Comparative Analysis of Herbaceous and Woody Cell Wall Digestibility by Pathogenic Fungi
Abstract
1. Introduction
2. Results and Discussion
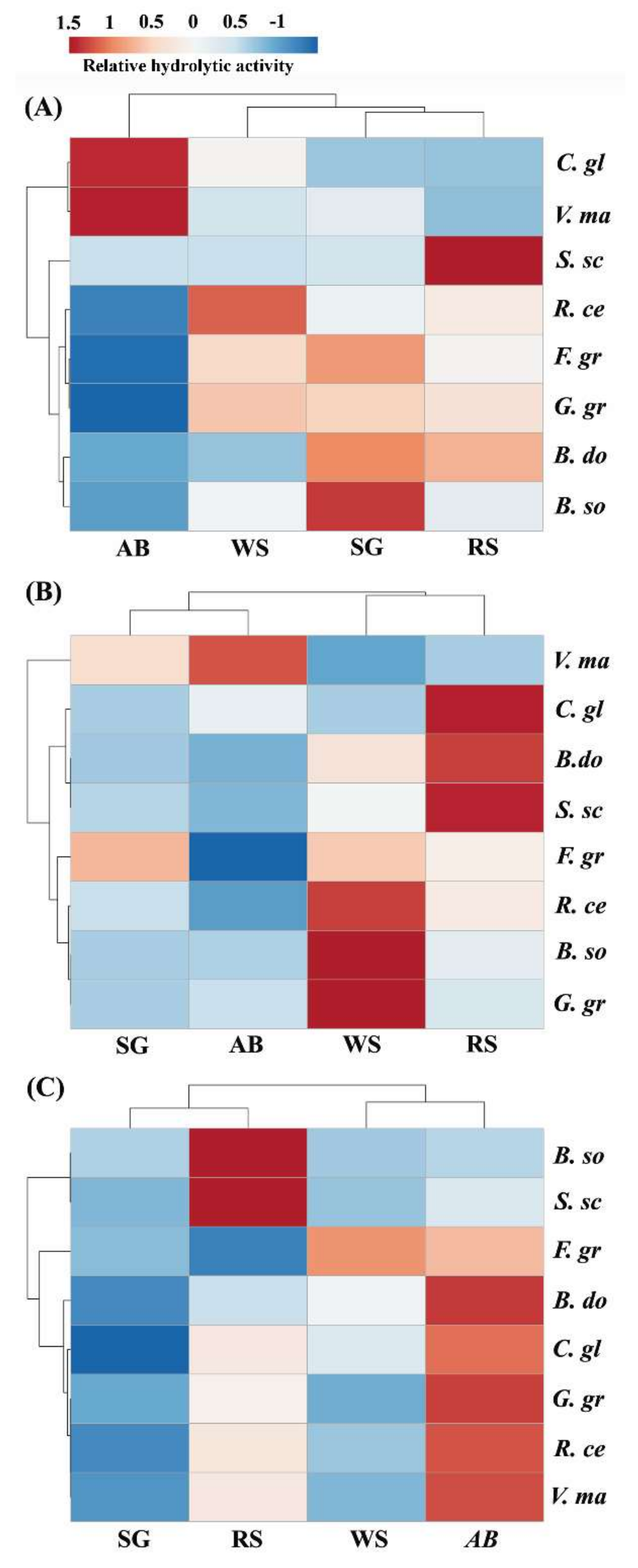
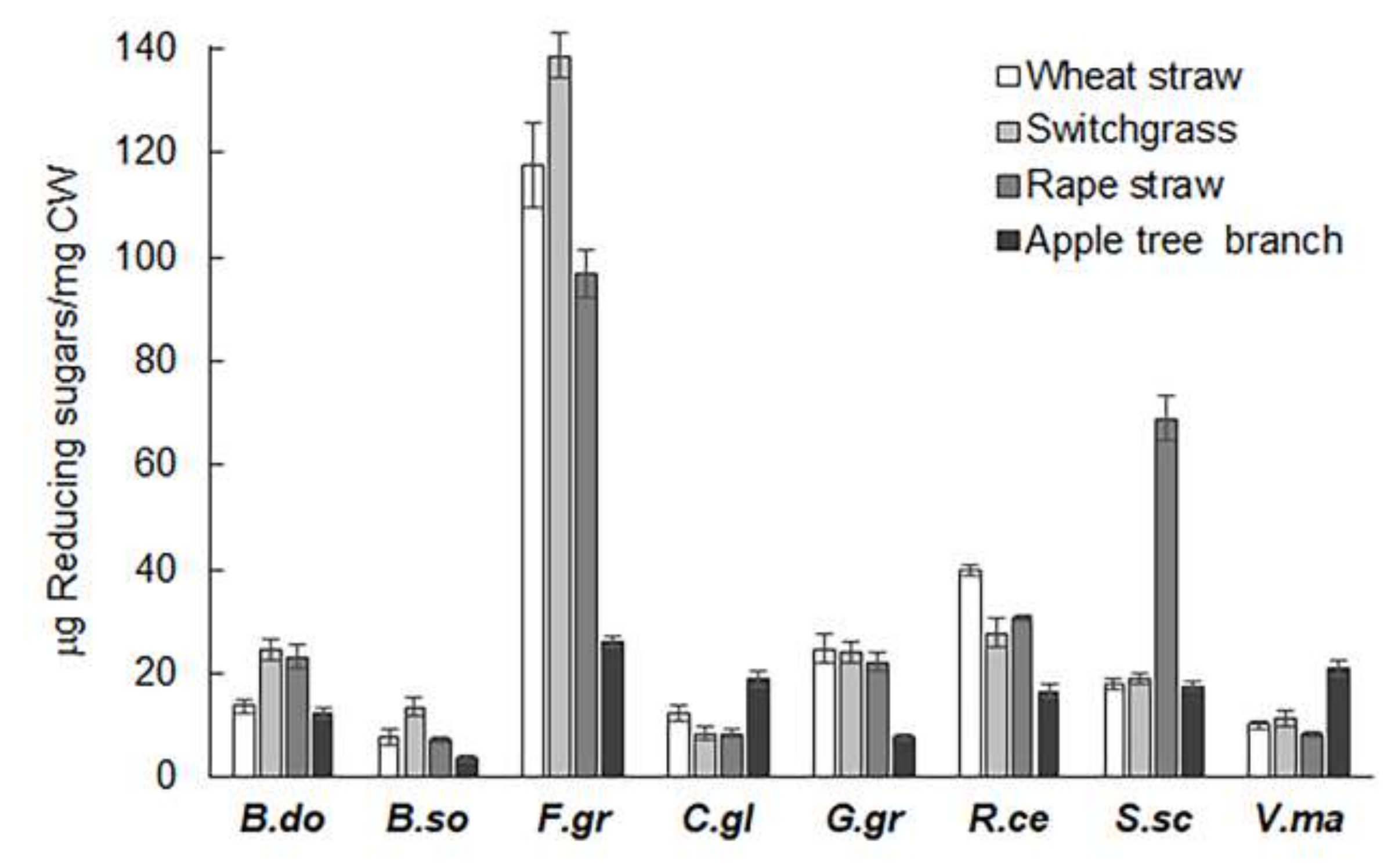
2.1. Extracts from Valsa mali, B. sorokiniana, and S. sclerotiorum Showed Significant Hydrolytic Preferences for Apple Tree Branch, Wheat Straw, and Rapeseed Straw, Respectively
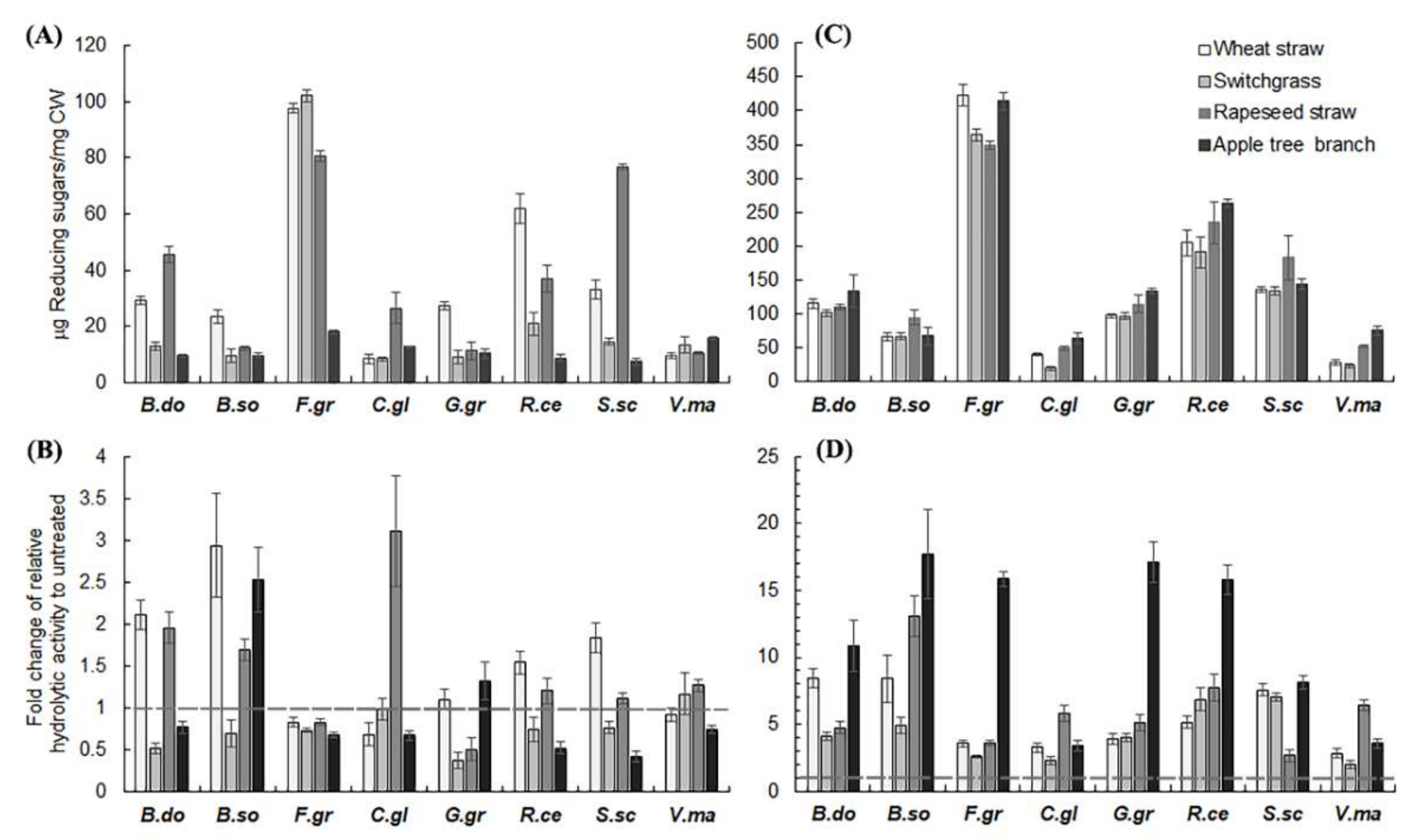
2.2. Hemicellulase Pretreatment Modulated Hydrolytic Preferences of Pathogenic Fungi for Biomass Type
2.3. Delignification by PAA Pretreatment Increased Enzymatic Hydrolysis of Crop Residues and the Increase Was Generally More Profound with Non-Host than Host Plant Biomass
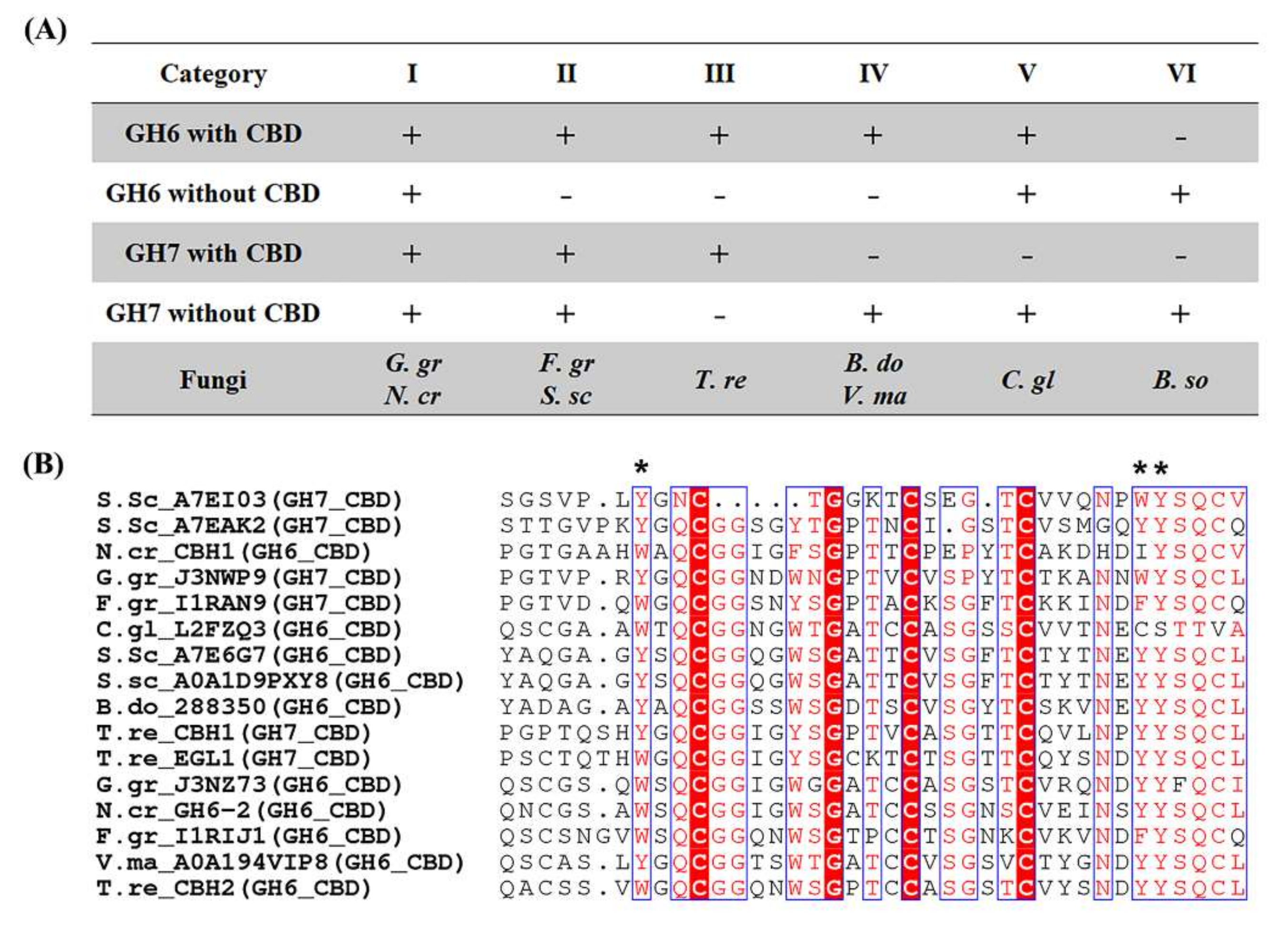
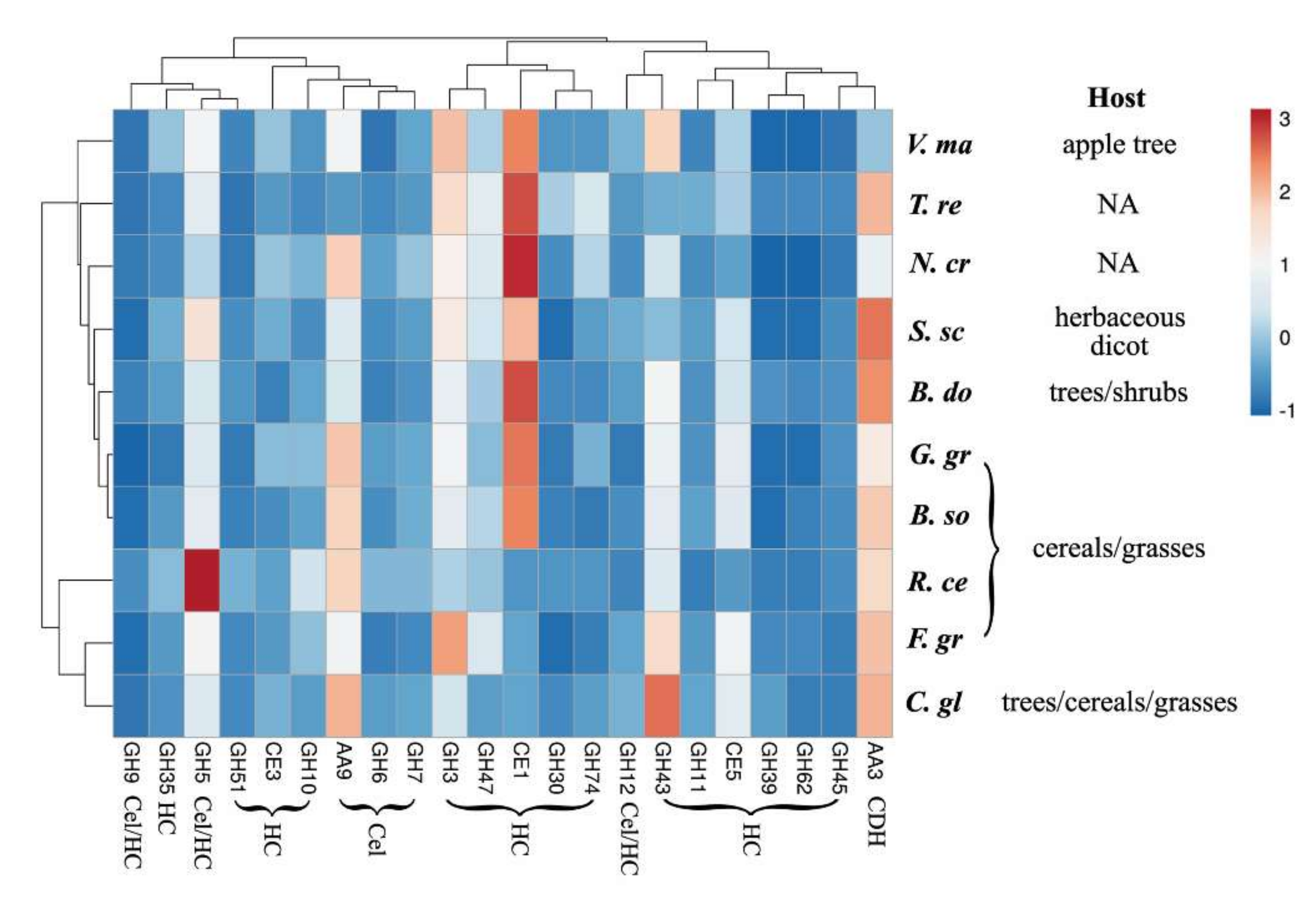
2.4. Cellulose Organization May Contribute to Enzymatic Digestibility of Plant Cell Walls by Pathogenic Fungi
3. Materials and Methods
3.1. Chemicals and Materials
3.2. Cultures and Growth Conditions
3.3. Microscale Enzymatic Saccharification
3.4. Hemicellulase and Peracetic Acid Pretreatment
3.5. Cell Wall Composition Analysis
3.6. FESEM Imaging
3.7. CAZyme Data Collection
3.8. GH6 and GH7 Gene Sequence Acquisition and Analysis
3.9. Data Analysis and Statistics
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Arthur, J.; Ragauskas, A.J.; Williams, C.K.; Davison, B.H.; Britovsek, G.; Cairney, J.; Eckert, C.A.; Frederick, W.J.; Hallett, J.P.; Leak, D.J.; et al. The path forward for biofuels and biomaterials. Science 2018, 311, 271–283. [Google Scholar] [CrossRef]
- Davis, R.; Tao, L.; Tan, E.; Biddy, M.; Beckham, G.; Scarlata, C.; Jacobson, J.; Cafferty, K.; Ross, J.; Lukas, J. Process Design and Economics for the Conversion of Lignocellulosic Biomass to Hydrocarbons: Dilute-Acid and Enzymatic Deconstruction of Biomass to Sugars and Biological Conversion of Sugars to Hydrocarbons; National Renewable Energy Laboratory (NREL): Golden, CO, USA, 2013. [Google Scholar]
- Humbird, D.; Davis, R.; Tao, L.; Kinchin, C.; Hsu, D.; Aden, A. Process Design and Economics for Biochemical Conversion of Lignocellulosic Biomass to Ethanol: Dilute-Acid Pretreatment and Enzymatic Hydrolysis of Corn Stover; National Renewable Energy Laboratory (NREL): Golden, CO, USA, 2011. [Google Scholar]
