Application of the Metabolomics Approach in Food Authentication
Abstract
:1. Introduction
2. Food Authentication
3. Metabolomics Approach
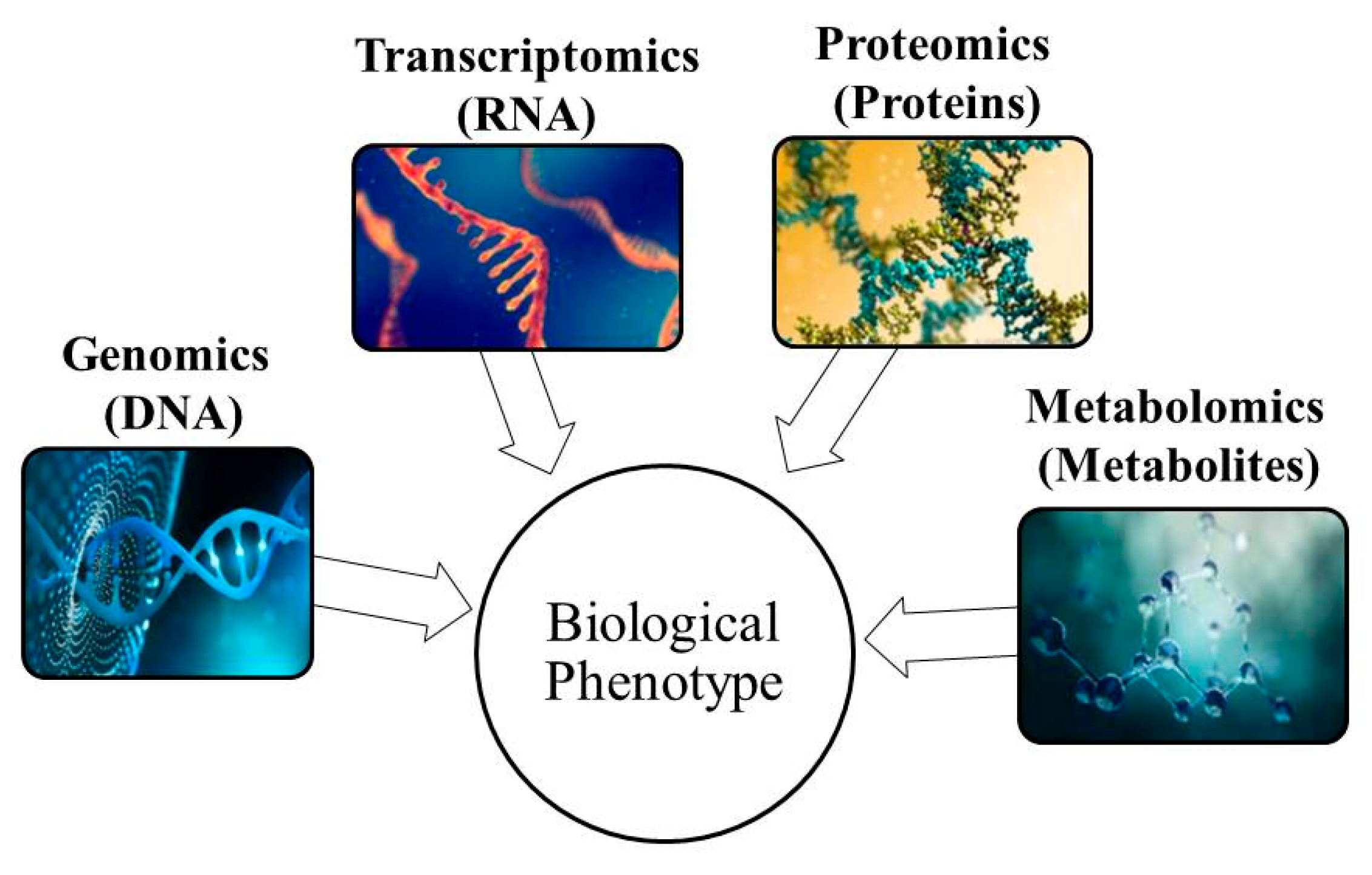
3.1. Principle
3.2. Challenges of Metabolomics Approach
3.3. Statistical Analysis in the Metabolomics Approach
4. Detection Technologies
4.1. High-Performance Liquid Chromatography (HPLC)
4.2. Fourier Transform Infrared (FTIR) Spectroscopy
4.3. Nuclear Magnetic Resonance (NMR) Spectroscopy
4.4. GC-MS and LC-MS
5. Food Authentication Using Metabolomics Approach
5.1. Meat and Fish/Seafood Products
5.2. Milk and Dairy Products
5.3. Fruit and Vegetable Product
5.4. Other Food Products
6. Conclusions and Recommendation
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Danezis, G.P.; Tsagkaris, A.S.; Camin, F.; Brusic, V.; Georgiou, C.A. Food authentication: Techniques, trends & emerging approaches. TrAC Trend. Anal. Chem. 2016, 85, 123–132. [Google Scholar] [CrossRef] [Green Version]
- Clish, C.B. Metabolomics: An emerging but powerful tool for precision medicine. Cold Spring Harbor. Mol. Case Stud. 2015, 1, a000588. [Google Scholar] [CrossRef] [Green Version]
- Kim, S.; Kim, J.; Yun, E.J.; Kim, K.H. Food metabolomics: From farm to human. Curr. Opin. Biotechnol. 2016, 37, 16–23. [Google Scholar] [CrossRef] [PubMed]
- Yan, M.; Xu, G. Current and future perspectives of functional metabolomics in disease studies—A review. Anal. Chim. Acta 2018, 1037, 41–54. [Google Scholar] [CrossRef]
- Liang, Q.; Wang, C.; Li, B.; Zhang, A.H. Metabolic fingerprinting to understand therapeutic effects and mechanisms of silybin on acute liver damage in rats. Pharmacogn. Mag. 2015, 11, 586–593. [Google Scholar] [CrossRef] [Green Version]
- Riedl, J.; Esslinger, S.; Fauhl-Hassek, C. Review of validation and reporting of non-targeted fingerprinting approaches for food authentication. Anal. Chim. Acta 2015, 885, 17–32. [Google Scholar] [CrossRef]
- Erban, A.; Fehrle, I.; Martinez-Seidel, F.; Brigante, F.; Más, A.L.; Baroni, V.; Wunderlin, D.; Kopka, J. Discovery of food identity markers by metabolomics and machine learning technology. Sci. Rep. 2019, 9, 1–19. [Google Scholar] [CrossRef] [Green Version]
- Cubero-Leon, E.; Peñalver, R.; Maquet, A. Review on metabolomics for food authentication. Food Res. Int. 2014, 60, 95–107. [Google Scholar] [CrossRef]
- Preti, R. Core-shell columns in high-performance liquid chromatography: Food analysis applications. Int. J. Anal. Chem. 2016, 2016, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Su, W.H.; Arvanitoyannis, I.S.; Sun, D.-W. Trends in Food Authentication. In Modern Techniques for Food Authentication, 2nd ed.; Elsevier Inc.: Amsterdam, The Netherlands, 2018; pp. 731–758. [Google Scholar] [CrossRef]
- Su, W.H.; He, H.J.; Sun, D.W. Non-destructive and rapid evaluation of staple foods quality by using spectroscopic techniques: A review. Crit. Rev. Food Sci. Nutr. 2017, 57, 1039–1051. [Google Scholar] [CrossRef]
- Kamal, M.; Karoui, R. Analytical methods coupled with chemometric tools for determining the authenticity and detecting the adulteration of dairy products: A review. Trend. Food Sci. Technol. 2015, 46, 27–48. [Google Scholar] [CrossRef]
- Rohman, A.; Man, Y.B.C. Fourier transforms infrared (FTIR) spectroscopy for analysis of extra virgin olive oil adulterated with palm oil. Food Res. Int. 2010, 43, 886–892. [Google Scholar] [CrossRef]
