Predictive Model for Drug-Induced Liver Injury Using Deep Neural Networks Based on Substructure Space
Abstract
:1. Introduction
2. Results
2.1. Preparation of Data Sets and Applicability Domain
2.2. Development and Validation of DNN-Based DILI-Prediction Model
2.3. Performance Comparison of the Best DNN Model with Machine Learning Models
2.4. Performance Test of the DNN Model on the External Data Sets from Prior Studies
2.5. DILI Prediction on Drugs That Have Case Reports on Liver Injury
3. Discussion
4. Materials and Methods
4.1. Data Set Preparation and Curation
4.2. Model Development and Architecture of the DNN Model
4.3. Model Training
4.4. Applicability Domain and Model Evaluation
4.5. Evaluation Metrics
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Sample Availability
References
- MacDonald, J.S.; Robertson, R.T. Toxicity Testing in the 21st Century: A View from the Pharmaceutical Industry. Toxicol. Sci. 2009, 110, 40–46. [Google Scholar] [CrossRef] [Green Version]
- Hay, M.; Thomas, D.W.; Craighead, J.L.; Economides, C.; Rosenthal, J. Clinical development success rates for investigational drugs. Nat. Biotechnol. 2014, 32, 40–51. [Google Scholar] [CrossRef] [PubMed]
- Harrison, R.K. Phase II and phase III failures: 2013–2015. Nat. Rev. Drug Discov. 2016, 15, 817–818. [Google Scholar] [CrossRef] [PubMed]
- Chi, L.H.; Burrows, A.D.; Anderson, R.L. Can preclinical drug development help to predict adverse events in clinical trials? Drug Discov. Today 2021, in press. [Google Scholar] [CrossRef]
- Reuben, A.; Koch, D.G.; Lee, W.M.; Group, A.L.F.S. Drug-induced acute liver failure: Results of a U.S. multicenter, prospective study. Hepatology 2010, 52, 2065–2076. [Google Scholar] [CrossRef] [Green Version]
- Larrey, D. Epidemiology and individual susceptibility to adverse drug reactions affecting the liver. Semin. Liver Dis. 2002, 22, 145–155. [Google Scholar] [CrossRef]
- Fung, M.; Thornton, A.; Mybeck, K.; Wu, J.H.; Hornbuckle, K.; Muniz, E. Evaluation of the Characteristics of Safety Withdrawal of Prescription Drugs from Worldwide Pharmaceutical Markets-1960 to 1999. Drug Inf. J. 2001, 35, 293–317. [Google Scholar] [CrossRef]
- Chen, M.; Vijay, V.; Shi, Q.; Liu, Z.; Fang, H.; Tong, W. FDA-approved drug labeling for the study of drug-induced liver injury. Drug Discov. Today 2011, 16, 697–703. [Google Scholar] [CrossRef]
