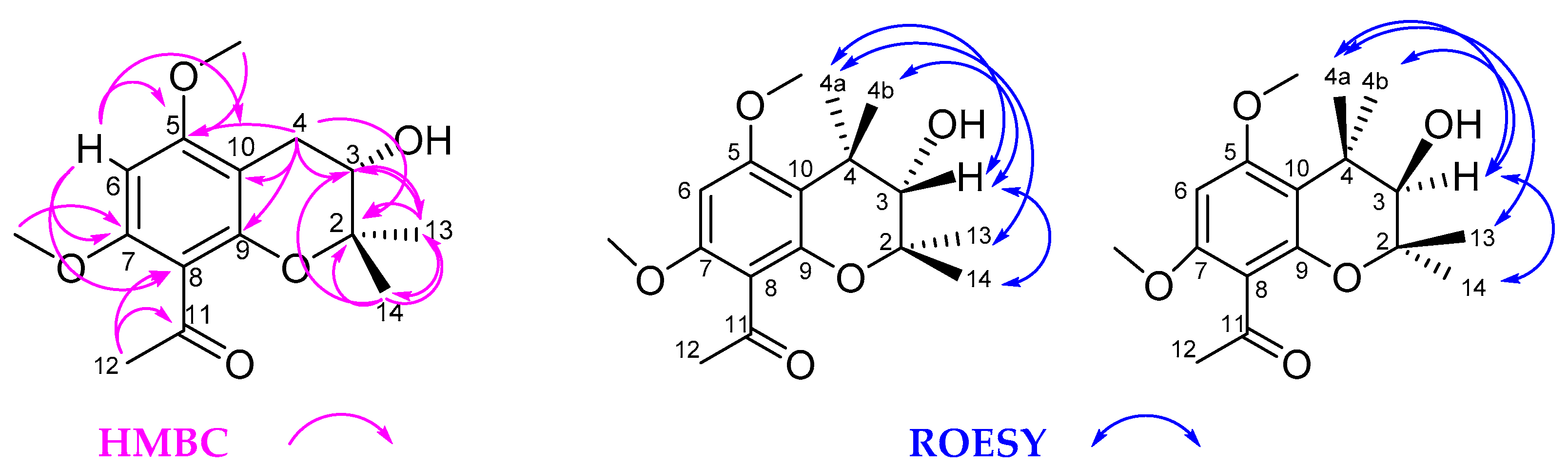

| No. | 1 | 2 | ||||
|---|---|---|---|---|---|---|
| δC | Type | δH (J in Hz) | δC | Type | δH (J in Hz) | |
| 1 | 203.0 | C | - | 203.1 | C | |
| 2 | 32.9 | CH3 | 2.56 s | 32.9 | CH3 | 2.57 s |
| 1′ | 105.0 | C | - | 104.7 | C | - |
| 2′ | 163.1 | C | - | 162.3 | C | - |
| 3′ | 106.0 | C | - | 108.5 | C | - |
| 4′ | 164.1 | C | - | 163.3 | C | - |
| 5′ | 87.1 | CH | 6.21 s | 87.3 | CH | 6.23 s |
| 6′ | 161.8 | C | - | 161.8 | C | - |
| 1″ | 28.6 | CH2 | 2.62 dd (13.0/6.6) | 20.8 | CH2 | 3.13 d (7.2) |
| 2.71 dd (13.0/7.7) | ||||||
| 2″ | 73.5 | CH | 4.14 m | 122.6 | CH | 5.07 t (7.2/1) |
| 3″ | 148.1 | C | - | 130.3 | C | - |
| 4″ | 109.7 | CH2 | 4.51 m | 25.5 | CH3 | 1.59 s |
| 4.54 m | ||||||
| 5″ | 16.9 | CH3 | 1.69 s | 17.6 | CH3 | 1.68 s |
| OCH3 at C-4′ | 55.8 | CH3 | 3.87 s | 56.0 | CH3 | 3.90 s |
| OCH3 at C-6′ | 55.9 | CH3 | 3.92 s | 55.9 | CH3 | 3.92 s |
| OH at C-2′ | - | 13.98 s | - | - | 13.95 s | |
| OH at C-2″ | - | 4.63 d | - | - | ||
| No. | 3 | 4 | ||||
|---|---|---|---|---|---|---|
| δC | Type | δH (J in Hz) | δC | Type | δH (J in Hz) | |
| 2 | 80.9 | C | - | 77.7 | C | - |
| 3 | 123.0 | CH | 5.38 d (10.1) | 69.3 | CH | 3.77 t (5.2/5.5) |
| 4a | 116.8 | CH | 6.59 d (10.1) | 26.2 | CH | 2.62 dd (17.1/5.5) |
| 4b | 2.84 dd (17.1/5.2) | |||||
| 5 | 161.2 | C | - | 159.7 | C | - |
| 6 | 91.9 | CH | 5.99 s | 88.1 | CH | 6.07 s |
| 7 | 166.7 | C | - | 156.8 | C | - |
| 8 | 106.0 | C | - | 113.8 | C | - |
| 9 | 156.7 | C | - | 151.5 | C | - |
| 10 | 102.7 | C | - | 100.9 | C | - |
| 11 | 202.9 | C | - | 201.6 | C | - |
| 12 | 33.1 | CH3 | 2.66 s | 32.7 | CH3 | 2.47 s |
| 13 a | 26.7 | CH3 | 1.43 s | 22.0 | CH3 | 1.33 s |
| 14 a | 41.7 | CH2 | 1.79 m | 24.8 | CH3 | 1.31 s |