- Klein-Marcuschamer, D.; Oleskowicz-Popiel, P.; Simmons, B.A.; Blanch, H.W. The challenge of enzyme cost in the production of lignocellulosic biofuels. Biotechnol. Bioeng. 2011, 109, 1083–1087. [Google Scholar] [CrossRef] [PubMed]
- Ding, S.Y.; Liu, Y.S.; Zeng, Y.; Himmel, M.E.; Baker, J.O.; Bayer, E.A. How does plant cell wall nanoscale architecture correlate with enzymatic digestibility? Science 2012, 338, 1055–1060. [Google Scholar] [CrossRef]
- Wilson, D.B. Microbial diversity of cellulose hydrolysis. Curr. Opin. Microbiol. 2011, 14, 259–263. [Google Scholar] [CrossRef] [PubMed]
- Somerville, C.; Youngs, H.; Taylor, C.; Davis, S.C.; Long, S.P. Feedstocks for lignocellulosic biofuels. Science 2010, 329, 790–792. [Google Scholar] [CrossRef] [PubMed]
- Fan, C.; Yu, H.; Qin, S.; Li, Y.; Alam, A.; Xu, C.; Fan, D.; Zhang, Q.; Wang, Y.; Zhu, W.; et al. Brassinosteroid overproduction improves lignocellulose quantity and quality to maximize bioethanol yield under green-like biomass process in transgenic poplar. Biotechnol. Biofuels 2020, 13, 1–17. [Google Scholar] [CrossRef]
- Pauly, M.; Keegstra, K. Plant cell wall polymers as precursors for biofuels. Curr. Opin. Plant Biol. 2010, 13, 304–311. [Google Scholar] [CrossRef]
- Busse-Wicher, M.; Li, A.; Silveira, R.L.; Pereira, C.S.; Tryfona, T.; Gomes, T.C.F.; Skaf, M.S.; Dupree, P. Evolution of xylan substitution patterns in gymnosperms and angiosperms: Implications for xylan interaction with cellulose. Plant Physiol. 2016, 171, 2418–2431. [Google Scholar] [CrossRef]
- Scheller, H.V.; Ulvskov, P. Hemicelluloses. Annu. Rev. Plant Biol. 2010, 61, 263–289. [Google Scholar] [CrossRef] [PubMed]
- Terrett, O.M.; Dupree, P. Covalent interactions between lignin and hemicelluloses in plant secondary cell walls. Curr. Opin. Biotechnol. 2019, 56, 97–104. [Google Scholar] [CrossRef] [PubMed]
- Grantham, N.; Wurman-Rodrich, J.; Terrett, O.; Lyczakowski, J.; Stott, K.; Iuga, D.; Simmons, T.; Durand-Tardif, M.; Brown, S.; Dupree, R.; et al. An even pattern of xylan substitution is critical for interaction with cellulose in plant cell walls. Nat. Plants 2017, 3, 859–865. [Google Scholar] [CrossRef] [PubMed]