- Chen, M.; Suzuki, A.; Borlak, J.; Andrade, R.J.; Lucena, M.I. Drug-induced liver injury: Interactions between drug properties and host factors. J. Hepatol. 2015, 63, 503–514. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kaplowitz, N. Idiosyncratic drug hepatotoxicity. Nat. Rev. Drug Discov. 2005, 4, 489–499. [Google Scholar] [CrossRef]
- Chen, M.; Suzuki, A.; Thakkar, S.; Yu, K.; Hu, C.; Tong, W. DILIrank: The largest reference drug list ranked by the risk for developing drug-induced liver injury in humans. Drug Discov. Today 2016, 21, 648–653. [Google Scholar] [CrossRef]
- Liew, C.Y.; Lim, Y.C.; Yap, C.W. Mixed learning algorithms and features ensemble in hepatotoxicity prediction. J. Comput.-Aided Mol. Des. 2011, 25, 855–871. [Google Scholar] [CrossRef]
- Kotsampasakou, E.; Montanari, F.; Ecker, G.F. Predicting drug-induced liver injury: The importance of data curation. Toxicology 2017, 389, 139–145. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Cheng, F.; Li, W.; Liu, G.; Lee, P.W.; Tang, Y. In silico Prediction of Drug Induced Liver Toxicity Using Substructure Pattern Recognition Method. Mol. Inform. 2016, 35, 136–144. [Google Scholar] [CrossRef] [PubMed]
- Ai, H.; Chen, W.; Zhang, L.; Huang, L.; Yin, Z.; Hu, H.; Zhao, Q.; Zhao, J.; Liu, H. Predicting Drug-Induced Liver Injury Using Ensemble Learning Methods and Molecular Fingerprints. Toxicol. Sci. 2018, 165, 100–107. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hoofnagle, J.H.; Serrano, J.; Knoben, J.E.; Navarro, V.J. LiverTox: A website on drug-induced liver injury. Hepatology 2013, 57, 873–874. [Google Scholar] [CrossRef]
- Dargan, S.; Kumar, M.; Ayyagari, M.R.; Kumar, G. A Survey of Deep Learning and Its Applications: A New Paradigm to Machine Learning. Arch. Comput. Methods Eng. 2020, 27, 1071–1092. [Google Scholar] [CrossRef]
- Purushotham, S.; Tripathy, B.K. Evaluation of Classifier Models Using Stratified Tenfold Cross Validation Techniques BT—Global Trends in Information Systems and Software Applications; Krishna, P.V., Babu, M.R., Ariwa, E., Eds.; Springer: Berlin/Heidelberg, Germany, 2012; pp. 680–690. [Google Scholar]