| 1′ | 23.0 | CH2 | 2.10 m | - | - | - |
| 2′ | 123.6 | CH | 5.09 t (7.1/1.5) | - | - | - |
| 3′ | 132.9 | C | - | - | - | - |
| 4′ a | 25.9 | CH3 | 1.57 s | - | - | - |
| 5′ a | 17.3 | CH3 | 1.66 s | - | - | - |
| OCH3 at C-5 | 55.7 | CH3 | 3.83 s | 55.7 | CH3 | 3.84 s |
| OCH3 at C-7 | - | - | - | 56.2 | CH3 | 3.80 s |
| OH at C-7 | - | - | 13.84 s | - | - | - |
| Nuclear Shrinkage Assay | MTT Assay | |||
|---|---|---|---|---|
| IC50 [µM] | pIC50 ± SEM | IC50 [µM] | pIC50 ± SEM | |
| Compound 2 | 30.0 | 4.52 ± 0.11 | >100 | <4 |
| Cisplatin | 4.65 | 5.33 ± 0.07 | 6.42 | 5.19 ± 0.01 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Le, K.-T.; Bandolik, J.J.; Kassack, M.U.; Wood, K.R.; Paetzold, C.; Appelhans, M.S.; Passreiter, C.M. New Acetophenones and Chromenes from the Leaves of Melicope barbigera A. Gray. Molecules 2021, 26, 688. https://doi.org/10.3390/molecules26030688
Le K-T, Bandolik JJ, Kassack MU, Wood KR, Paetzold C, Appelhans MS, Passreiter CM. New Acetophenones and Chromenes from the Leaves of Melicope barbigera A. Gray. Molecules. 2021; 26(3):688. https://doi.org/10.3390/molecules26030688
Chicago/Turabian StyleLe, Kim-Thao, Jan J. Bandolik, Matthias U. Kassack, Kenneth R. Wood, Claudia Paetzold, Marc S. Appelhans, and Claus M. Passreiter. 2021. "New Acetophenones and Chromenes from the Leaves of Melicope barbigera A. Gray" Molecules 26, no. 3: 688. https://doi.org/10.3390/molecules26030688
APA StyleLe, K.-T., Bandolik, J. J., Kassack, M. U., Wood, K. R., Paetzold, C., Appelhans, M. S., & Passreiter, C. M. (2021). New Acetophenones and Chromenes from the Leaves of Melicope barbigera A. Gray. Molecules, 26(3), 688. https://doi.org/10.3390/molecules26030688