- Kettunen, J.; Tukiainen, T.; Sarin, A.P.; Ortega-Alonso, A.; Tikkanen, E.; Lyytikäinen, L.P.; Kangas, A.J.; Soininen, P.; Würtz, P.; Silander, K.; et al. Genome-wide association study identifies multiple loci influencing human serum metabolite levels. Nat. Genet. 2012, 44, 269–276. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Turi, K.N.; Romick-Rosendale, L.; Ryckman, K.K.; Hartert, T.V. A review of metabolomics approaches and their application in identifying causal pathways of childhood asthma. J. Allergy Clin. Immunol. 2018, 141, 1191–1201. [Google Scholar] [CrossRef] [Green Version]
- Li, S.; Tian, Y.; Jiang, P.; Lin, Y.; Liu, X.; Yang, H. Recent advances in the application of metabolomics for food safety control and food quality analyses. Crit. Rev. Food Sci. Nutr. 2020, 61, 1–22. [Google Scholar] [CrossRef]
- Pinu, F.R.; Goldansaz, S.A.; Jaine, J. Translational metabolomics: Current challenges and future opportunities. Metabolites 2019, 9, 108. [Google Scholar] [CrossRef] [Green Version]
- Gertsman, I.; Barshop, B.A. Promises and pitfalls of untargeted metabolomics. J. Inherit. Metabol. Dis. 2018, 41, 355–366. [Google Scholar] [CrossRef]
- Dunn, W.B.; Erban, A.; Weber, R.J.M.; Creek, D.J.; Brown, M.; Breitling, R.; Hankemeier, T.; Goodacre, R.; Neumann, S.; Kopka, J.; et al. Mass appeal: Metabolite identification in mass spectrometry-focused untargeted metabolomics. Metabolomics 2013, 9, 44–66. [Google Scholar] [CrossRef] [Green Version]
- Roberts, L.D.; Souza, A.L.; Gerszten, R.E.; Clish, C.B. Targeted metabolomics. Curr. Protoc. Mol. Biol. 2012, 1, 1–24. [Google Scholar] [CrossRef]
- Fan, T.W.M.; Lane, A.N. Applications of NMR spectroscopy to systems biochemistry. Progr. Nucl. Magnet. Reason. Spectrosc. 2016, 92–93, 18–53. [Google Scholar] [CrossRef] [Green Version]
- Tognarelli, J.M.; Dawood, M.; Shariff, M.I.F.; Grover, V.P.B.; Crossey, M.M.E.; Cox, I.J.; Taylor-Robinson, S.D.; McPhail, M.J.W. Magnetic Resonance Spectroscopy: Principles and Techniques: Lessons for Clinicians. J. Clin. Exp. Hepatol. 2015, 5, 320–328. [Google Scholar] [CrossRef] [Green Version]
- Schrimpe-Rutledge, A.C.; Codreanu, S.G.; Sherrod, S.D.; McLean, J.A. Untargeted metabolomics strategies—Challenges and emerging directions. J. Am. Soc. Mass Spectrom. 2016, 27, 1897–1905. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jacob, M.; Malkawi, A.; Albast, N.; Al Bougha, S.; Lopata, A.; Dasouki, M.; Rahman, A.M.A. A targeted metabolomics approach for clinical diagnosis of inborn errors of metabolism. Anal. Chim. Acta 2018, 1025, 141–153. [Google Scholar] [CrossRef]
- Patti, G.J.; Yanes, O.; Siuzdak, G. Metabolomics: The apogee of the omics trilogy. Nat. Rev. Mol. Cell Biol. 2012, 13, 263–269. [Google Scholar] [CrossRef]
- Pinu, F.R. Grape and wine metabolomics to develop new insights using untargeted and targeted approaches. Fermentation 2018, 4, 92. [Google Scholar] [CrossRef] [Green Version]
- Villas-Bôas, S.G.; Rasmussen, S.; Lane, G.A. Metabolomics or metabolite profiles? Trend. Biotechnol. 2005, 8, 385–386. [Google Scholar] [CrossRef] [PubMed]
- Kell, D.B.; Oliver, S.G. The metabolome 18 years on: A concept comes of age. Metabolomics 2016, 12, 1–8. [Google Scholar] [CrossRef]
- Pinu, F.R.; Beale, D.J.; Paten, A.M.; Kouremenos, K.; Swarup, S.; Schirra, H.J.; Wishart, D. Systems biology and multi-omics integration: Viewpoints from the metabolomics research community. Metabolites 2019, 9, 76. [Google Scholar] [CrossRef] [Green Version]