- Gupta, M.; Rawal, T.B.; Dupree, P.; Smith, J.C.; Petridis, L. Spontaneous rearrangement of acetylated xylan on hydrophilic cellulose surfaces. Cellulose 2021, 28, 3327–3345. [Google Scholar] [CrossRef]
- Tavares, E.Q.P.; De Souza, A.; Buckeridge, M.S. How endogenous plant cell-wall degradation mechanisms can help achieve higher efficiency in saccharification of biomass. J. Exp. Bot. 2015, 66, 4133–4143. [Google Scholar] [CrossRef]
- Choi, J.-H.; Park, S.-Y.; Kim, J.-H.; Cho, S.-M.; Jang, S.-K.; Hong, C.; Choi, I.-G. Selective deconstruction of hemicellulose and lignin with producing derivatives by sequential pretreatment process for biorefining concept. Bioresour. Technol. 2019, 291, 121913. [Google Scholar] [CrossRef] [PubMed]
- Han, L.; Wang, X.; Spiertz, H.; Yang, L.; Zhou, Y.; Liu, J.; Xie, G. Spatio-temporal availability of field crop residues for biofuel production in Northwest and Southwest China. Bioenergy Res. 2014, 8, 402–414. [Google Scholar] [CrossRef]
- Zhu, K.; Liu, Z.; Tan, X.; Lin, J.; Xu, D. Study on the ecological potential of Chinese straw resources available for bioenergy producing based on soil protection functions. Biomass Bioenergy 2018, 116, 26–38. [Google Scholar] [CrossRef]
- Zhang, Y.; Tian, J.; Shen, Z.; Wang, W.; Ni, H.; Liu, S.; Cao, J. Emission characteristics of PM2.5 and trace gases from household wood burning in Guanzhong plain, Northwest China. Aerosol Sci. Eng. 2018, 2, 130–140. [Google Scholar] [CrossRef]
- Wang, X.; Zang, R.; Yin, Z.; Kang, Z.; Huang, L. Delimiting cryptic pathogen species causing apple Valsa canker with multilocus data. Ecol. Evol. 2014, 4, 1369–1380. [Google Scholar] [CrossRef]
- Dong, X.-L.; Cheng, Z.-Z.; Leng, W.-F.; Li, B.-H.; Xu, X.-M.; Lian, S.; Wang, C.-X. Progression of symptoms caused by Botryosphaeria dothidea on apple branches. Phytopathology 2021. [Google Scholar] [CrossRef]
- Marsberg, A.; Kemler, M.; Jami, F.; Nagel, J.H.; Postma-Smidt, A.; Naidoo, S.; Wingfield, M.J.; Crous, P.W.; Spatafora, J.W.; Hesse, C.N.; et al. Botryosphaeria dothidea: A latent pathogen of global importance to woody plant health. Mol. Plant Pathol. 2017, 18, 477–488. [Google Scholar] [CrossRef]
