Columnar Aggregates of Azobenzene Stars: Exploring Intermolecular Interactions, Structure, and Stability in Atomistic Simulations
Abstract
1. Introduction
2. Materials and Methods
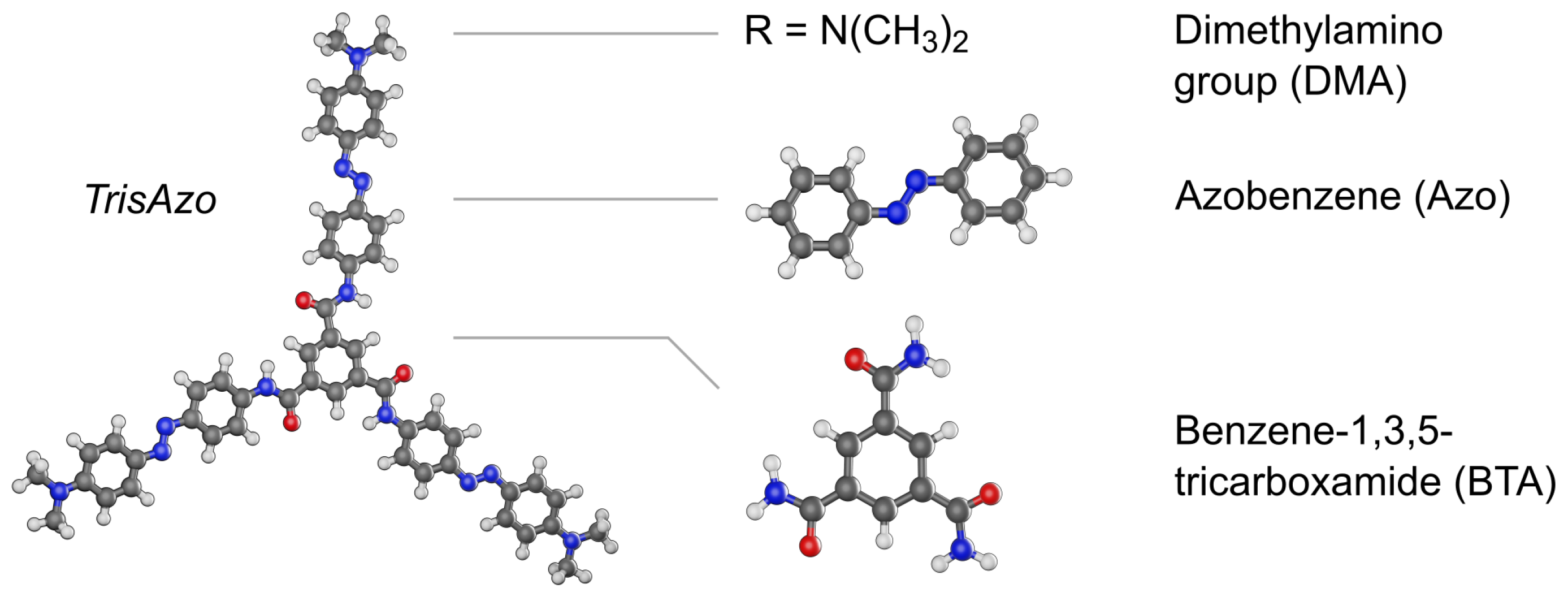
2.1. Object of Study: Supramolecular Aggregates of TrisAzo
2.2. Simulation Model
2.3. Calculation of the Intermolecular Energies
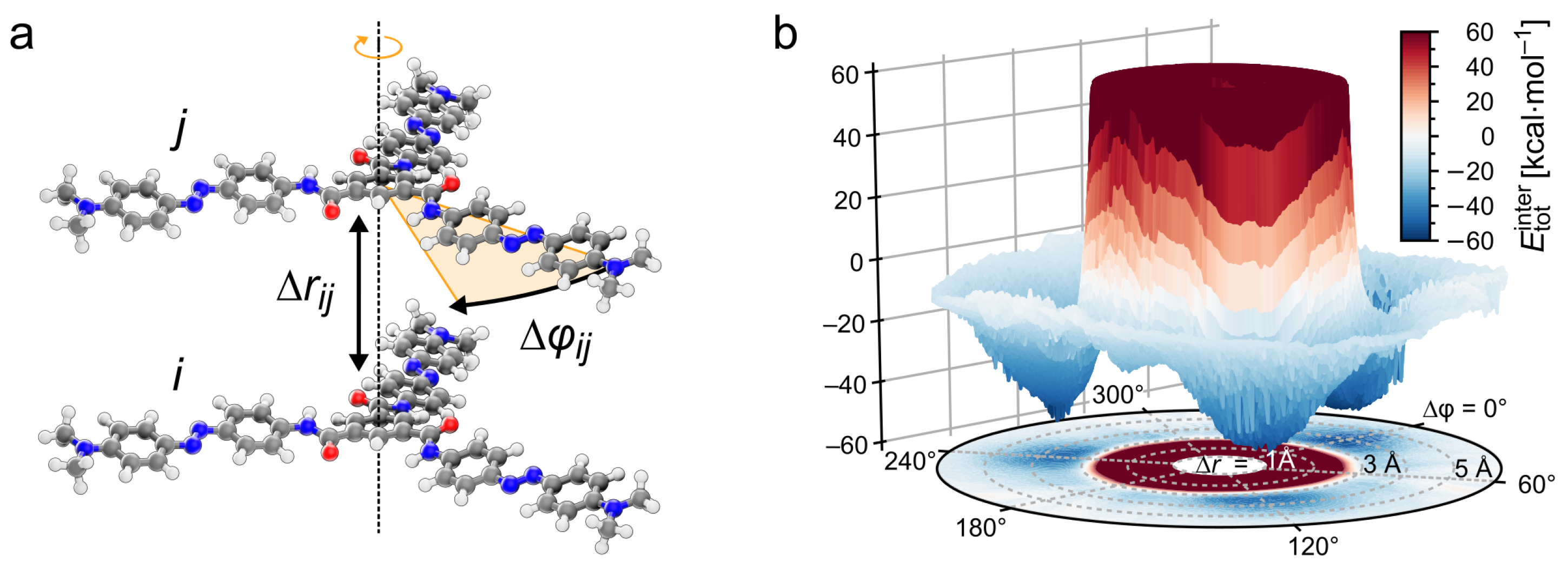
2.4. Calculation of the Intermolecular Energy Landscape of a TrisAzo Dimer
3. Results and Discussion
3.1. Intermolecular Energy of a TrisAzo Dimer