- Schenker, B.; Agarwal, M. Cross-validated structure selection for neural networks. Comput. Chem. Eng. 1996, 20, 175–186. [Google Scholar] [CrossRef]
- Srivastava, N.; Geoffrey, H.; Alex, K.; Ilya, S.; Ruslan, S. Dropout: A simple way to prevent neural networks from over-fitting. J. Mach. Learn. Res. 2014, 15, 1929–1958. [Google Scholar]
- Xu, J.J.; Henstock, P.V.; Dunn, M.C.; Smith, A.R.; Chabot, J.R.; de Graaf, D. Cellular Imaging Predictions of Clinical Drug-Induced Liver Injury. Toxicol. Sci. 2008, 105, 97–105. [Google Scholar] [CrossRef] [Green Version]
- Greene, N.; Fisk, L.; Naven, R.T.; Note, R.R.; Patel, M.L.; Pelletier, D.J. Developing Structure−Activity Relationships for the Prediction of Hepatotoxicity. Chem. Res. Toxicol. 2010, 23, 1215–1222. [Google Scholar] [CrossRef] [PubMed]
- Thakkar, S.; Li, T.; Liu, Z.; Wu, L.; Roberts, R.; Tong, W. Drug-induced liver injury severity and toxicity (DILIst): Binary classification of 1279 drugs by human hepatotoxicity. Drug Discov. Today 2020, 25, 201–208. [Google Scholar] [CrossRef]
- Rao, R.B.; Fung, G.; Rosales, R. On the Dangers of Cross-Validation. An Experimental Evaluation. In Proceedings of the 2008 SIAM International Conference on Data Mining (SDM); Society for Industrial and Applied Mathematics: Philadelphia, PA, USA, 2008; pp. 588–596, ISBN 978-0-89871-654-2. [Google Scholar]
- Mulliner, D.; Schmidt, F.; Stolte, M.; Spirkl, H.-P.; Czich, A.; Amberg, A. Computational Models for Human and Animal Hepatotoxicity with a Global Application Scope. Chem. Res. Toxicol. 2016, 29, 757–767. [Google Scholar] [CrossRef] [Green Version]
- Angheleanu, R.; Swart, J.N. Flucloxacillin-induced liver injury in elderly patient. BMJ Case Rep. 2021, 14, e241071. [Google Scholar] [CrossRef]
- Crepin, S.; Godet, B.; Carrier, P.; Villeneuve, C.; Merle, L.; Laroche, M.-L. Probable drug-induced liver injury associated with aliskiren: Case report and review of adverse event reports from pharmacovigilance databases. Am. J. Health-Syst. Pharm. 2014, 71, 643–647. [Google Scholar] [CrossRef]
- Lee, M.J.; Berry, P.; D’Errico, F.; Miquel, R.; Kulasegaram, R. A case of rilpivirine drug-induced liver injury. Sex. Transm. Infect. 2020, 96, 618–619. [Google Scholar] [CrossRef] [Green Version]