- Ajay Kumar, G. Colletotrichum gloeosporioides: Biology, pathogenicity and management in India. J. Plant Physiol. Pathol. 2014, 2, 2. [Google Scholar]
- Boland, G.; Hall, R. Index of plant hosts of Sclerotinia sclerotiorum. Can. J. Plant Pathol. 1994, 16, 93–108. [Google Scholar] [CrossRef]
- Fajolu, O.L.; Wadl, P.A.; Vu, A.L.; Gwinn, K.D.; Scheffler, B.E.; Trigiano, R.N.; Ownley, B.H. Development and characterization of simple sequence repeats for Bipolaris sorokiniana and cross transferability to related species. Mycologia 2013, 105, 1164–1173. [Google Scholar] [CrossRef]
- Kumar, J.; Schäfer, P.; Hückelhoven, R.; Langen, G.; Baltruschat, H.; Stein, E.; Nagarajan, S.; Kogel, K.-H. Bipolaris sorokiniana, a cereal pathogen of global concern: Cytological and molecular approaches towards better control. Mol. Plant Pathol. 2002, 3, 185–195. [Google Scholar] [CrossRef] [PubMed]
- Bryan, G.T.; Labourdette, E.; Melton, R.; Nicholson, P.; Daniels, M.J.; Osbourn, A.E. DNA polymorphism and host range in the take-all fungus, Gaeumannomyces graminis. Mycol. Res. 1999, 103, 319–327. [Google Scholar] [CrossRef]
- Harris, L.J.; Balcerzak, M.; Johnston, A.; Schneiderman, D.; Ouellet, T. Host-preferential Fusarium graminearum gene expression during infection of wheat, barley, and maize. Fungal Biol. 2016, 120, 111–123. [Google Scholar] [CrossRef]
- Collins, S.B. Characterization of Fusarium Species and Alternaria Alternata, and Their Effects on Switchgrass Health and Chemical Components; University of Tennessee: Knoxville, TN, USA, 2016. [Google Scholar]
- Freeman, J.; Ward, E. Gaeumannomyces graminis, the take-all fungus and its relatives. Mol. Plant Pathol. 2004, 5, 235–252. [Google Scholar] [CrossRef]
- Etheridge, J.V.; Davey, L.; Christian, D.G. First report of Rhizoctonia cerealis causing sharp eyespot in Panicum virgatum in the UK. Plant Pathol. 2001, 50, 807. [Google Scholar] [CrossRef][Green Version]
- Hamada, M.S.; Yin, Y.; Chen, H.; Ma, Z. The escalating threat of Rhizoctonia cerealis, the causal agent of sharp eyespot in wheat. Pest Manag. Sci. 2011, 67, 1411–1419. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Ehrhardt, D.W.; Somerville, C.R. Mutations of cellulose synthase (CESA1) phosphorylation sites modulate anisotropic cell expansion and bidirectional mobility of cellulose synthase. Proc. Natl. Acad. Sci. USA 2010, 107, 17188–17193. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Jia, H.; Zhao, H.; Liu, D.; Liu, Y.; Liu, B.; Bauer, S.; Somerville, C.R. Anisotropic cell expansion is affected through the bidirectional mobility of cellulose synthase complexes and phosphorylation at two critical residues on CESA3. Plant Physiol. 2016, 171, 242–250. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Li, J.; Zhao, H.; Liu, B.; Pomorski, T.G.; Chen, S.; Liesche, J. Novel tool to quantify cell wall porosity relates wall structure to cell growth and drug uptake. J. Cell Biol. 2019, 218, 1408–1421. [Google Scholar] [CrossRef] [PubMed]
- Pu, Y.; Hu, F.; Huang, F.; Davison, B.H.; Ragauskas, A.J. Assessing the molecular structure basis for biomass recalcitrance during dilute acid and hydrothermal pretreatments. Biotechnol. Biofuels 2013, 6, 15. [Google Scholar] [CrossRef]
- Medve, J.; Lee, D.; Tjerneld, F. Ion-exchange chromatographic purification and quantitative analysis of Trichoderma reesei cellulases cellobiohydrolase I, II and endoglucanase II by fast protein liquid chromatography. J. Chromatogr. A 1998, 808, 153–165. [Google Scholar] [CrossRef]
- Yin, Z.; Liu, H.; Li, Z.; Ke, X.; Dou, D.; Gao, X.; Song, N.; Dai, Q.; Wu, Y.; Xu, J.; et al. Genome sequence of Valsa canker pathogens uncovers a potential adaptation of colonization of woody bark. New Phytol. 2015, 208, 1202–1216. [Google Scholar] [CrossRef] [PubMed]
- King, B.C.; Waxman, K.D.; Nenni, N.V.; Walker, L.P.; Bergstrom, G.C.; Gibson, D.M. Arsenal of plant cell wall degrading enzymes reflects host preference among plant pathogenic fungi. Biotechnol. Biofuels 2011, 4, 4. [Google Scholar] [CrossRef] [PubMed]
- Yu, C.; Li, T.; Shi, X.; Saleem, M.; Li, B.; Liang, W.; Wang, C. Deletion of Endo-β-1,4-xylanase vmxyl1 impacts the virulence of Valsa mali in apple tree. Front. Plant Sci. 2018, 9, 663. [Google Scholar] [CrossRef]
- Xu, M.; Gao, X.; Chen, J.; Yin, Z.; Feng, H.; Huang, L. The feruloyl esterase genes are required for full pathogenicity of the apple tree canker pathogen Valsa mali. Mol. Plant Pathol. 2017, 19, 1353–1363. [Google Scholar] [CrossRef]