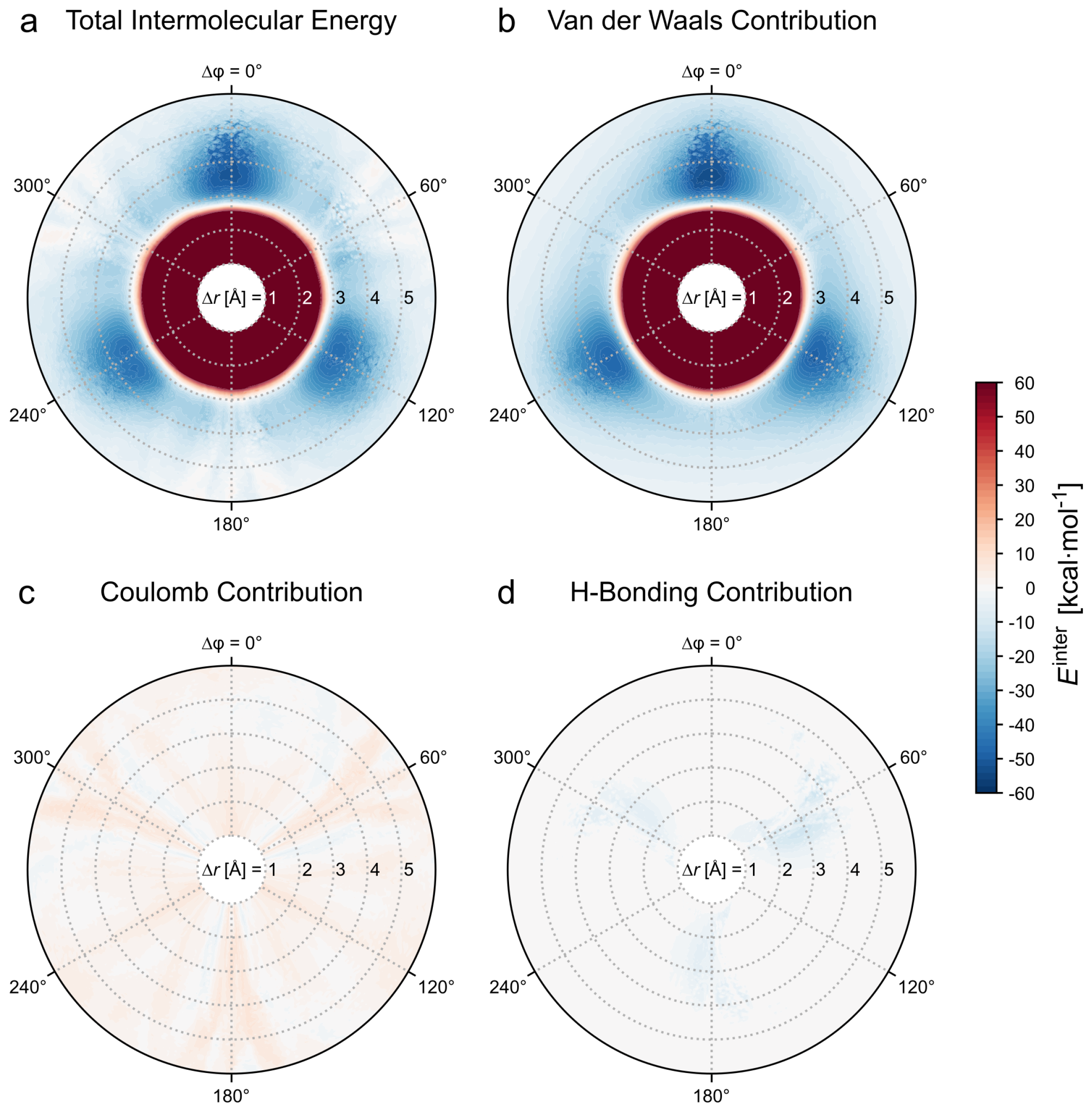
3.1.1. Total Intermolecular Energy of the Dimer
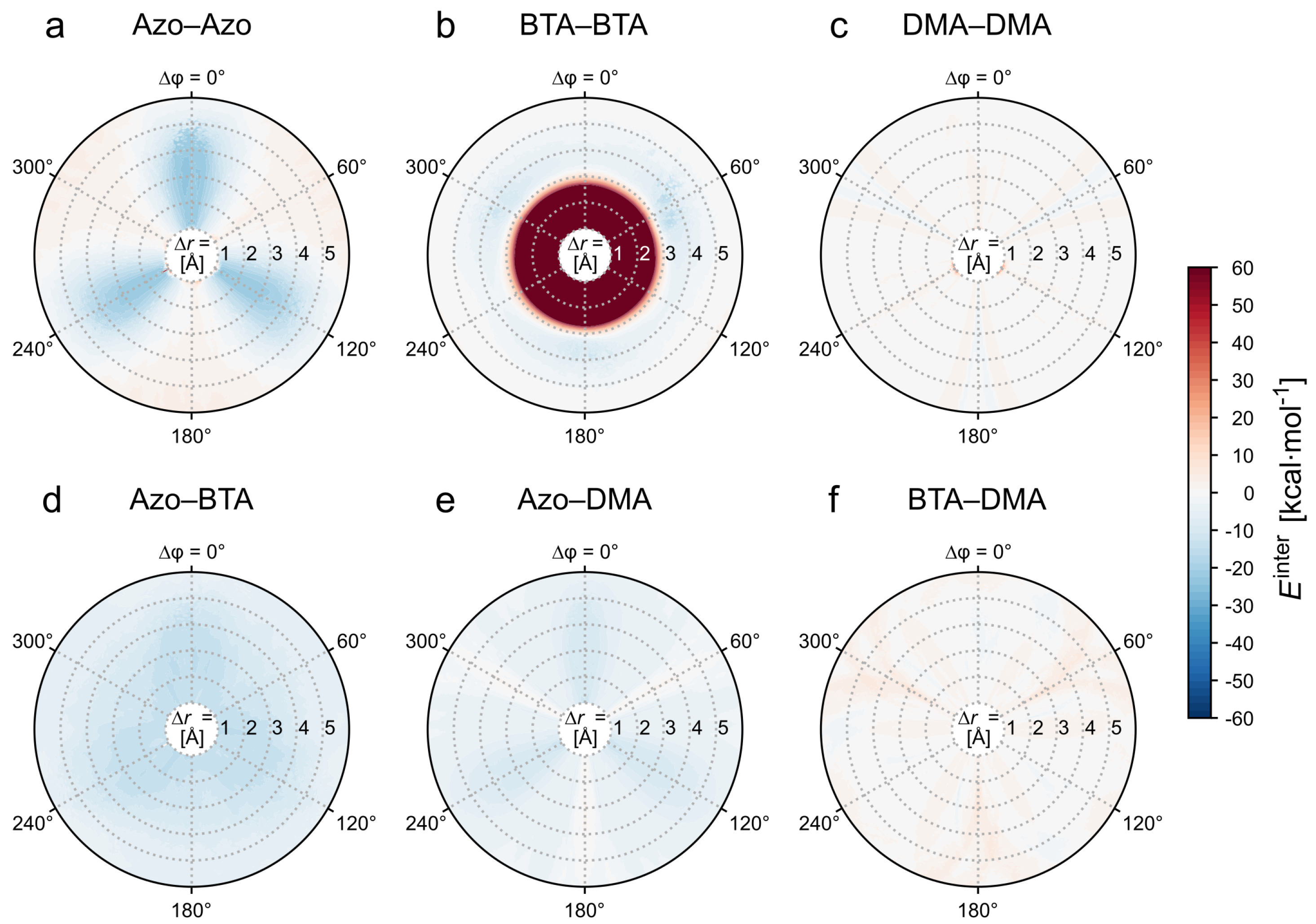
3.1.2. Decomposition of the Intermolecular Energy of the Dimer
- the different types of non-covalent interactions, and
- the interactions between different parts of the molecule.
- — van der Waals interactions,
- — Coulomb interactions,
- — explicit hydrogen bonds.
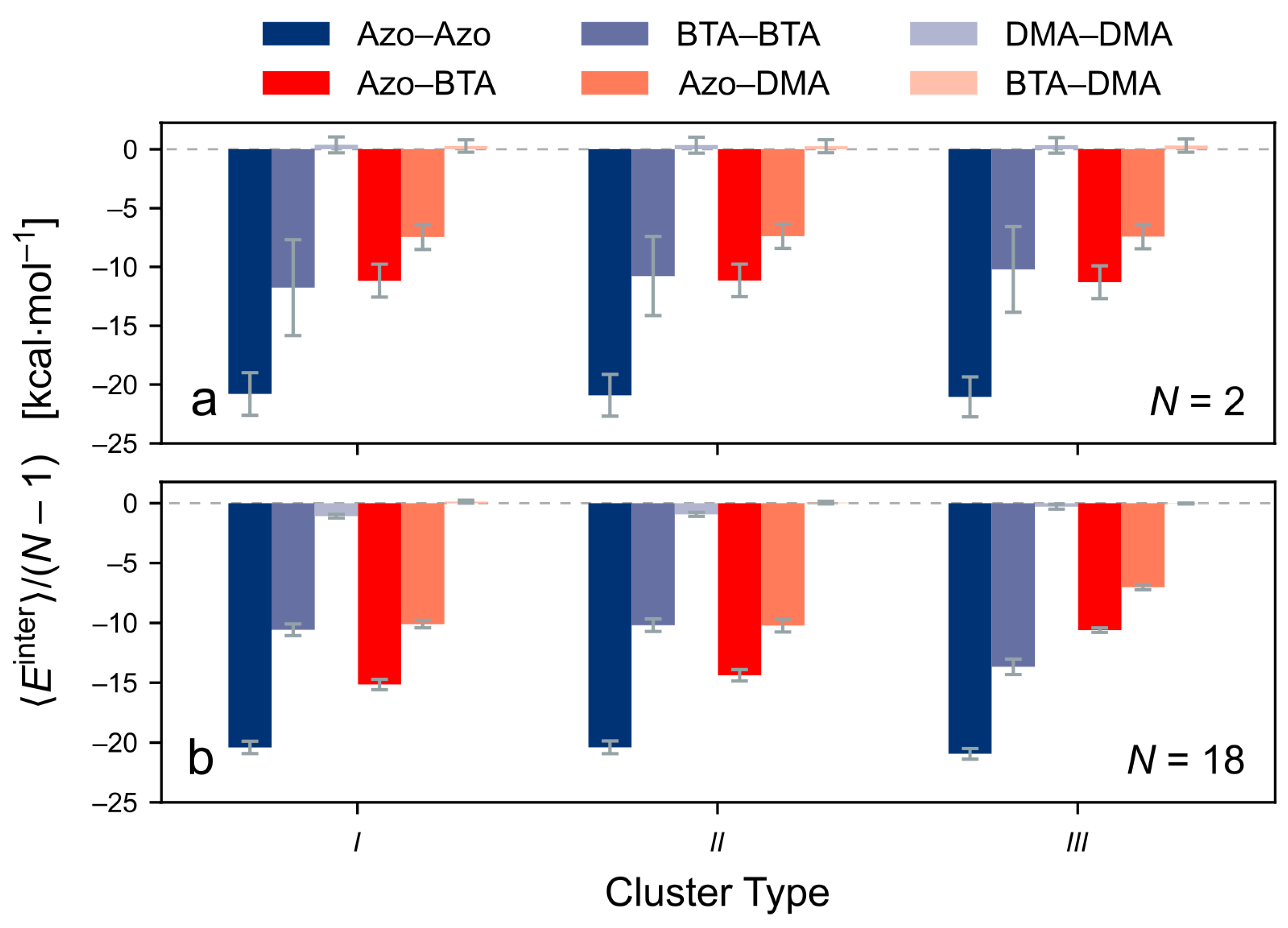
- — Azo–Azo,
- — BTA–BTA,
- — DMA–DMA,
- — Azo–BTA,
- — Azo–DMA,
- — BTA–DMA.