- Muroya, S.; Ueda, S.; Komatsu, T.; Miyakawa, T.; Ertbjerg, P. Meatabolomics: Muscle and meat metabolomics in domestic animals. Metabolites 2020, 10, 188. [Google Scholar] [CrossRef]
- Von, B.C.; Brockmeyer, J.; Humpf, H.U. Meat authentication: A new HPLC- MS/MS based method for the fast and sensitive detection of horse and pork in highly processed food. J. Agric. Food Chem. 2014, 62, 9428–9435. [Google Scholar] [CrossRef]
- Stephan, N.; Halama, A.; Mathew, S.; Hayat, S.; Bhagwat, A.; Mathew, L.S.; Diboun, I.; Malek, J.; Suhre, K. A comprehensive metabolomic data set of the date palm fruit. Data Brief. 2018, 18, 1313–1321. [Google Scholar] [CrossRef]
- Salzano, A.; Manganiello, G.; Neglia, G.; Vinale, F.; De Nicola, D.; D’Occhio, M.; Campanile, G. A preliminary study on metabolome profiles of buffalo milk and corresponding mozzarella cheese: Safeguarding the authenticity and traceability of protected status buffalo dairy products. Molecules 2020, 25, 304. [Google Scholar] [CrossRef] [Green Version]
- Scano, P.; Murgia, A.; Pirisi, F.M.; Caboni, P. A gas chromatography-mass spectrometry-based metabolomic approach for the characterization of goat milk compared with cow milk. J. Dairy Sci. 2014, 97, 6057–6066. [Google Scholar] [CrossRef]
- Wang, L.; Meeus, I.; Rombouts, C.; Van Meulebroek, L.; Vanhaecke, L.; Smagghe, G. Metabolomics-based biomarker discovery for bee health monitoring: A proof of concept study concerning nutritional stress in Bombus terrestris. Sci. Rep. 2019, 9, 11423. [Google Scholar] [CrossRef]
- Cambiaghi, A.; Ferrario, M.; Masseroli, M. Analysis of metabolomic data: Tools, current strategies and future challenges for omics data integration. Brief. Bioinform. 2017, 18, 498–510. [Google Scholar] [CrossRef] [Green Version]
- Khakimov, B.; Gürdeniz, G.; Engelsen, S.B. Trends in the application of chemometrics to foodomics studies. Acta Aliment. 2015, 1, 4–31. [Google Scholar] [CrossRef] [Green Version]
- Nollet, L.M.L. High pressure liquid chromatography (HPLC) in food authentication. Food Authent. Traceabil. 2013, 218–238. [Google Scholar] [CrossRef]
- Noman, A.; AL-Bukhaiti, W.Q.; Ammar, A.; Abed, S.M.; Mahdi, A.A.; Al-ansi, W.A. HPLC technique used in food analysis–Review. Int. J. Agric. Innov. Res. 2016, 5, 181–188. [Google Scholar]
- Sylvestre, N.J.; Fofana, I.; Hadjadj, Y.; Beroual, A. Review of physicochemical- based diagnostic techniques for assessing insulation condition in aged transformers. Energies 2016, 9, 367. [Google Scholar] [CrossRef]
- Rodriguez-Saona, L.E.; Allendorf, M.E. Use of FTIR for rapid authentication and detection of adulteration of food. Annu. Rev. Food Sci. Technol. 2011, 2, 467–483. [Google Scholar] [CrossRef] [PubMed]
- Georgouli, K.; Martinez Del Rincon, J.; Koidis, A. Continuous statistical modeling for rapid detection of adulteration of extra virgin olive oil using mid-infrared and Raman spectroscopic data. Food Chem. 2017, 217, 735–742. [Google Scholar] [CrossRef] [Green Version]
- Rohman, A.; Sismindari, E.Y.; Che Man, Y.B. Analysis of pork adulteration in beef meatball using Fourier transform infrared (FTIR) spectroscopy. Meat Sci. 2011, 88, 91–95. [Google Scholar] [CrossRef]