- Noda, J.; Brito, N.; González, C. The botrytis cinerea xylanase xyn11a contributes to virulence with its necrotizing activity, not with its catalytic activity. Bmc Plant Biol. 2010, 10, 38. [Google Scholar] [CrossRef]
- Nguyen, Q.B.; Itoh, K.; Van Vu, B.; Tosa, Y.; Nakayashiki, H. Simultaneous silencing of endo-β-1,4 xylanase genes reveals their roles in the virulence of Magnaporthe oryzae. Mol. Microbiol. 2011, 81, 1008–1019. [Google Scholar] [CrossRef]
- Thaker, A.; Mehta, K.; Patkar, R. Feruloyl esterase Fae1 is required specifically for host colonisation by the rice-blast fungus Magnaporthe oryzae. Res. Sq. 2021. [Google Scholar] [CrossRef] [PubMed]
- Berglund, J.; Mikkelsen, D.; Flanagan, B.M.; Dhital, S.; Gaunitz, S.; Henriksson, G.; Lindström, M.E.; Yakubov, G.E.; Gidley, M.J.; Vilaplana, F. Wood hemicelluloses exert distinct biomechanical contributions to cellulose fibrillar networks. Nat. Commun. 2020, 11, 4692. [Google Scholar] [CrossRef] [PubMed]
- Alemdar, A.; Sain, M. Isolation and characterization of nanofibers from agricultural residues—Wheat straw and soy hulls. Bioresour. Technol. 2008, 99, 1664–1671. [Google Scholar] [CrossRef] [PubMed]
- Xu, B.; Escamilla-Treviño, L.L.; Sathitsuksanoh, N.; Shen, Z.; Shen, H.; Zhang, Y.-H.P.; Dixon, R.A.; Zhao, B. Silencing of 4-coumarate:coenzyme A ligase in switchgrass leads to reduced lignin content and improved fermentable sugar yields for biofuel production. New Phytol. 2011, 192, 611–625. [Google Scholar] [CrossRef] [PubMed]
- Lu, X.; Zhang, Y.; Angelidaki, I. Optimization of H2SO4-catalyzed hydrothermal pretreatment of rapeseed straw for bioconversion to ethanol: Focusing on pretreatment at high solids content. Bioresour. Technol. 2009, 100, 3048–3053. [Google Scholar] [CrossRef]
- Lyu, S.; Du, G.; Yang, Y. Apple branch decomposition and nutrient turnover in the orchard soil. Bioresources 2017, 12, 3108–3121. [Google Scholar] [CrossRef][Green Version]
- Jiang, B.; Yu, J.; Luo, X.; Zhu, Y.; Jin, Y. A strategy to improve enzymatic saccharification of wheat straw by adding water-soluble lignin prepared from alkali pretreatment spent liquor. Process. Biochem. 2018, 71, 147–151. [Google Scholar] [CrossRef]