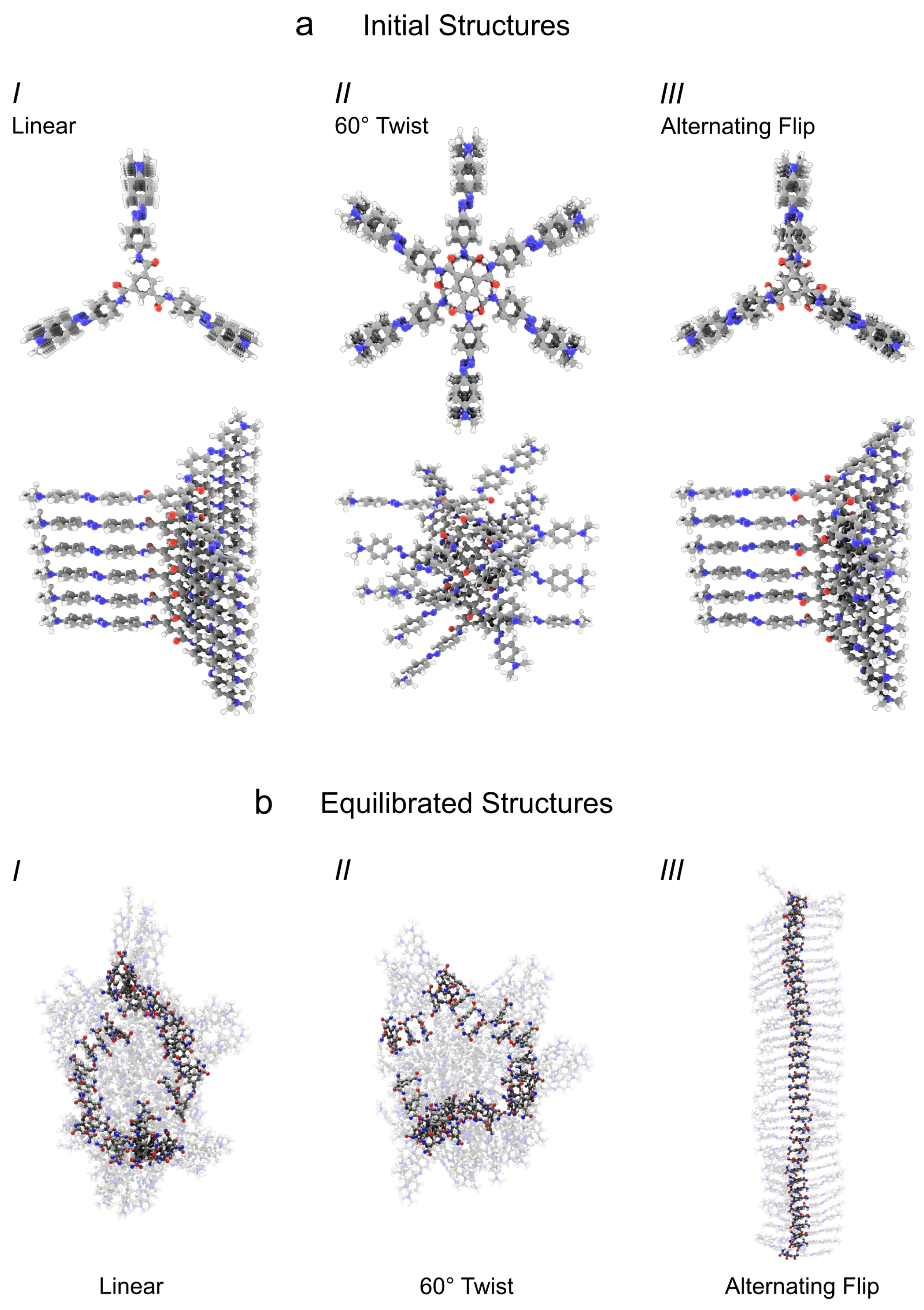
3.2. Pre-Assembly of Columnar TrisAzo Clusters
- I
- Linear arrangement—Columns are built from centrally aligned monomers (). Such an arrangement corresponds to a concatenation of TrisAzo dimers at the minimum of their intermolecular energy (Figure 3a). This structure maximizes the alignment between the Azo groups but may be unfavorable for hydrogen bond formation, for which consecutive in-plane rotations by between the BTAs would be optimal [14,15,18].
- II
- twist—In this case, columnar aggregates are built from centrally aligned TrisAzo molecules with an in-plane rotation of between neighboring pairs. This is expected to maximize the formation of hydrogen bonds and may possibly enable the strongest binding between the BTA cores, as found for columnar stacks of BTAs [14,15,18]. However, this rotation leads to a maximum displacement between the Azo arms of consecutive molecules. According to the dimer results (Section 3.1), this geometry is energetically less favorable than type I, but by considering it the impact of H-bonds on the binding can be examined.
- III
- Alternating (flipped) linear arrangement—Here, every second molecule in the column is flipped by , i.e., rotated around an axis perpendicular to the stacking axis. After the flip, the rotation angle is slightly adjusted to maximize the alignment of the Azo arms. This is equivalent to rotating all three Azo arms of every second molecule by in cluster type I. Such arm rotations have only a small energy barrier [15,16], and are, thus, occurring frequently for free TrisAzo molecules. In this cluster arrangement, all three hydrogen bonds between adjacent molecules are expected to form. The amides in the neighboring BTA cores are arranged in such a manner, that a simple rotation of these groups results in their H-bonding. At the same time, the Azo groups are aligned as well, which is expected to yield a strong binding. The difference between this arrangement and type I can be seen most clearly from the positioning of the amide groups, e.g., the amide oxygens (red), in Figure 5a.