- Vardin, H.; Tay, A.; Ozen, B.; Mauer, L. Authentication of pomegranate juice concentrate using FTIR spectroscopy and chemometrics. Food Chem. 2008, 108, 742–748. [Google Scholar] [CrossRef] [Green Version]
- Rohman, A. The employment of Fourier transforms infrared spectroscopy coupled with chemometrics techniques for traceability and authentication of meat and meat products. J. Adv. Vet. Anim. Res. 2019, 6, 9–17. [Google Scholar] [CrossRef]
- Valand, R.; Tanna, S.; Lawson, G.; Bengtström, L. A review of Fourier Transform Infrared (FTIR) spectroscopy used in food adulteration and authenticity investigations. Food Addit. Contam. Part A Chem. Anal. Control. Expos. Risk Assess. 2020, 37, 19–38. [Google Scholar] [CrossRef]
- Markley, J.L.; Brüschweiler, R.; Edison, A.; Eghbalnia, H.R.; Powers, R.; Raftery, D.; Wishart, D.S. The future of NMR-based metabolomics. Curr. Opin. Biotechnol. 2016, 43, 34–40. [Google Scholar] [CrossRef] [Green Version]
- Miggiels, P.; Wouters, B.; van Westen, G.J.; Dubbelman, A.C.; Hankemeier, T. Novel technologies for metabolomics: More for less. TrAC Trend. Anal. Chem. 2019, 120, 115323. [Google Scholar] [CrossRef]
- Franca, A.S.; Oliveira, L.S. Potential uses of Fourier transform infrared spectroscopy (FTIR) in food processing and engineering. Food Eng. 2011, 16, 211–227. [Google Scholar]
- Hatzakis, E. Nuclear Magnetic Resonance (NMR) Spectroscopy in Food Science: A Comprehensive Review. Comprehens. Rev. Food Sci. Food Saf. 2019, 18, 189–220. [Google Scholar] [CrossRef] [Green Version]
- Cao, G.; Li, K.; Guo, J.; Lu, M.; Hong, Y.; Cai, Z. Mass spectrometry for analysis of changes during food storage and processing. J. Agric. Food Chem. 2020, 68, 6956–6966. [Google Scholar] [CrossRef] [PubMed]
- Witjaksono, G.; Alva, S. Applications of Mass Spectrometry to the Analysis of Adulterated Food. Mass Spectrom. Future Percep. Appl. 2019. [Google Scholar] [CrossRef] [Green Version]
- Pitt, J.J. Principles and applications of liquid chromatography-mass spectrometry in clinical biochemistry. Clin. Biochem. Rev. 2009, 30, 19–34. [Google Scholar] [PubMed]
- Schütz, D.; Achten, E.; Creydt, M.; Riedl, J.; Fischer, M. Non-targeted LC-MS metabolomics approach towards an authentication of the geographical origin of grain maize (Zea mays L.) samples. Foods 2021, 10, 2160. [Google Scholar] [CrossRef] [PubMed]
- Böhme, K.; Calo-Mata, P.; Barros-Velázquez, J.; Ortea, I. Recent applications of omics-based technologies to main topics in food authentication. TrAC Trend. Anal. Chem. 2019, 110, 221–232. [Google Scholar] [CrossRef]
- Hajslova, J.; Cajka, T.; Vaclavik, L. Challenging applications offered by direct analysis in real time (DART) in food-quality and safety analysis. TrAC Trend. Anal. Chem. 2011, 30, 204–218. [Google Scholar] [CrossRef]
- Senyuva, H.Z.; Gökmen, V.; Sarikaya, E.A. Future perspectives in Orbitrap™-high-resolution mass spectrometry in food analysis: A review. Food Addit. Contam. Part A 2015, 32, 1568–1606. [Google Scholar] [CrossRef]
- Giannoukos, K.; Giannoukos, S.; Lagogianni, C.; Tsitsigiannis, D.I.; Taylor, S. Analysis of volatile emissions from grape berries infected with Aspergillus carbonarius using hyphenated and portable mass spectrometry. Sci. Rep. 2020, 10, 21179. [Google Scholar] [CrossRef]