- Ng, Q.X.; Yong, C.S.K.; Loke, W.; Yeo, W.S.; Soh, A.Y. Sen Escitalopram-induced liver injury: A case report and review of literature. World J. Hepatol. 2019, 11, 719–724. [Google Scholar] [CrossRef] [PubMed]
- Tan, Y.; Ye, Y.; Zhou, X. Nilotinib-induced liver injury: A case report. Medicine 2020, 99, e22061. [Google Scholar] [CrossRef] [PubMed]
- de la Torre-Aláez, M.; Iñarrairaegui, M. Drug Liver Injury Induced by Olmesartan Mediated by Autoimmune-Like Mechanism: A Case Report. Eur. J. Case Rep. Intern. Med. 2020, 7, 1407. [Google Scholar] [CrossRef]
- Pérez Palacios, D.; Giráldez Gallego, Á.; Carballo Rubio, V.; Solà Fernández, A.; Pascasio Acevedo, J.M. Drug-induced liver injury due to mesterolone: A case report. Gastroenterol. Hepatol. 2019, 42, 629–630. [Google Scholar] [CrossRef]
- Yu, H.; Zhang, W.; Shen, C.; Zhang, H.; Zhang, H.; Zhang, Y.; Zou, D.; Gong, X. Liver dysfunction induced by Levothyroxine Sodium Tablets (Euthyrox®) in a hypothyroid patient with Hashimoto’s thyroiditis: Case report and literature review. Endocr. J. 2019, 66, 769–775. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Atallah, E.; Wijayasiri, P.; Cianci, N.; Abdullah, K.; Mukherjee, A.; Aithal, G.P. Zanubrutinib-induced liver injury: A case report and literature review. BMC Gastroenterol. 2021, 21, 244. [Google Scholar] [CrossRef] [PubMed]
- Duzenli, T.; Tanoglu, A.; Akyol, T.; Kara, M.; Yazgan, Y. Drug-induced Liver Injury Caused by Phenprobamate: Strong Probability Due to Repeated Toxicity. Euroasian J. Hepato-Gastroenterol. 2019, 9, 49–51. [Google Scholar]
- Anastasia, E.J.; Rosenstein, R.S.; Bergsman, J.A.; Parra, D. Use of apixaban after development of suspected rivaroxaban-induced hepatic steatosis; a case report. Blood Coagul. Fibrinolysis Int. J. Haemost. Thromb. 2015, 26, 699–702. [Google Scholar] [CrossRef]
- Shavadia, J.S.; Sharma, A.; Gu, X.; Neaton, J.; DeLeve, L.; Holmes, D.; Home, P.; Eckel, R.H.; Watkins, P.B.; Granger, C.B. Determination of fasiglifam-induced liver toxicity: Insights from the data monitoring committee of the fasiglifam clinical trials program. Clin. Trials 2019, 16, 253–262. [Google Scholar] [CrossRef]
- Verma, N.; Kumar, P.; Mitra, S.; Taneja, S.; Dhooria, S.; Das, A.; Duseja, A.; Dhiman, R.K.; Chawla, Y. Drug idiosyncrasy due to pirfenidone presenting as acute liver failure: Case report and mini-review of the literature. Hepatol. Commun. 2018, 2, 142–147. [Google Scholar] [CrossRef]