- Qiu, J.; Tian, D.; Shen, F.; Hu, J.; Zeng, Y.; Yang, G.; Zhang, Y.; Deng, S.; Zhang, J. Bioethanol production from wheat straw by phosphoric acid plus hydrogen peroxide (PHP) pretreatment via simultaneous saccharification and fermentation (SSF) at high solid loadings. Bioresour. Technol. 2018, 268, 355–362. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Wang, M.L.; Tonnis, B.; Habteselassie, M.; Liao, X.; Huang, Q. Fungal pretreatment of switchgrass for improved saccharification and simultaneous enzyme production. Bioresour. Technol. 2013, 135, 39–45. [Google Scholar] [CrossRef]
- Zhang, D.; Yang, Q.; Zhu, J.; Pan, X. Sulfite (SPORL) pretreatment of switchgrass for enzymatic saccharification. Bioresour. Technol. 2012, 129, 127–134. [Google Scholar] [CrossRef] [PubMed]
- Karagöz, P.; Rocha, I.V.; Özkan, M.; Angelidaki, I. Alkaline peroxide pretreatment of rapeseed straw for enhancing bioethanol production by same vessel saccharification and co-fermentation. Bioresour. Technol. 2012, 104, 349–357. [Google Scholar] [CrossRef] [PubMed]
- Zeng, Y.; Zhao, S.; Yang, S.; Ding, S.-Y. Lignin plays a negative role in the biochemical process for producing lignocellulosic biofuels. Curr. Opin. Biotechnol. 2014, 27, 38–45. [Google Scholar] [CrossRef] [PubMed]
- Penttilä, P.A.; Altgen, M.; Awais, M.; Österberg, M.; Rautkari, L.; Schweins, R. Bundling of cellulose microfibrils in native and polyethylene glycol-containing wood cell walls revealed by small-angle neutron scattering. Sci. Rep. 2020, 10, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Wilson, T.H.; Kumar, M.; Turner, S.R. The molecular basis of plant cellulose synthase complex organisation and assembly. Biochem. Soc. Trans. 2021, 49, 379–391. [Google Scholar] [CrossRef] [PubMed]
- Carroll, A.; Specht, C.D. Understanding plant cellulose synthases through a comprehensive investigation of the cellulose synthase family sequences. Front. Plant Sci. 2011, 2, 5. [Google Scholar] [CrossRef] [PubMed]
- Payne, C.M.; Knott, B.C.; Mayes, H.B.; Hansson, H.; Himmel, M.E.; Sandgren, M.; Stahlberg, J.; Beckham, G.T. Fungal cellulases. Chem. Rev. 2015, 115, 1308–1448. [Google Scholar] [CrossRef] [PubMed]