- Jjunju, F.P.; Giannoukos, S.; Marshall, A.; Taylor, S. In-situ analysis of essential fragrant oils using a portable mass spectrometer. Int. J. Anal. Chem. 2019, 2019, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Li, L.F.; Chen, C.H.; Ren, Y.; Hendricks, P.I.; Cooks, R.G.; Ouyang, Z. Mini 12, Miniature mass spectrometer for clinical and other applications-introduction and characterization. Anal. Chem. 2014, 86, 2909–2916. [Google Scholar] [CrossRef]
- García-Reyes, J.F.; Mazzoti, F.; Harper, J.D.; Charipar, N.A.; Oradu, S.; Ouyang, Z.; Sindona, G.; Cooks, R.G. Direct olive oil analysis by low-temperature plasma (LTP) ambient ionization mass spectrometry. Rapid Commun. Mass Spectrom. 2009, 23, 3057–3062. [Google Scholar] [CrossRef]
- Gamboa-Becerra, R.; Montero-Vargas, J.M.; Martínez-Jarquín, S.; Gálvez-Ponce, E.; Moreno-Pedraza, A.; Winkler, R. Rapid classification of coffee products by data mining models from direct electrospray and plasma-based mass spectrometry analyses. Food Anal. Methods 2017, 10, 1359–1368. [Google Scholar] [CrossRef]
- Huang, G.; Ouyang, Z.; Cooks, R.G. High-throughput trace melamine analysis in complex mixtures. Chem. Commun. 2009, 5, 556–558. [Google Scholar] [CrossRef]
- Huang, G.; Xu, W.; Visbal-Onufrak, M.A.; Ouyang, Z.; Cooks, R.G. Direct analysis of melamine in complex matrices using a handheld mass spectrometer. Analyst 2010, 135, 705–711. [Google Scholar] [CrossRef] [PubMed]
- Ibáñez, C.; García-Cañas, V.; Valdés, A.; Simó, C. Novel MS-based approaches and applications in food metabolomics. TrAC Trend. Anal. Chem. 2013, 52, 100–111. [Google Scholar] [CrossRef]
- Campmajó, G.; Núñez, N.; Núñez, O. The Role of Liquid Chromatography-Mass Spectrometry in Food Integrity and Authenticity. In Mass Spectrometry—Future Perceptions and Applications; Kamble, G.S., Ed.; IntechOpen: London, UK, 2019; pp. 3–20. Available online: https://www.intechopen.com/chapters/66149 (accessed on 14 December 2020). [CrossRef] [Green Version]
- Kurniawati, E.; Rohman, A.; Triyana, K. Analysis of lard in meatball broth using Fourier transforms infrared spectroscopy and chemometrics. Meat Sci. 2014, 96, 94–98. [Google Scholar] [CrossRef]
- Sidwick, K.L.; Johnson, A.E.; Adam, C.D.; Pereira, L.; Thompson, D.F. Use of liquid chromatography quadrupole time-of-flight mass spectrometry and metabonomic profiling to differentiate between normally slaughtered and dead on arrival poultry meat. Anal. Chem. 2017, 89, 12131–12136. [Google Scholar] [CrossRef]
- Cheng, J.H.; Dai, Q.; Sun, D.W.; Zeng, X.A.; Liu, D.; Pu, H.B. Applications of non-destructive spectroscopic techniques for fish quality and safety evaluation and inspection. Trend. Food Sci. Technol. 2013, 34, 18–31. [Google Scholar] [CrossRef]
- Gudjónsdóttir, M.; Arason, S.; Rustad, T. The effects of pre-salting methods on water distribution and protein denaturation of dry salted and rehydrated cod—A low-field NMR study. J. Food Eng. 2011, 104, 23–29. [Google Scholar] [CrossRef]
- Sánchez-Alonso, I.; Martinez, I.; Sánchez-Valencia, J.; Careche, M. Estimation of freezing storage time and quality changes in hake (Merluccius merluccius, L.) by low field NMR. Food Chem. 2012, 135, 1626–1634. [Google Scholar] [CrossRef] [Green Version]
- Cozzolino, D.; Murray, I. A review on the application of infrared technologies to determine and monitor the composition and other quality characteristics in raw fish, fish products, and seafood. Appl. Spectrosc. Rev. 2012, 47, 207–218. [Google Scholar] [CrossRef]