- Barbara, M.; Dhingra, S.; Mindikoglu, A.L. Ligandrol (LGD-4033)-Induced Liver Injury. ACG Case Rep. J. 2020, 7, e00370. [Google Scholar] [CrossRef]
- Dinis-Oliveira, R.J.; Vieira, D.N. Acute liver failure requiring transplantation: A possible link to ulipristal acetate treatment? Basic Clin. Pharmacol. Toxicol. 2021, 129, 278–282. [Google Scholar] [CrossRef]
- Barbedo, J.G.A. Impact of dataset size and variety on the effectiveness of deep learning and transfer learning for plant disease classification. Comput. Electron. Agric. 2018, 153, 46–53. [Google Scholar] [CrossRef]
- Xu, Y.; Dai, Z.; Chen, F.; Gao, S.; Pei, J.; Lai, L. Deep Learning for Drug-Induced Liver Injury. J. Chem. Inf. Model. 2015, 55, 2085–2093. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Tong, W.; Roberts, R.; Liu, Z.; Thakkar, S. DeepDILI: Deep Learning-Powered Drug-Induced Liver Injury Prediction Using Model-Level Representation. Chem. Res. Toxicol. 2021, 34, 550–565. [Google Scholar] [CrossRef]
- Mayr, A.; Klambauer, G.; Unterthiner, T.; Hochreiter, S. DeepTox: Toxicity Prediction using Deep Learning. Front. Environ. Sci. 2016, 3, 80. [Google Scholar] [CrossRef] [Green Version]
- Salas-Eljatib, C.; Fuentes-Ramirez, A.; Gregoire, T.G.; Altamirano, A.; Yaitul, V. A study on the effects of unbalanced data when fitting logistic regression models in ecology. Ecol. Indic. 2018, 85, 502–508. [Google Scholar] [CrossRef]
- Kang, S.; Brinker, A.; Jones, S.C.; Dimick-Santos, L.; Avigan, M.I. An Evaluation of Postmarketing Reports of Serious Idiosyncratic Liver Injury Associated with Ulipristal Acetate for the Treatment of Uterine Fibroids. Drug Saf. 2020, 43, 1267–1276. [Google Scholar] [CrossRef]
- Lindh, M.; Hallberg, P.; Yue, Q.-Y.; Wadelius, M. Clinical factors predicting drug-induced liver injury due to flucloxacillin. Drug Healthc. Patient Saf. 2018, 10, 95–101. [Google Scholar] [CrossRef] [Green Version]
- Kaku, K.; Enya, K.; Nakaya, R.; Ohira, T.; Matsuno, R. Efficacy and safety of fasiglifam (TAK-875), a G protein-coupled receptor 40 agonist, in Japanese patients with type 2 diabetes inadequately controlled by diet and exercise: A randomized, double-blind, placebo-controlled, phase III trial. Diabetes Obes. Metab. 2015, 17, 675–681. [Google Scholar] [CrossRef] [Green Version]