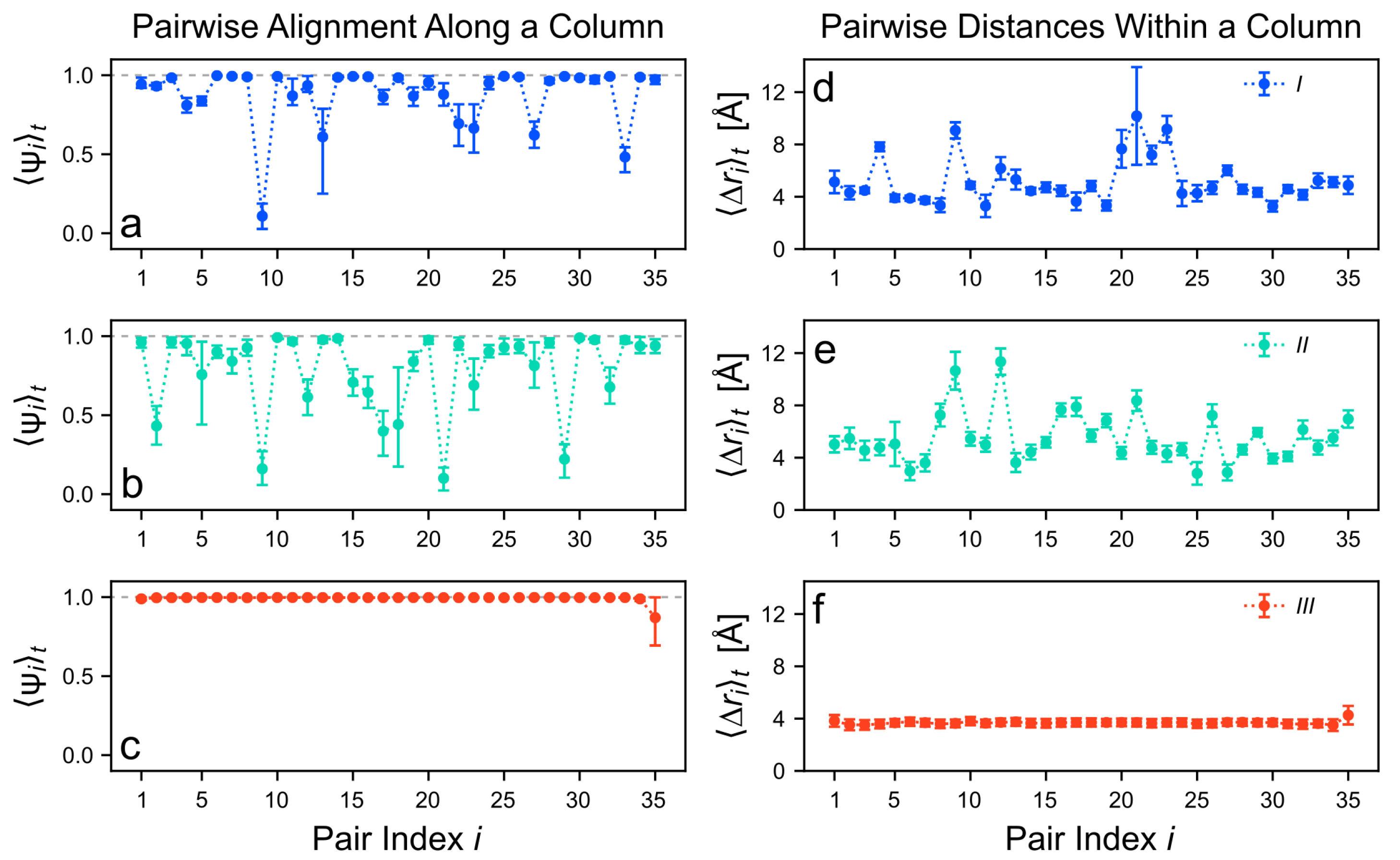
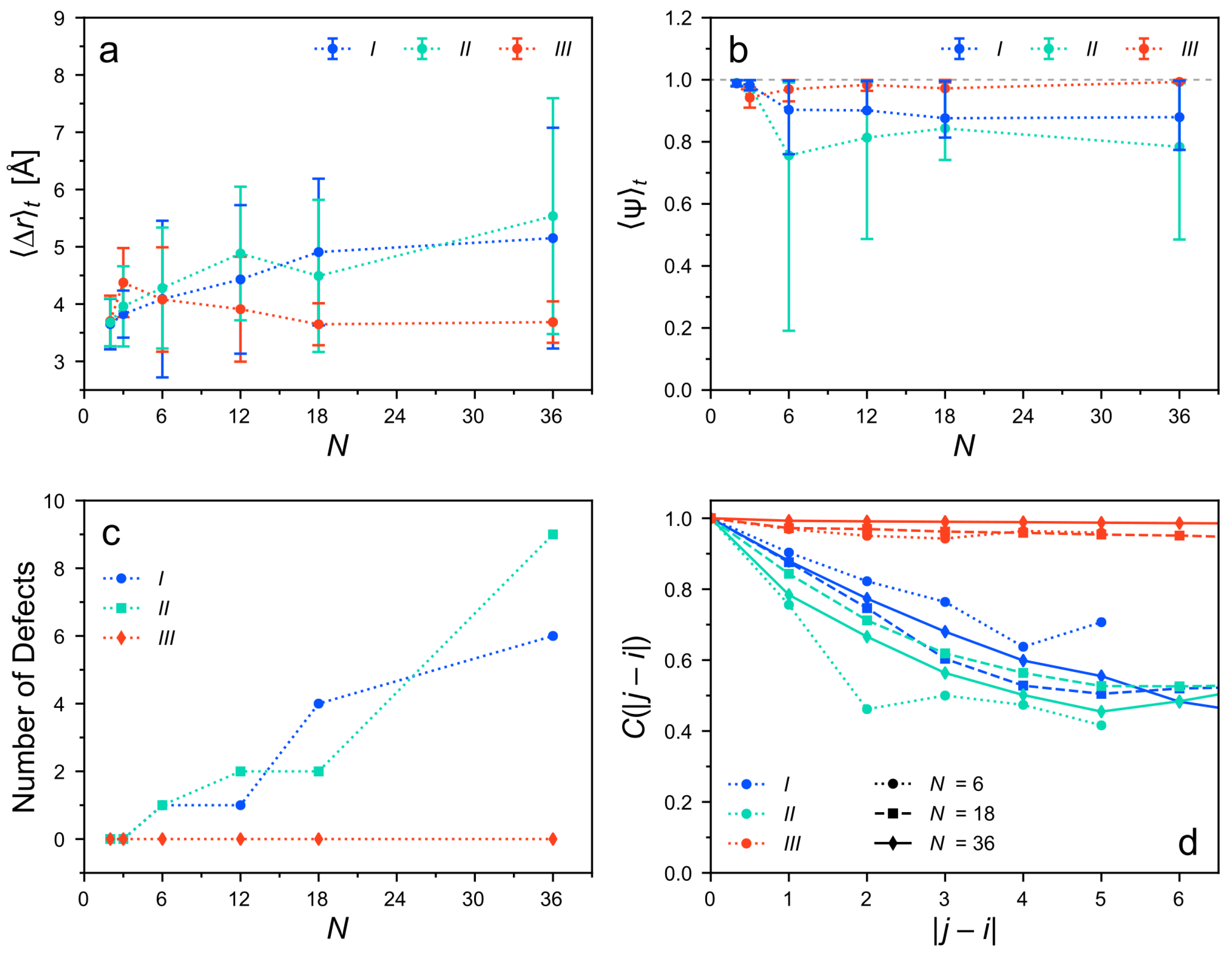
3.3. Structure of the Clusters after Equilibration
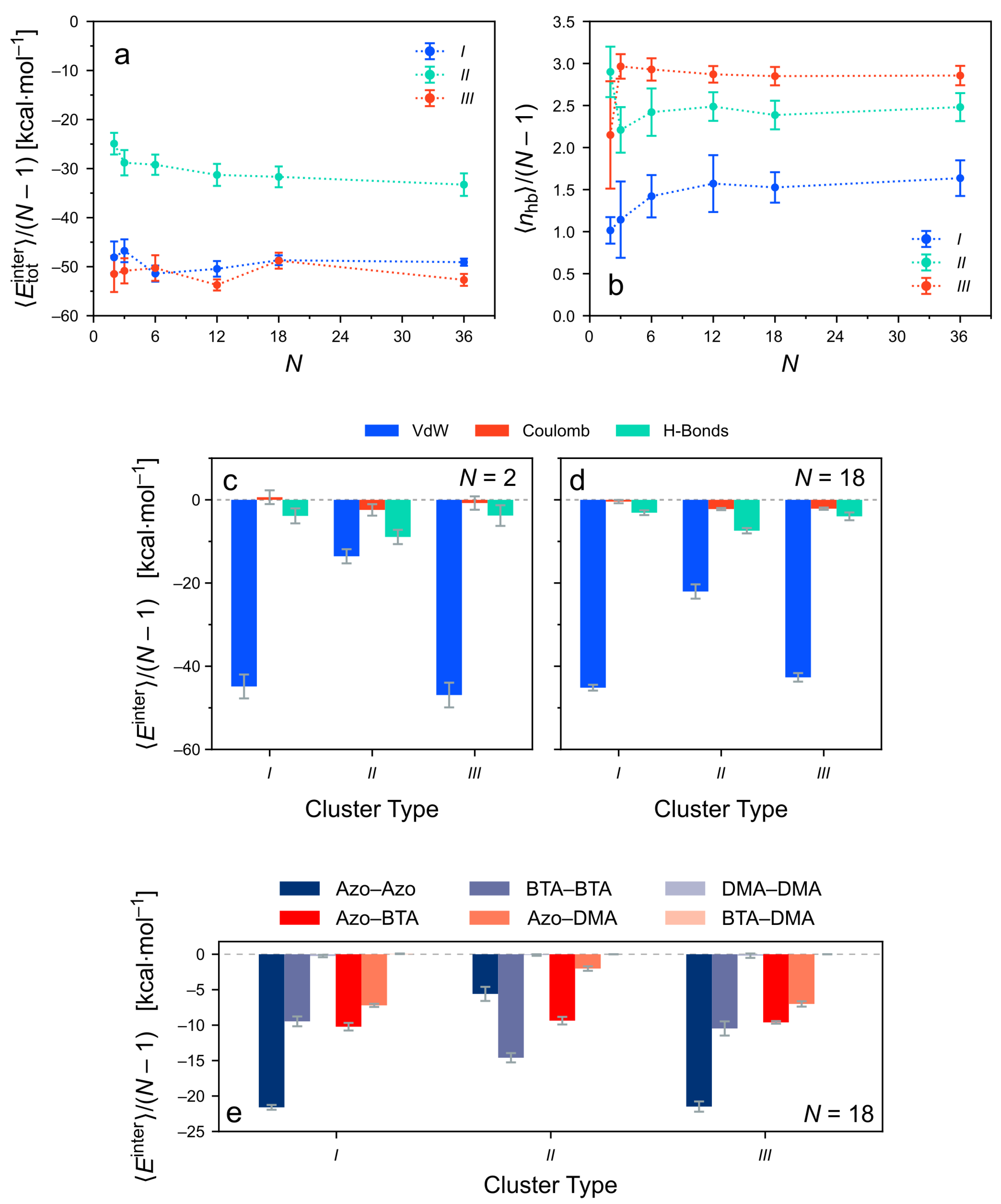
3.4. Intermolecular Energy of the Clusters after Equilibration
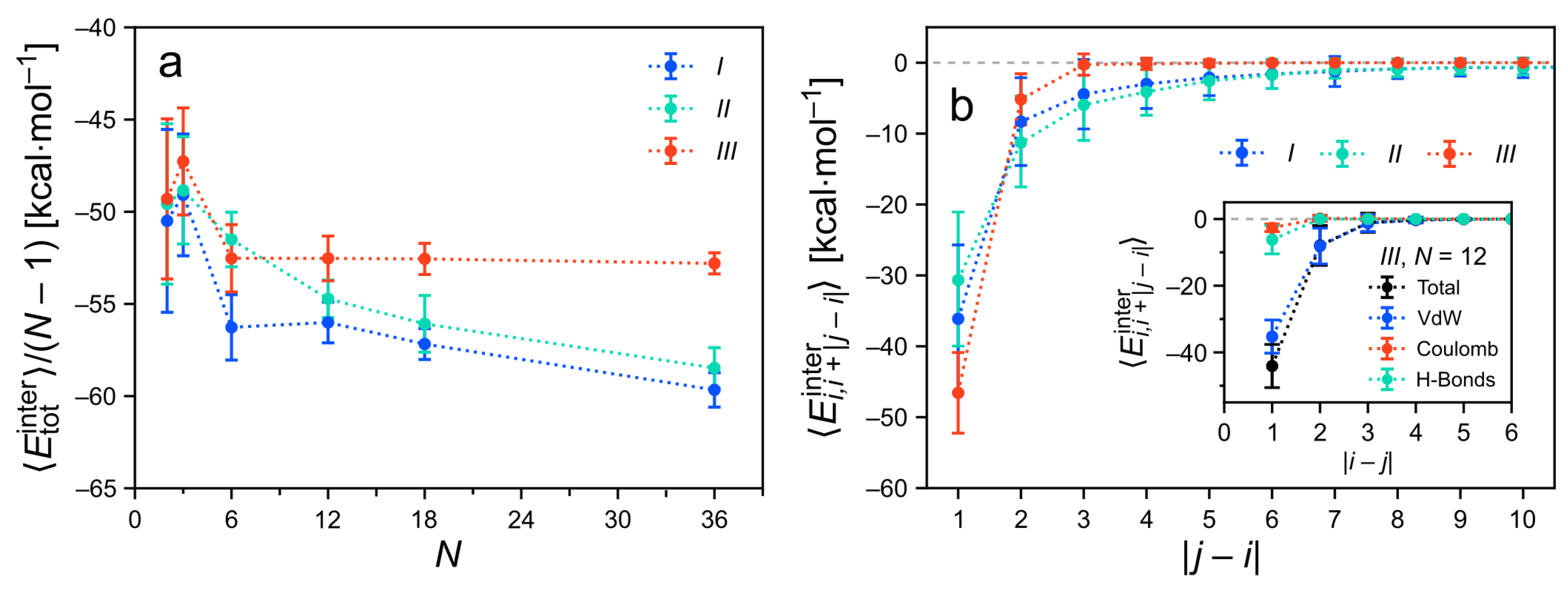