- Takakura, Y.; Sakamoto, T.; Hirai, S.; Masuzawa, T.; Wakabayashi, H.; Nishimura, T. Characterization of the key aroma compounds in beef extract using aroma extract dilution analysis. Meat Sci. 2014, 97, 27–31. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Yang, Y.; Pan, D.; He, J.; Cao, J.; Wang, H.; Ertbjerg, P. Metabolite profile based on 1H NMR of broiler chicken breasts affected by wooden breast myodegeneration. Food Chem. 2020, 310, 125852. [Google Scholar] [CrossRef]
- Xing, T.; Zhao, X.; Xu, X.; Li, J.; Zhang, L.; Gao, F. Physiochemical properties, protein and metabolite profiles of muscle exudate of chicken meat affected by wooden breast myopathy. Food Chem. 2020, 316, 126271. [Google Scholar] [CrossRef] [PubMed]
- Welzenbach, J.; Neuhoff, C.; Looft, C.; Schellander, K.; Tholen, E.; Große-Brinkhaus, C. Different statistical approaches to investigate porcine muscle metabolome profiles to highlight new biomarkers for pork quality assessment. PLoS ONE 2016, 11, e0149758. [Google Scholar] [CrossRef]
- Lytou, A.E.; Nychas, G.J.E.; Panagou, E.Z. Effect of pomegranate based marinades on the microbiological, chemical, and sensory quality of chicken meat: A metabolomics approach. Int. J. Food MicroBiol. 2018, 267, 42–53. [Google Scholar] [CrossRef] [PubMed]
- Standal, I.B.; Axelson, D.E.; Aursand, M. 13C NMR as a tool for authentication of different gadoid fish species with emphasis on phospholipid profiles. Food Chem. 2010, 121, 608–615. [Google Scholar] [CrossRef]
- Jinadasa, B.K.K.K.; Jayasinghe, G.D.T.M.; Ahmad, S.B.N. Validation of high-performance liquid chromatography (HPLC) method for quantitative analysis of histamine in fish and fishery products. Cogent Chem. 2016, 2, 1156806. [Google Scholar] [CrossRef]
- Chatterjee, N.S.; Chevallier, O.P.; Wielogorska, E.; Black, C.; Elliott, C.T. Simultaneous authentication of species identity and geographical origin of shrimps: Untargeted metabolomics to recurrent biomarker ions. J. Chromatogr. A 2019, 1599, 75–84. [Google Scholar] [CrossRef]
- Yang, Y.; Zheng, N.; Zhao, X.; Zhang, Y.; Han, R.; Yang, J.; Zhao, S.; Li, S.; Guo, T.; Zang, C.; et al. Metabolomic biomarkers identify differences in milk produced by Holstein cows and other minor dairy animals. J. Proteom. 2016, 136, 174–182. [Google Scholar] [CrossRef]
- Sundekilde, U.K.; Frederiksen, P.D.; Clausen, M.R.; Larsen, L.B.; Bertram, H.C. Relationship between the metabolite profile and technological properties of bovine milk from two dairy breeds elucidated by NMR-based metabolomics. J. Agric. Food Chem. 2011, 59, 7360–7367. [Google Scholar] [CrossRef]
- Trimigno, A.; Lyndgaard, C.B.; Atladóttir, G.A.; Aru, V.; Engelsen, S.B.; Clemmensen, L.K.H. An NMR metabolomics approach to investigate factors affecting the yoghurt fermentation process and quality. Metabolites 2020, 10, 293. [Google Scholar] [CrossRef] [PubMed]
- Sundekilde, U.; Larsen, L.; Bertram, H. NMR-based milk metabolomics. Metabolites 2013, 3, 204–222. [Google Scholar] [CrossRef] [PubMed]
- Klein, M.S.; Almstetter, M.F.; Schlamberger, G.; Nürnberger, N.; Dettmer, K.; Oefner, P.J.; Gronwald, W. Nuclear magnetic resonance and mass spectrometry-based milk metabolomics in dairy cows during early and late lactation. J. Dairy Sci. 2010, 93, 1539–1550. [Google Scholar] [CrossRef] [PubMed]
- Monakhova, Y.B.; Kuballa, T.; Leitz, J.; Andlauer, C.; Lachenmeier, D.W. NMR spectroscopy as a screening tool to validate nutrition labeling of milk, lactose-free milk, and milk substitutes based on soy and grains. Dairy Sci. Technol. 2012, 92, 109–120. [Google Scholar] [CrossRef] [Green Version]