- Ali, M.; Al-Hendy, A. Selective progesterone receptor modulators for fertility preservation in women with symptomatic uterine fibroids. Biol. Reprod. 2017, 97, 337–352. [Google Scholar] [CrossRef] [Green Version]
- Bouchard, P.; Chabbert-Buffet, N.; Fauser, B.C.J.M. Selective progesterone receptor modulators in reproductive medicine: Pharmacology, clinical efficacy and safety. Fertil. Steril. 2011, 96, 1175–1189. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Chen, J.; Cheng, T.; Gindulyte, A.; He, J.; He, S.; Li, Q.; Shoemaker, B.A.; Thiessen, P.A.; Yu, B.; et al. PubChem in 2021: New data content and improved web interfaces. Nucleic Acids Res. 2016, 47, D1388–D1395. [Google Scholar] [CrossRef]
- LeCun, Y.; Bengio, Y.; Hinton, G. Deep learning. Nature 2015, 521, 436–444. [Google Scholar] [CrossRef]

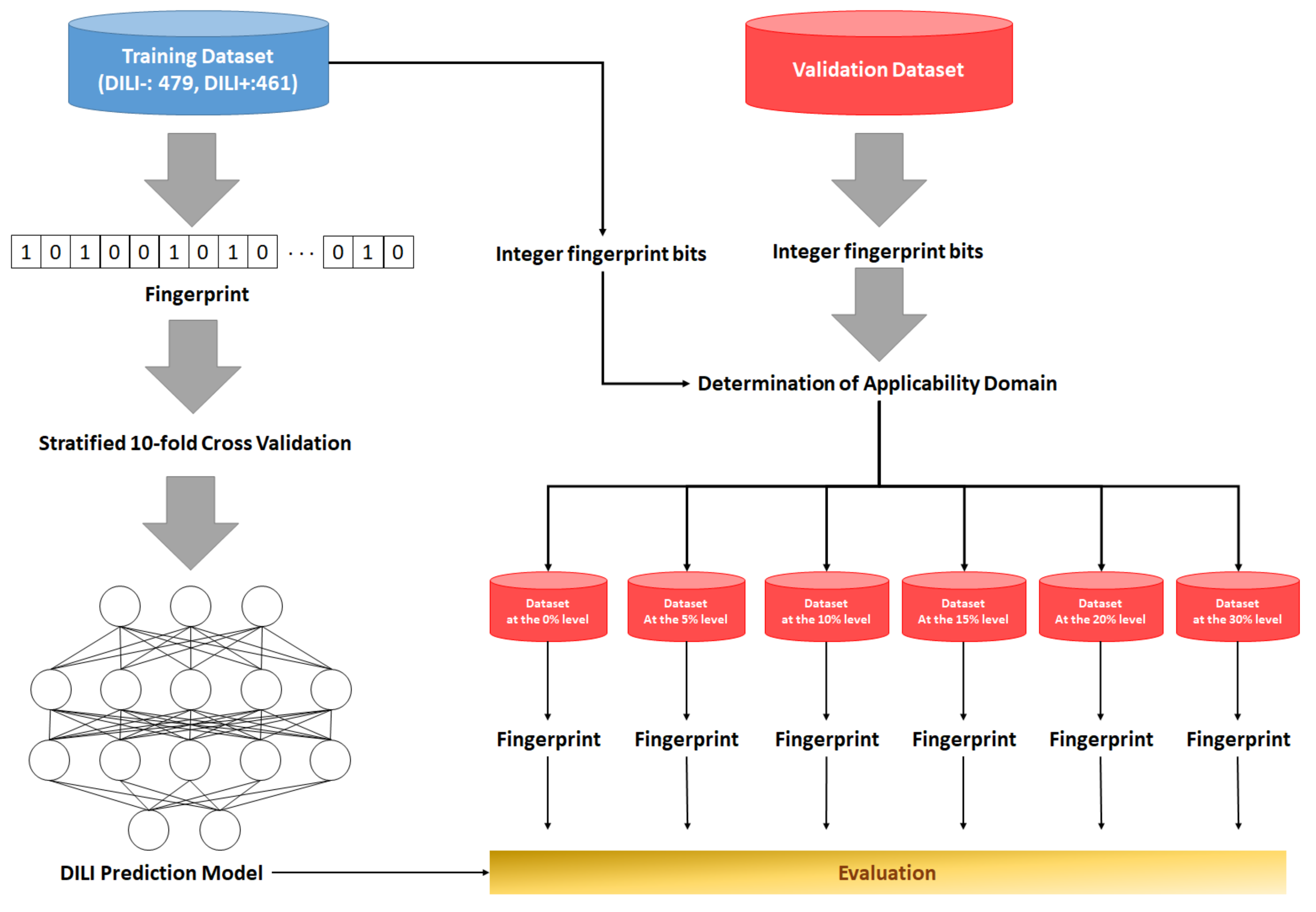
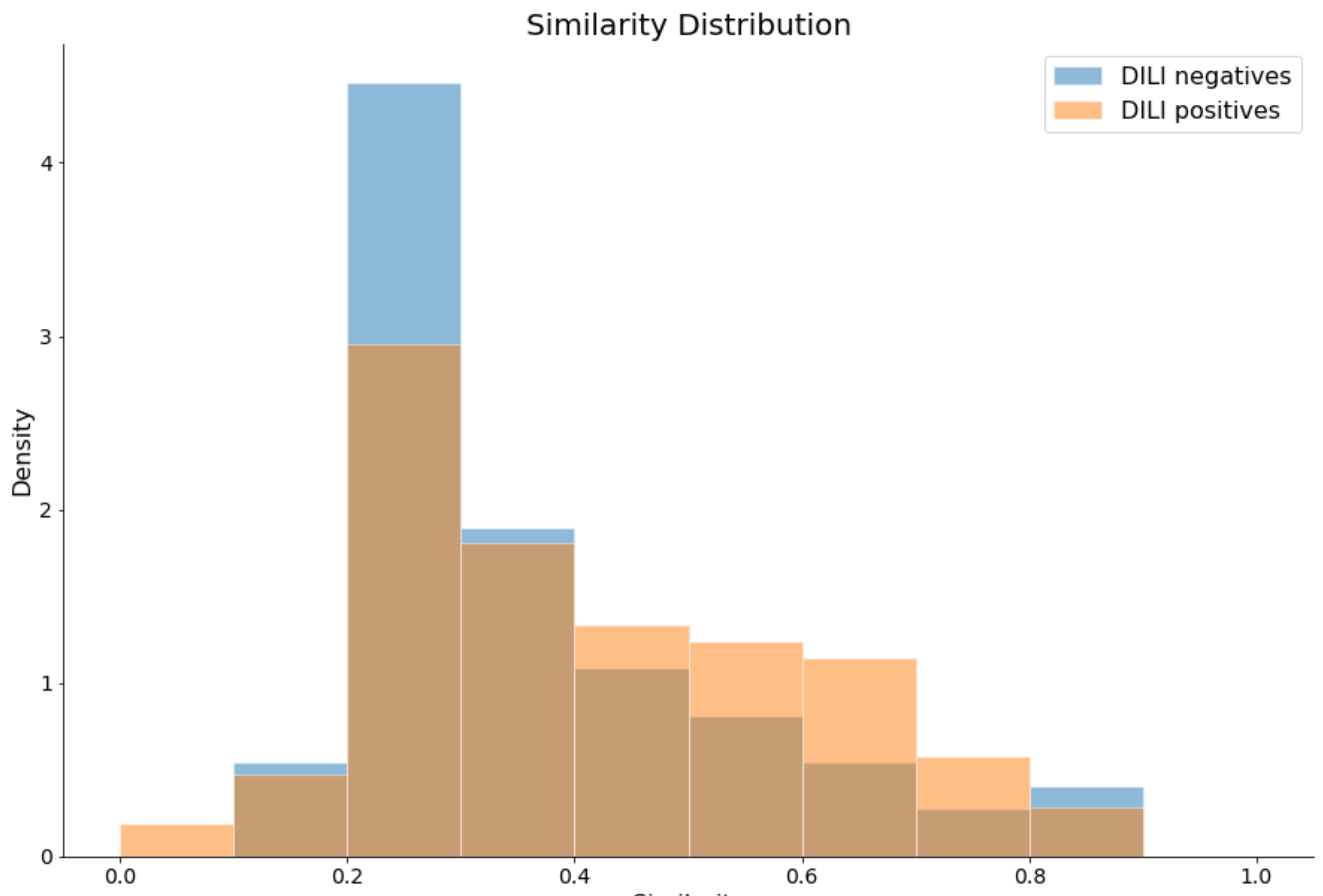
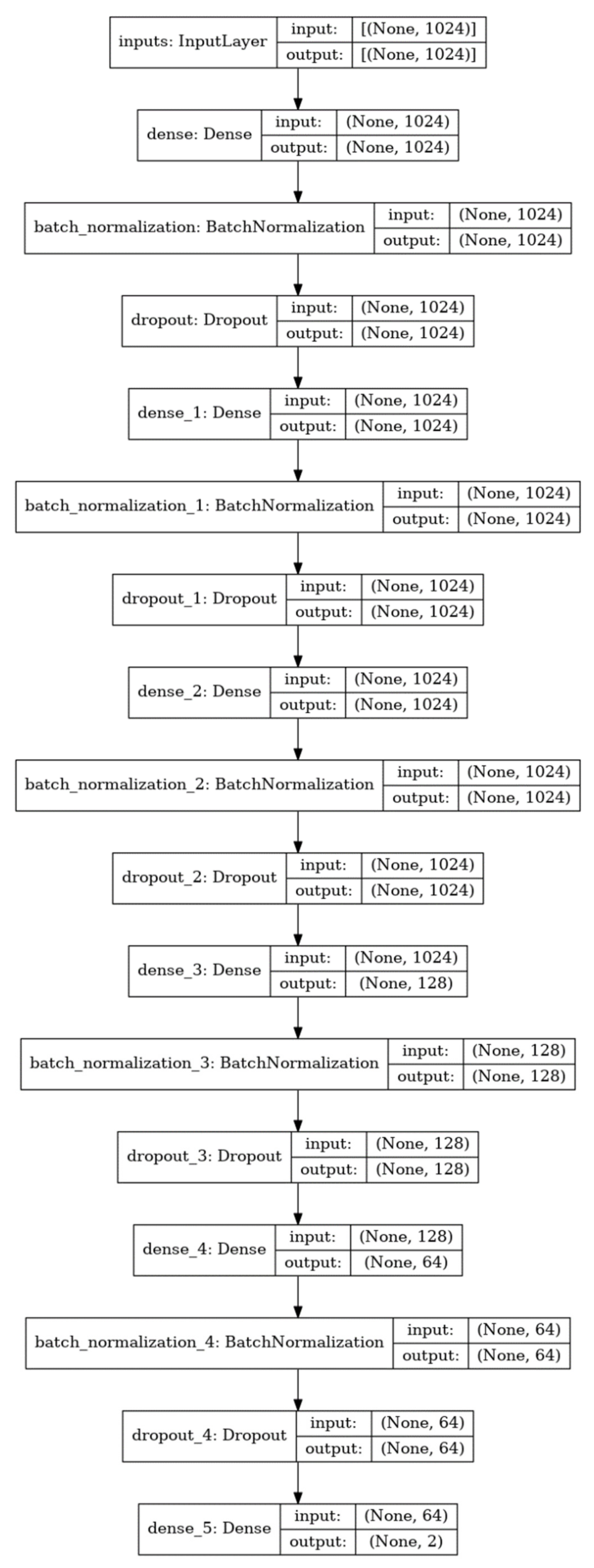
| Data Sets | DILI-Negatives | DILI-Positives | Total | |
|---|---|---|---|---|
| Training data set | DILIrank data set | 245 | 439 | 684 |
| LiverTox data set | 234 | 22 | 256 | |
| SUM | 479 | 461 | 940 | |
| Validation data set | Greene data set | 64 | 92 | 156 |
| Xu data set | 10 | 13 | 23 | |
| SUM | 74 | 105 | 179 |
| Iteration | ACC | Best Loss |
|---|---|---|
| 1 | 0.873 ± 0.0957 | 0.2271 |
| 2 | 0.864 ± 0.1074 | 0.1713 |
| 3 | 0.945 ± 0.0834 | 0.1264 |
| 4 | 0.918 ± 0.0725 | 0.1066 |
| 5 | 0.939 ± 0.0778 | 0.1276 |
| 6 | 0.940 ± 0.0859 | 0.0837 |
| 7 | 0.835 ± 0.1557 | 0.1858 |
| 8 | 0.927 ± 0.0788 | 0.1102 |
| 9 | 0.889 ± 0.0943 | 0.1258 |
| 10 | 0.814 ± 0.1928 | 0.2245 |
| Endurance Level | ECFP4 | ECFP6 | ||||||
| ACC | SE | SP | F1 | ACC | SE | SP | F1 | |
| 0% | 0.731 | 0.714 | 0.750 | 0.741 | 0.750 | 0.500 | 1.000 | 0.667 |
| 5% | 0.667 | 0.778 | 0.524 | 0.724 | 0.706 | 0.667 | 0.750 | 0.706 |
| 10% | 0.648 | 0.744 | 0.500 | 0.719 | 0.615 | 0.667 | 0.556 | 0.651 |
| 15% | 0.642 | 0.742 | 0.488 | 0.706 | 0.608 | 0.607 | 0.609 | 0.630 |
| 20% | 0.632 | 0.763 | 0.434 | 0.706 | 0.571 | 0.571 | 0.571 | 0.593 |
| 30% | 0.607 | 0.758 | 0.397 | 0.686 | 0.540 | 0.507 | 0.591 | 0.574 |