3.4.1. Total Intermolecular Energy of the Clusters
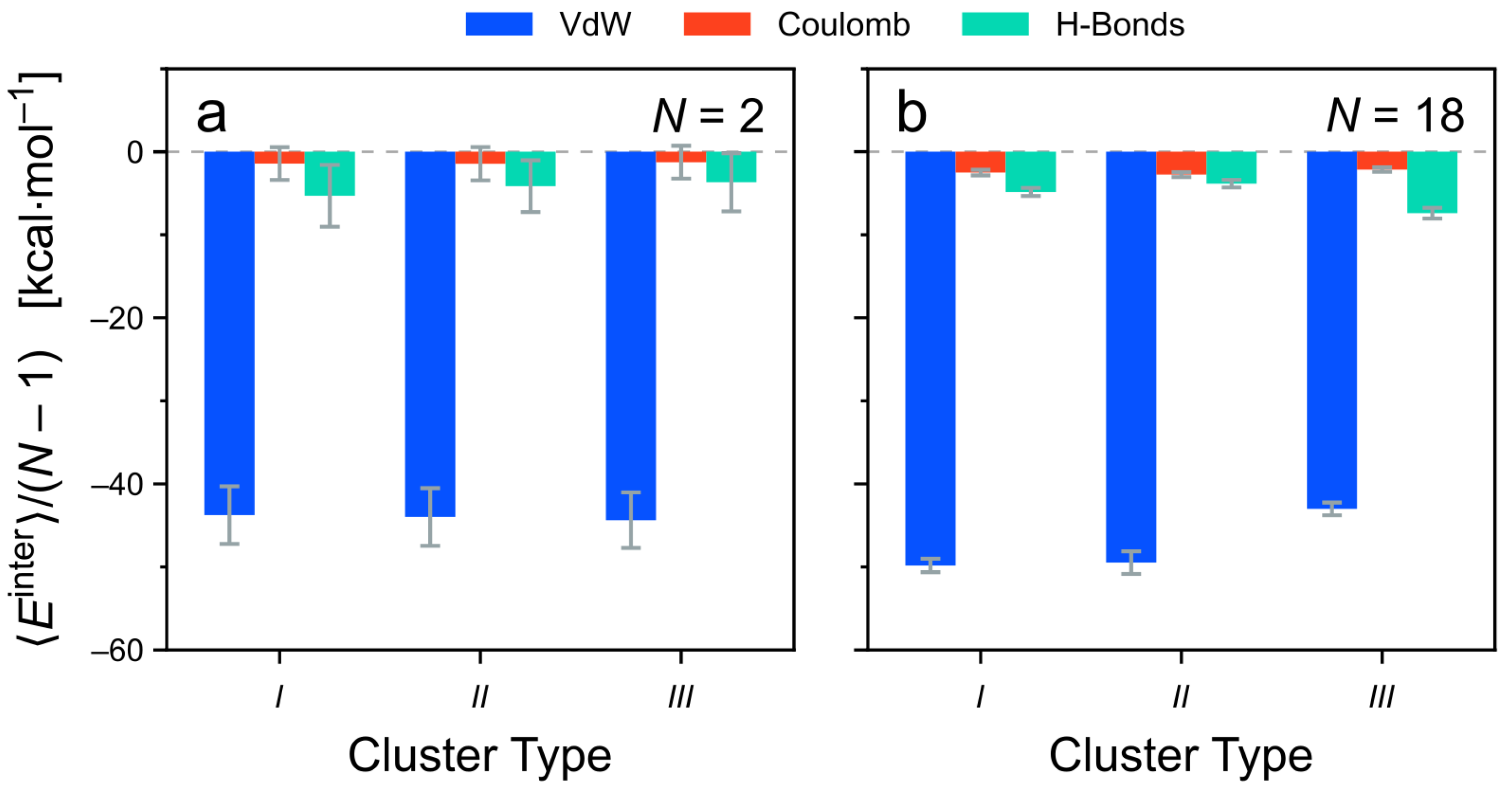
3.4.2. Decomposition of the Intermolecular Energy of the Clusters
- the contributions of each monomer;
- the types of non-covalent interactions; and
- the interactions between different parts of the molecule.