- Suzuki, Y.; Nakashita, R. Authentication and Traceability of Fruits and Vegetables. In Comprehensive Analytical Chemistry, 1st ed.; Elsevier: Amsterdam, The Netherlands, 2013; Volume 60. [Google Scholar] [CrossRef]
- Zhang, J.; Yu, Q.; Cheng, H.; Ge, Y.; Liu, H.; Ye, X.; Chen, Y. Metabolomic approach for the authentication of berry fruit juice by liquid chromatography quadrupole time-of-flight mass spectrometry coupled to chemometrics. J. Agric. Food Chem. 2018, 66, 8199–8208. [Google Scholar] [CrossRef]
- Åkerström, A.; Jaakola, L.; Bång, U.; Jäderlund, A. Effects of altitude-related factors and geographical origin on anthocyanidin concentrations in fruits of Vaccinium myrtillus L. (Bilberries). J. Agric. Food Chem. 2010, 58, 11939–11945. [Google Scholar] [CrossRef]
- Soto-hernández, M.; Ibarra-estrada, E.; Barrientos-priego, A.F. Metabolomic approaches for the characterization of fruits: A case study on avocado. Curr. Top. PhytoChem. 2016, 13, 79–89. [Google Scholar]
- Anjaritha, A.A.; Ridwani, S.; Dwivany, F.M.; Putri, S.P.; Fukusaki, E. A metabolomics-based approach for the evaluation of off-tree ripening conditions and different postharvest treatments in mangosteen (Garcinia mangostana). Metabolomics 2019, 15, 1–16. [Google Scholar] [CrossRef]
- Abreu, A.C.; Fernández, I. NMR metabolomics applied on the discrimination of variables influencing tomato (Solanum lycopersicum). Molecules 2020, 25, 3738. [Google Scholar] [CrossRef]
- Vlaic, R.A.; Mureșan, A.E.; Mureșan, C.C.; Petruț, G.S.; Mureșan, V.; Muste, S. Quantitative analysis by HPLC and FT-MIR prediction of individual sugars from the plum fruit harvested during growth and fruit development. Agronomy 2018, 8, 306. [Google Scholar] [CrossRef] [Green Version]
- Epriliati, I.; Kerven, G.; D’Arcy, B.; Gidley, M.J. Chromatographic analysis of diverse fruit components using HPLC and UPLC. Anal. Meth. 2010, 2, 1606–1613. [Google Scholar] [CrossRef]
- Jayaprakasha, G.K.; Patil, B.S. A metabolomics approach to identify and quantify the phytochemicals in watermelons by quantitative 1HNMR. Talanta 2016, 153, 268–277. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tamura, Y.; Mori, T.; Nakabayashi, R.; Kobayashi, M.; Saito, K.; Okazaki, S.; Kusano, M. Metabolomic evaluation of the quality of leaf lettuce grown in practical plant factory to capture metabolite signature. Front. Plant Sci. 2018, 9, 665. [Google Scholar] [CrossRef]
- Hrbek, V.; Rektorisova, M.; Chmelarova, H.; Ovesna, J.; Hajslova, J. Authenticity assessment of garlic using a metabolomic approach based on high-resolution mass spectrometry. J. Food Compos. Anal. 2018, 67, 19–28. [Google Scholar] [CrossRef]
- Chudzinska, M.; Baralkiewicz, D. Estimation of honey authenticity by multi-elements characteristics using inductively coupled plasma-mass spectrometry (ICP-MS) combined with chemometrics. Food Chem. Toxicol. 2010, 48, 284–290. [Google Scholar] [CrossRef]
- Ouchemoukh, S.; Schweitzer, P.; Bachir, B.M.; Djoudad-Kadji, H.; Louaileche, H. HPLC sugar profiles of Algerian honey. Food Chem. 2010, 121, 561–568. [Google Scholar] [CrossRef]
- Kenjerić, D.; Mandić, M.L.; Primorac, L.; Bubalo, D.; Perl, A. Flavonoid profile of Robinia honey produced in Croatia. Food Chem. 2007, 102, 683–690. [Google Scholar] [CrossRef]