| Endurance Level | FCFP4 | FCFP6 | ||||||
| ACC | SE | SP | F1 | ACC | SE | SP | F1 | |
| 0% | 0.548 | 0.556 | 0.538 | 0.609 | 0.650 | 0.600 | 0.700 | 0.556 |
| 5% | 0.553 | 0.635 | 0.452 | 0.630 | 0.618 | 0.632 | 0.600 | 0.611 |
| 10% | 0.552 | 0.663 | 0.390 | 0.640 | 0.614 | 0.677 | 0.538 | 0.613 |
| 15% | 0.531 | 0.638 | 0.379 | 0.622 | 0.586 | 0.640 | 0.513 | 0.626 |
| 20% | 0.515 | 0.616 | 0.375 | 0.601 | 0.575 | 0.652 | 0.471 | 0.629 |
| 30% | 0.517 | 0.606 | 0.392 | 0.600 | 0.551 | 0.629 | 0.449 | 0.604 |
| Endurance Level | Naive Bayesian | SVM | RF | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ACC | SE | SP | F1 | ACC | SE | SP | F1 | ACC | SE | SP | F1 | |
| 0% | 0.538 | 0.643 | 0.417 | 0.600 | 0.577 | 0.571 | 0.583 | 0.593 | 0.577 | 0.500 | 0.667 | 0.560 |
| 5% | 0.604 | 0.778 | 0.381 | 0.689 | 0.583 | 0.630 | 0.524 | 0.630 | 0.604 | 0.667 | 0.524 | 0.655 |
| 10% | 0.606 | 0.767 | 0.357 | 0.702 | 0.592 | 0.651 | 0.500 | 0.659 | 0592 | 0.651 | 0.500 | 0.659 |
| 15% | 0.606 | 0.803 | 0.302 | 0.711 | 0.615 | 0.667 | 0.535 | 0.677 | 0.578 | 0.652 | 0.465 | 0.652 |
| 20% | 0.602 | 0.825 | 0.264 | 0.714 | 0.602 | 0.688 | 0.472 | 0.675 | 0.564 | 0.675 | 0.396 | 0.651 |
| 30% | 0.589 | 0.810 | 0.279 | 0.697 | 0.601 | 0.684 | 0.485 | 0.667 | 0.571 | 0.674 | 0.426 | 0.646 |
| References | Level * (%) | Training Data Set Size | ACC | SE | SP | AUC |
|---|---|---|---|---|---|---|
| Liew et al. (entire data set) [12] | 0% | 114 (68+/46−) | 0.789 | 0.838 | 0.717 | 0.853 |
| 100% | 187 (105+/82−) | 0.642 | 0.724 | 0.537 | 0.742 | |
| valBLACK | 0% | 38 | 0.974 | 0.955 | 1.000 | 0.955 |
| (22+/16−) | (0.809) | (0.957) | (0.667) | (0.924) | ||
| 100% | 47 (23+/24−) | 0.830 | 0.957 | 0.708 | 0.937 | |
| valPAIR | 0% | 14 | 0.500 | 0.857 | 0.143 | 0.551 |
| (7+/7−) | (0.550) | (0.800) | (0.300) | (0.450) | ||
| 100% | 20 (10+/10−) | 0.450 | 0.700 | 0.200 | 0.525 | |
| valRANDOM | 0% | 62 | 0.742 | 0.769 | 0.696 | 0.836 |
| (39+/23−) | (0.750) | (0.819) | (0.646) | (0.595) | ||
| 100% | 120 (72+/48−) | 0.600 | 0.653 | 0.521 | 0.687 | |
| Zhang et al. [14] | 0% | 80 | 0.950 | 1.000 | 0.926 | 0.957 |
| (53+/27−) | (0.750) | (0.932) | (0.379) | (0.667) | ||
| 100% | 85 (57+/28−) | 0.941 | 0.982 | 0.857 | 0.952 | |