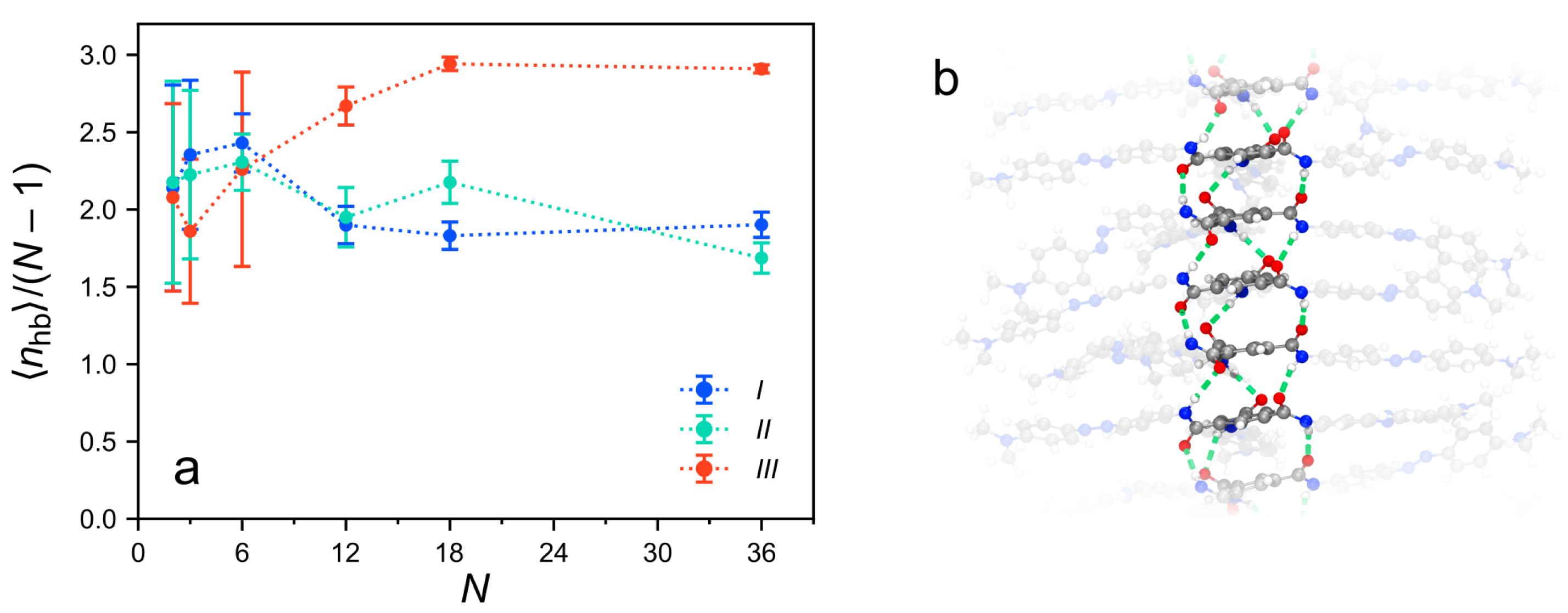
3.5. The Role of Hydrogen Bonding
3.6. Rationalizing the Structural Differences of the Considered Cluster Types
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
Abbreviations
| Azo | Azobenzene |
| BTA | Benzene-1,3,5-tricarboxamide |
| COM | Center of mass |
| DMA | Dimethylamine / Dimethylamino group |
| DMSO | Dimethylsulfoxide |
| DFT | Density functional theory |
| hb | Hydrogen bonding |
| LAMMPS | Large-scale atomic/molecular massively parallel simulator |
| LJ | Lennard–Jones |
| MD | Molecular dynamics |
| PPPM | Particle-particle-particle-mesh Fourier-based Ewald summation method |
| VdW | Van der Waals |
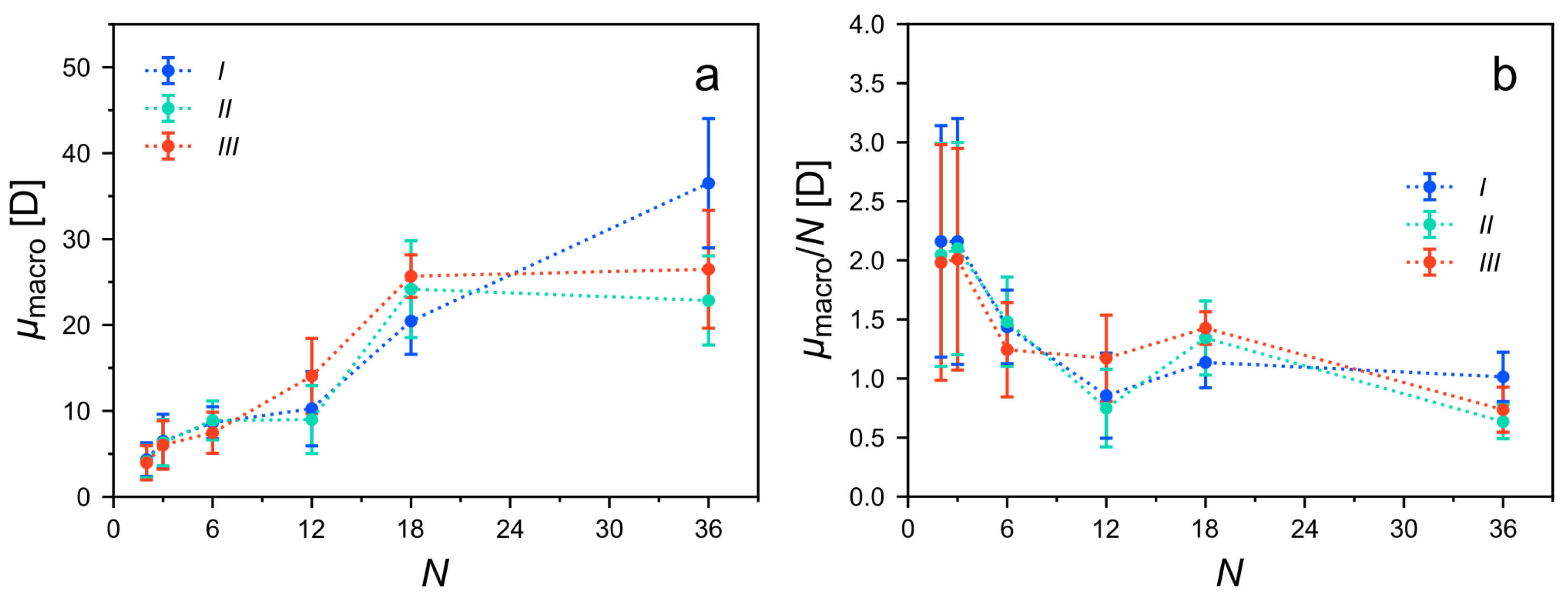
Appendix A. Macrodipoles
References
- Lehn, J.M. Supramolecular Chemistry—Scope and Perspectives Molecules, Supermolecules, and Molecular Devices (Nobel Lecture). Angew. Chem. Int. Ed. 1988, 27, 89–112. [Google Scholar] [CrossRef]
- Pieters, B.J.G.E.; van Eldijk, M.B.; Nolte, R.J.M.; Mecinović, J. Natural supramolecular protein assemblies. Chem. Soc. Rev. 2016, 45, 24–39. [Google Scholar] [CrossRef] [PubMed]
- Chiesa, G.; Kiriakov, S.; Khalil, A.S. Protein assembly systems in natural and synthetic biology. BMC Biol. 2020, 18, 35. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Li, H.; Xu, B. Biological functions of supramolecular assemblies of small molecules in the cellular environment. RSC Chem. Biol. 2021, 2, 289–305. [Google Scholar] [CrossRef]
- Savyasachi, A.J.; Kotova, O.; Shanmugaraju, S.; Bradberry, S.J.; Ó’Máille, G.M.; Gunnlaugsson, T. Supramolecular Chemistry: A Toolkit for Soft Functional Materials and Organic Particles. Chem 2017, 3, 764–811. [Google Scholar] [CrossRef]
- Williams, G.T.; Haynes, C.J.E.; Fares, M.; Caltagirone, C.; Hiscock, J.R.; Gale, P.A. Advances in applied supramolecular technologies. Chem. Soc. Rev. 2021, 50, 2737–2763. [Google Scholar] [CrossRef]
- Buerkle, L.E.; Rowan, S.J. Supramolecular gels formed from multi-component low molecular weight species. Chem. Soc. Rev. 2012, 41, 6089–6102. [Google Scholar] [CrossRef]
- De Silva, A.P.; Moody, T.S.; Wright, G.D. Fluorescent PET (Photoinduced Electron Transfer) sensors as potent analytical tools. Analyst 2009, 134, 2385–2393. [Google Scholar] [CrossRef]
- Shi, C.-Y.; Zhang, Q.; Tian, H.; Qu, D.-H. Supramolecular adhesive materials from small-molecule self-assembly. SmartMat 2020, 1, e1012. [Google Scholar] [CrossRef]
- Jin, X.; Zhu, L.; Xue, B.; Zhu, X.; Yan, D. Supramolecular nanoscale drug-delivery system with ordered structure. Natl. Sci. Rev. 2019, 6, 1128–1137. [Google Scholar] [CrossRef]
- Varela-Aramburu, S.; Morgese, G.; Su, L.; Schoenmakers, S.M.C.; Perrone, M.; Leanza, L.; Perego, C.; Pavan, G.M.; Palmans, A.R.A.; Meijer, E.W. Exploring the Potential of Benzene-1,3,5-tricarboxamide Supramolecular Polymers as Biomaterials. Biomacromolecules 2020, 21, 4105–4115. [Google Scholar] [CrossRef]
- Uhlenheuer, D.A.; Petkau, K.; Brunsveld, L. Combining supramolecular chemistry with biology. Chem. Soc. Rev. 2010, 39, 2817–2826. [Google Scholar] [CrossRef] [PubMed]
- Kulkarni, C.; Reddy, S.K.; George, S.J.; Balasubramanian, S. Cooperativity in the stacking of benzene-1,3,5-tricarboxamide: The role of dispersion. Chem. Phys. Lett. 2011, 515, 226–230. [Google Scholar] [CrossRef]
- Cantekin, S.; de Greef, T.F.A.; Palmans, A.R.A. Benzene-1,3,5-tricarboxamide: A versatile ordering moiety for supramolecular chemistry. Chem. Soc. Rev. 2012, 41, 6125–6137. [Google Scholar] [CrossRef] [PubMed]
- Bejagam, K.K.; Fiorin, G.; Klein, M.L.; Balasubramanian, S. Supramolecular Polymerization of Benzene-1,3,5-tricarboxamide: A Molecular Dynamics Simulation Study. J. Phys. Chem. B 2014, 118, 5218–5228. [Google Scholar] [CrossRef]
- Bejagam, K.K.; Kulkarni, C.; George, S.J.; Balasubramanian, S. External electric field reverses helical handedness of a supramolecular columnar stack. Chem. Commun. 2015, 51, 16049–16052. [Google Scholar] [CrossRef]
- Garzoni, M.; Baker, M.B.; Leenders, C.M.A.; Voets, I.K.; Albertazzi, L.; Palmans, A.R.A.; Meijer, E.W.; Pavan, G.M. Effect of H-Bonding on Order Amplification in the Growth of a Supramolecular Polymer in Water. J. Am. Chem. Soc. 2016, 138, 13985–13995. [Google Scholar] [CrossRef]
- Banach, E.; Invernizzi, C.; Baudin, M.; Neier, R.; Carnevale, D. Columnar self-assembly of N,N′,N′′-trihexylbenzene-1,3,5-tricarboxamides investigated by means of NMR spectroscopy and computational methods in solution and the solid state. Phys. Chem. Chem. Phys. 2017, 19, 5525–5539. [Google Scholar] [CrossRef]
- Li, C.; Tan, J.; Guan, Z.; Zhang, Q. A Three-Armed Polymer with Tunable Self-Assembly and Self-Healing Properties Based on Benzene-1,3,5-tricarboxamide and Metal–Ligand Interactions. Macromol. Rapid Commun. 2019, 40, 1800909. [Google Scholar] [CrossRef] [PubMed]
- De Windt, L.N.J.; Fernández, Z.; Fernández-Míguez, M.; Freire, F.; Palmans, A.R.A. Elucidating the Supramolecular Copolymerization of N- and C-Centered Benzene-1,3,5-Tricarboxamides: The Role of Parallel and Antiparallel Packing of Amide Groups in the Copolymer Microstructure. Chem. Eur. J. 2021. [Google Scholar] [CrossRef]
- Bochicchio, D.; Salvalaglio, M.; Pavan, G.M. Into the Dynamics of a Supramolecular Polymer at Submolecular Resolution. Nat. Commun. 2017, 8, 147. [Google Scholar] [CrossRef]
- Ter Huurne, G.M.; Chidchob, P.; Long, A.; Martinez, A.; Palmans, A.R.A.; Vantomme, G. Controlling the Length of Cooperative Supramolecular Polymers with Chain Cappers. Chem. Eur. J. 2020, 26, 9964–9970. [Google Scholar] [CrossRef] [PubMed]