- Jerković, I.; Marijanović, Z.; Kezić, J.; Gugić, M. Headspace, volatile and semi-volatile organic compounds diversity and radical scavenging activity of ultrasonic solvent extracts from Amorpha fruticosa honey samples. Molecules 2009, 14, 2717–2728. [Google Scholar] [CrossRef] [Green Version]
- Jumhawan, U.; Putri, S.P.; Yusianto, B.T.; Fukusaki, E. Quantification of coffee blends for authentication of Asian palm civet coffee (Kopi Luwak) via metabolomics: A proof of concept. J. Biosci. Bioeng. 2016, 122, 79–84. [Google Scholar] [CrossRef]
- Chen, Q.; Zhao, J.; Lin, H. Study on discrimination of Roast green tea (Camellia sinensis L.) according to geographical origin by FT-NIR spectroscopy and supervised pattern recognition. SpectroChim. Acta Part A Mol. BioMol. Spectrosc. 2009, 72, 845–850. [Google Scholar] [CrossRef]
- Fraser, K.; Lane, G.A.; Otter, D.E.; Hemar, Y.; Quek, S.Y.; Harrison, S.J.; Rasmussen, S. Analysis of metabolic markers of tea origin by UHPLC and high-resolution mass spectrometry. Food Res. Int. 2013, 53, 827–835. [Google Scholar] [CrossRef]
| Commodity | Issues | References |
|---|---|---|
| Fruits and vegetables |
| [10] |
| Grain |
| [11] |
| Milk and dairy |
| [12] |
| Oil and fat |
| [10,13] |
| Meat and fish |
| [10] |
| Features | Untargeted Metabolomics | Targeted Metabolomics |
|---|---|---|
| Benefits |
|
|
| Limitations |
|
|
| Instruments | Benefits | Limitations | References |
|---|---|---|---|
| HPLC |
|
| [37] |
| FTIR |
|
| [43] |
| Food Type | Factors Analyzed | Instrument Used | References |
|---|---|---|---|
| Beef | Flavor | GC-MS | [73] |
| Chicken | Wooden breast disorder (muscle abnormalities) | H-NMR | [74,75] |
| Pig | Drip loss (SNP) | GC-MS, LC-MS | [76] |
| Chicken | Marinade type, storage time, microbial load, sensory | GC-MS | [77] |
| Beef | Pork adulteration | FTIR | [41] |
| Fish | Muscle lipid | C-NMR | [78] |
| Fish | Histamine | HPLC | [79] |
| Shrimp | Species and geographical origin | [80] |
| Food Type | Factors Analyzed | Instrument Used | References |
|---|---|---|---|
| Milk |
| NMR, LC-MS | [81] |
| GC-MS | [34] | |
| NMR | [84] | |
| NMR, GC-MS | [85] | |
| NMR | [86] | |
| Yogurt |
| NMR | [83] |
| Cheese |
| GC-MS | [33] |
| Food Type | Factors Analyzed | Instrument Used | References |
|---|---|---|---|
| Mangosteen fruit |
| GC-MS | [91] |
| Palm fruit |
| GC-MS, LC-MS | [32] |
| Plum fruit |
| HPLC, FT-MIR | [93] |
| Fruits |
| HPLC | [94] |
| Garlic |
| HPLC-HRMS | [95] |
| Watermelon |
| NMR | [96] |
| Lettuce |
| GC-MS, LC-MS | [97] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Selamat, J.; Rozani, N.A.A.; Murugesu, S. Application of the Metabolomics Approach in Food Authentication. Molecules 2021, 26, 7565. https://doi.org/10.3390/molecules26247565
Selamat J, Rozani NAA, Murugesu S. Application of the Metabolomics Approach in Food Authentication. Molecules. 2021; 26(24):7565. https://doi.org/10.3390/molecules26247565
Chicago/Turabian StyleSelamat, Jinap, Nur Amalyn Alyaa Rozani, and Suganya Murugesu. 2021. "Application of the Metabolomics Approach in Food Authentication" Molecules 26, no. 24: 7565. https://doi.org/10.3390/molecules26247565
APA StyleSelamat, J., Rozani, N. A. A., & Murugesu, S. (2021). Application of the Metabolomics Approach in Food Authentication. Molecules, 26(24), 7565. https://doi.org/10.3390/molecules26247565