| Ai et al. [15] | 0% | 84 | 0.881 | 0.905 | 0.810 | 0.920 |
| (63+/21−) | (0.843) | (0.869) | (0.754) | (0.904) | ||
| 100% | 121 (94+/27−) | 0.893 | 0.904 | 0.852 | 0.911 | |
| Kotsampasakou et al. [13] | 0% | 151 | 0.636 | 0.595 | 0.687 | 0.672 |
| (84+/67−) | (0.600) | (0.670) | (0.520) | (0.642) | ||
| 100% | 973 (524+/449−) | 0.585 | 0.635 | 0.526 | 0.605 |
| Drugs | CID | Endurance Levels | Prediction | Prediction Probability |
|---|---|---|---|---|
| Flucloxacillin [26] | 21,319 | 6.7% | DILI-positive | 0.999 |
| Aliskiren [27] | 5,493,444 | 7.0% | DILI-positive | 0.999 |
| Rilpivirine [28] | 6,451,164 | 5.0% | DILI-positive | 0.994 |
| Escitalopram [29] | 146,570 | 5.0% | DILI-positive | 0.989 |
| Nilotinib [30] | 644,241 | 7.6% | DILI-positive | 0.982 |
| Olmesartan [31] | 158,781 | 6.3% | DILI-positive | 0.974 |
| Mesterolone [32] | 15,020 | 4.1% | DILI-positive | 0.971 |
| Levothyroxine [33] | 5819 | 3.9% | DILI-positive | 0.965 |
| Zanubrutinib [34] | 135,565,884 | 6.4% | DILI-positive | 0.922 |
| Phenprobamate [35] | 4770 | 2.8% | DILI-positive | 0.896 |
| Apixaban [36] | 10,182,969 | 5.5% | DILI-positive | 0.804 |
| Fasiglifam [37] | 24,857,286 | 7.0% | DILI-positive | 0.660 |
| Pirfenidone [38] | 40,632 | 2.6% | DILI-positive | 0.608 |
| Ligandrol [39] | 44,137,686 | 4.6% | DILI-negative | 0.378 |
| Ulipristal acetate [40] | 130,904 | 6.5% | DILI-negative | 0.036 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kang, M.-G.; Kang, N.S. Predictive Model for Drug-Induced Liver Injury Using Deep Neural Networks Based on Substructure Space. Molecules 2021, 26, 7548. https://doi.org/10.3390/molecules26247548
Kang M-G, Kang NS. Predictive Model for Drug-Induced Liver Injury Using Deep Neural Networks Based on Substructure Space. Molecules. 2021; 26(24):7548. https://doi.org/10.3390/molecules26247548
Chicago/Turabian StyleKang, Myung-Gyun, and Nam Sook Kang. 2021. "Predictive Model for Drug-Induced Liver Injury Using Deep Neural Networks Based on Substructure Space" Molecules 26, no. 24: 7548. https://doi.org/10.3390/molecules26247548
APA StyleKang, M.-G., & Kang, N. S. (2021). Predictive Model for Drug-Induced Liver Injury Using Deep Neural Networks Based on Substructure Space. Molecules, 26(24), 7548. https://doi.org/10.3390/molecules26247548