- Shikata, T.; Ogata, D.; Hanabusa, K. Viscoelastic Behavior of Supramolecular Polymeric Systems Consisting of N,N′,N′′-Tris(3,7-dimethyloctyl)benzene-1,3,5-tricarboxamide and n-Alkanes. J. Phys. Chem. B 2004, 108, 508–514. [Google Scholar] [CrossRef]
- Shi, N.E.; Dong, H.; Yin, G.; Xu, Z.; Li, S.H. A Smart Supramolecular Hydrogel Exhibiting pH-Modulated Viscoelastic Properties. Adv. Funct. Mater. 2007, 17, 1837–1843. [Google Scholar] [CrossRef]
- Broaders, K.E.; Pastine, S.J.; Grandhe, S.; Fréchet, J.M.J. Acid-degradable solid-walled microcapsules for pH-responsive burst-release drug delivery. Chem. Commun. 2011, 47, 665–667. [Google Scholar] [CrossRef] [PubMed]
- Besenius, P.; Heynens, J.L.M.; Straathof, R.; Nieuwenhuizen, M.M.L.; Bomans, P.H.H.; Terreno, E.; Aime, S.; Strijkers, G.J.; Nicolay, K.; Meijer, E.W. Paramagnetic self-assembled nanoparticles as supramolecular MRI contrast agents. Contrast Media Mol. Imaging 2012, 7, 356–361. [Google Scholar] [CrossRef] [PubMed]
- Urbanavičiūtė, I.; Cornelissen, T.D.; Meng, X.; Sijbesma, R.P.; Kemerink, M. Physical reality of the Preisach model for organic ferroelectrics. Nat. Commun. 2018, 9, 4409. [Google Scholar] [CrossRef]
- Stals, P.J.M.; Smulders, M.M.J.; Martín-Rapún, R.; Palmans, A.R.A.; Meijer, E.W. Asymmetrically Substituted Benzene-1,3,5-Tricarboxamides: Self-Assembly and Odd–Even Effects in the Solid State and in Dilute Solution. Chem. Eur. J. 2009, 15, 2071–2080. [Google Scholar] [CrossRef]
- Danila, I.; Riobé, F.; Piron, F.; Puigmartí-Luis, J.; Wallis, J.D.; Linares, M.; Ågren, H.; Beljonne, D.; Amabilino, D.B.; Avarvari, N. Hierarchical Chiral Expression from the Nano- to Mesoscale in Synthetic Supramolecular Helical Fibers of a Nonamphiphilic C3-Symmetrical π-Functional Molecule. J. Am. Chem. Soc. 2011, 133, 8344–8353. [Google Scholar] [CrossRef]
- Bejagam, K.K.; Remsing, R.C.; Klein, M.L.; Balasubramanian, S. Understanding the self-assembly of amino ester-based benzene-1,3,5-tricarboxamides using molecular dynamics simulations. Phys. Chem. Chem. Phys. 2017, 19, 258–266. [Google Scholar] [CrossRef]
- Bochicchio, D.; Pavan, G.M. From Cooperative Self-Assembly to Water-Soluble Supramolecular Polymers Using Coarse-Grained Simulations. ACS Nano 2017, 11, 1000–1011. [Google Scholar] [CrossRef]
- Gerth, M.; Berrocal, J.A.; Bochicchio, D.; Pavan, G.M.; Voets, I.K. Discordant Supramolecular Fibres Reversibly Depolymerised by Temperature and Light. Chem. Eur. J. 2020, 27, 1829–1838. [Google Scholar] [CrossRef] [PubMed]
- Koch, M.; Saphiannikova, M.; Santer, S.; Guskova, O. Photoisomers of Azobenzene Star with a Flat Core: Theoretical Insights into Multiple States from DFT and MD Perspective. J. Phys. Chem. B 2017, 121, 8854–8867. [Google Scholar] [CrossRef] [PubMed]
- Yao, X.; Li, T.; Wang, J.; Ma, X.; Tian, H. Recent Progress in Photoswitchable Supramolecular Self-Assembling Systems. Adv. Opt. Mater. 2016, 4, 1322–1349. [Google Scholar] [CrossRef]
- Yagai, S.; Karatsu, T.; Kitamura, A. Photocontrollable Self-Assembly. Chem. Eur. J. 2005, 11, 4054–4063. [Google Scholar] [CrossRef]
- Yagai, S.; Kitamura, A. Recent advances in photoresponsive supramolecular self-assemblies. Chem. Soc. Rev. 2008, 37, 1520–1529. [Google Scholar] [CrossRef]
- Lee, S.; Oh, S.; Lee, J.; Malpani, Y.; Jung, Y.-S.; Kang, B.; Lee, J.Y.; Ozasa, K.; Isoshima, T.; Lee, S.Y.; et al. Stimulus-Responsive Azobenzene Supramolecules: Fibers, Gels, and Hollow Spheres. Langmuir 2013, 29, 5869–5877. [Google Scholar] [CrossRef]
- Malpani, Y.R.; Oh, S.; Lee, S.; Jung, Y.-S.; Kim, J.-M. Photoinduced Phase Transition of Azobenzene-Coupled Benzenetricarboxamide. Bull. Korean Chem. Soc. 2014, 35, 2563–2566. [Google Scholar] [CrossRef][Green Version]
- Lee, J.; Oh, S.; Pyo, J.; Kim, J.-M.; Je, J.H. A light-driven supramolecular nanowire actuator. Nanoscale 2015, 7, 6457–6461. [Google Scholar] [CrossRef] [PubMed]
- Baroncini, M.; Bergamini, G. Azobenzene: A Photoactive Building Block for Supramolecular Architectures. Chem. Rec. 2017, 17, 700–712. [Google Scholar] [CrossRef]
- Fihey, A.; Perrier, A.; Browne, W.R.; Jacquemin, D. Multiphotochromic Molecular Systems. Chem. Soc. Rev. 2015, 44, 3719–3759. [Google Scholar] [CrossRef]
- Galanti, A.; Santoro, J.; Mannancherry, R.; Duez, Q.; Diez-Cabanes, V.; Valášek, M.; De Winter, J.; Cornil, J.; Gerbaux, P.; Mayor, M.; et al. A New Class of Rigid Multi(azobenzene) Switches Featuring Electronic Decoupling: Unravelling the Isomerization in Individual Photochromes. J. Am. Chem. Soc. 2019, 141, 9273–9283. [Google Scholar] [CrossRef]
- Galanti, A.; Diez-Cabanes, V.; Santoro, J.; Valášek, M.; Minoia, A.; Mayor, M.; Cornil, J.; Samorì, P. Electronic Decoupling in C3-Symmetrical Light-Responsive Tris(Azobenzene) Scaffolds: Self-Assembly and Multiphotochromism. J. Am. Chem. Soc. 2018, 140, 16062–16070. [Google Scholar] [CrossRef] [PubMed]
- Gupta, D.; Gaur, A.K.; Kumar, P.; Kumar, H.; Mahadevan, A.; Devi, S.; Roy, S.; Venkataramani, S. Tuning of Bistability, Thermal Stability of the Metastable States, and Application Prospects in the C3-Symmetric Designs of Multiple Azo(hetero)arenes Systems. Chem. Eur. J. 2021, 27, 3463–3472. [Google Scholar] [CrossRef]
- Devi, S.; Bala, I.; Gupta, S.P.; Kumar, P.; Pal, S.K.; Venkataramani, S. Reversibly photoswitchable alkoxy azobenzenes connected benzenetricarboxamide discotic liquid crystals with perpetual long range columnar assembly. Org. Biomol. Chem. 2019, 17, 1947–1954. [Google Scholar] [CrossRef]
- Koch, M.; Saphiannikova, M.; Guskova, O. Do Columns of Azobenzene Stars Disassemble under Light Illumination? Langmuir 2019, 35, 14659–14669. [Google Scholar] [CrossRef]
- Savchenko, V.; Koch, M.; Pavlov, A.S.; Saphiannikova, M.; Guskova, O. Stacks of Azobenzene Stars: Self-Assembly Scenario and Stabilising Forces Quantified in Computer Modelling. Molecules 2019, 24, 4387. [Google Scholar] [CrossRef] [PubMed]
- Koch, M. The Influence of Light on a Three-Arm Azobenzene Star: A Computational Study. Ph.D. Dissertation, Technische Universität Dresden, Dresden, Germany, 2022. [Google Scholar]
- Mayo, S.L.; Olafson, B.D.; Goddard, W.A. DREIDING: A Generic Force Field for Molecular Simulations. J. Phys. Chem. 1990, 94, 8897–8909. [Google Scholar] [CrossRef]
- BIOVIA, Dassault Systèmes. BIOVIA Materials Studio 8.0.100.21; Dassault Systèmes: San Diego, CA, USA, 2014. [Google Scholar]
- Jewett, A.I.; Stelter, D.; Lambert, J.; Saladi, S.M.; Roscioni, O.M.; Ricci, M.; Autin, L.; Maritan, M.; Bashusqeh, S.M.; Keyes, T.; et al. Moltemplate: A Tool for Coarse-Grained Modeling of Complex Biological Matter and Soft Condensed Matter Physics. J. Mol. Biol. 2021, 433, 166841. [Google Scholar] [CrossRef]
- Berendsen, H.J.C.; Grigera, J.R.; Straatsma, T.P. The Missing Term in Effective Pair Potentials. J. Phys. Chem. 1987, 91, 6269–6271. [Google Scholar] [CrossRef]
- Martínez, J.M.; Martínez, L. Packing Optimization for Automated Generation of Complex System’s Initial Configurations for Molecular Dynamics and Docking. J. Comput. Chem. 2003, 24, 819–825. [Google Scholar] [CrossRef]
- Martínez, L.; Andrade, R.; Birgin, E.G.; Martínez, J.M. PACKMOL: A Package for Building Initial Configurations for Molecular Dynamics Simulations. J. Comput. Chem. 2009, 30, 2157–2164. [Google Scholar] [CrossRef]
- Plimpton, S. Fast Parallel Algorithms for Short-Range Molecular Dynamics. J. Comput. Phys. 1995, 117, 1–19. [Google Scholar] [CrossRef]
- Thompson, A.P.; Aktulga, H.M.; Berger, R.; Bolintineanu, D.S.; Brown, W.M.; Crozier, P.S.; in’t Veld, P.J.; Kohlmeyer, A.; Moore, S.G.; Nguyen, T.D.; et al. LAMMPS—A flexible simulation tool for particle-based materials modeling at the atomic, meso, and continuum scales. Comput. Phys. Commun. 2022, 271, 108171. [Google Scholar] [CrossRef]
- Lorentz, H.A. Ueber die Anwendung des Satzes vom Virial in der kinetischen Theorie der Gase. Ann. Phys. 1881, 248, 127–136. [Google Scholar] [CrossRef]
- Berthelot, D. Sur le mélange des gaz. C. R. Hebd. Séances Acad. Sci. 1898, 126, 1703–1706. [Google Scholar]
- Eastwood, J.W.; Hockney, R.W.; Lawrence, D.N. P3M3DP—The three-dimensional periodic particle-particle/particle-mesh program. Comput. Phys. Commun. 1980, 19, 215–261. [Google Scholar] [CrossRef]
- Hockney, R.W.; Eastwood, J.W. Computer Simulation Using Particles; CRC Press: Boca Raton, FL, USA, 1988. [Google Scholar] [CrossRef]
- Ryckaert, J.-P.; Ciccotti, G.; Berendsen, H.J.C. Numerical Integration of the Cartesian Equations of Motion of a System with Constraints: Molecular Dynamics of n-Alkanes. J. Comput. Phys. 1977, 23, 327–341. [Google Scholar] [CrossRef]
- Polak, E.; Ribiere, G. Note sur la convergence de méthodes de directions conjuguées. ESAIM Math. Modell. Numer. Anal. 1969, 3, 35–43. [Google Scholar] [CrossRef]
- Ilnytskyi, J.M.; Toshchevikov, V.; Saphiannikova, M. Modeling of the photo-induced stress in azobenzene polymers by combining theory and computer simulations. Soft Matter 2019, 15, 9894–9908. [Google Scholar] [CrossRef]
- Albuquerque, R.Q.; Timme, A.; Kress, R.; Senker, J.; Schmidt, H.-W. Theoretical Investigation of Macrodipoles in Supramolecular Columnar Stackings. Chem. Eur. J. 2013, 19, 1647–1657. [Google Scholar] [CrossRef]
- Semenov, A.N. Bond–Vector Correlation Functions in Dense Polymer Systems. Macromolecules 2010, 43, 9139–9154. [Google Scholar] [CrossRef]
- Michaud-Agrawal, N.; Denning, E.J.; Woolf, T.B.; Beckstein, O. MDAnalysis: A Toolkit for the Analysis of Molecular Dynamics Simulations. J. Comput. Chem. 2011, 32, 2319–2327. [Google Scholar] [CrossRef] [PubMed]
- Gowers, R.J.; Linke, M.; Barnoud, J.; Reddy, T.J.E.; Melo, M.N.; Seyler, S.L.; Domański, J.; Dotson, D.L.; Buchoux, S.; Kenney, I.M.; et al. MDAnalysis: A Python Package for the Rapid Analysis of Molecular Dynamics Simulations. In Proceedings of the 15th Python in Science Conference, Austin, TX, USA, 11–17 July 2016; pp. 98–105. [Google Scholar] [CrossRef]
- Korlepara, D.B.; Henderson, W.R.; Castellano, R.K.; Balasubramanian, S. Differentiating the mechanism of self-assembly in supramolecular polymers through computation. Chem. Commun. 2019, 55, 3773–3776. [Google Scholar] [CrossRef] [PubMed]
- Koch, M.; Saphiannikova, M.; Guskova, O. Cyclic Photoisomerization of Azobenzene in Atomistic Simulations: Modeling the Effect of Light on Columnar Aggregates of Azo Stars. Molecules 2021. under review. [Google Scholar]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Koch, M.; Saphiannikova, M.; Guskova, O. Columnar Aggregates of Azobenzene Stars: Exploring Intermolecular Interactions, Structure, and Stability in Atomistic Simulations. Molecules 2021, 26, 7598. https://doi.org/10.3390/molecules26247598
Koch M, Saphiannikova M, Guskova O. Columnar Aggregates of Azobenzene Stars: Exploring Intermolecular Interactions, Structure, and Stability in Atomistic Simulations. Molecules. 2021; 26(24):7598. https://doi.org/10.3390/molecules26247598
Chicago/Turabian StyleKoch, Markus, Marina Saphiannikova, and Olga Guskova. 2021. "Columnar Aggregates of Azobenzene Stars: Exploring Intermolecular Interactions, Structure, and Stability in Atomistic Simulations" Molecules 26, no. 24: 7598. https://doi.org/10.3390/molecules26247598
APA StyleKoch, M., Saphiannikova, M., & Guskova, O. (2021). Columnar Aggregates of Azobenzene Stars: Exploring Intermolecular Interactions, Structure, and Stability in Atomistic Simulations. Molecules, 26(24), 7598. https://doi.org/10.3390/molecules26247598