- Yan, S.; Wu, G. Implications of carbohydrate binding modules of cellulases summarized from visualization. In Proceedings of the 2020 22nd International Conference on Transparent Optical Networks (ICTON), Bari, Italy, 19–23 July 2020; pp. 1–7. [Google Scholar] [CrossRef]
- Christensen, S.J.; Krogh, K.B.R.M.; Spodsberg, N.; Borch, K.; Westh, P. A biochemical comparison of fungal GH6 cellobiohydrolases. Biochem. J. 2019, 476, 2157–2172. [Google Scholar] [CrossRef]
- Christensen, S.J.; Badino, S.F.; Cavaleiro, A.M.; Borch, K.; Westh, P. Functional analysis of chimeric TrCel6A enzymes with different carbohydrate binding modules. Protein Eng. Des. Sel. 2019, 32, 401–409. [Google Scholar] [CrossRef] [PubMed]
- Kubicek, C.P.; Starr, T.L.; Glass, N.L. Plant cell wall-degrading enzymes and their secretion in plant-pathogenic fungi. In Annual Review of Phytopathology; Van Alfen, N.K., Ed.; Annual Reviews: Palo Alto, CA, USA, 2014; Volume 52, pp. 427–451. [Google Scholar] [CrossRef]
- Sidar, A.; Albuquerque, E.D.; Voshol, G.P.; Ram, A.F.J.; Vijgenboom, E.; Punt, P.J. Carbohydrate binding modules: Diversity of domain architecture in amylases and cellulases from filamentous microorganisms. Front. Bioeng. Biotechnol. 2020, 8, 871. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Z.; Liu, H.; Wang, C.; Xu, J.-R. Erratum to: Comparative analysis of fungal genomes reveals different plant cell wall degrading capacity in fungi. BMC Genom. 2014, 15, 6. [Google Scholar] [CrossRef]
- Zarattini, M.; Corso, M.; Kadowaki, M.A.; Monclaro, A.; Magri, S.; Milanese, I.; Jolivet, S.; de Godoy, M.O.; Hermans, C.; Fagard, M.; et al. LPMO-oxidized cellulose oligosaccharides evoke immunity in Arabidopsis conferring resistance towards necrotrophic fungus B. cinerea. Commun. Biol. 2021, 4, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Mewis, K.; Lenfant, N.; Lombard, V.; Henrissat, B. Dividing the large glycoside hydrolase family 43 into subfamilies: A motivation for detailed enzyme characterization. Appl. Env. Microbiol. 2016, 82, 1686–1692. [Google Scholar] [CrossRef] [PubMed]
- Méndez-Líter, J.A.; de Eugenio, L.I.; Nieto-Domínguez, M.; Prieto, A.; Martínez, M.J. Hemicellulases from Penicillium and Talaromyces for lignocellulosic biomass valorization: A review. Bioresour. Technol. 2020, 324, 124623. [Google Scholar] [CrossRef]
- King, B.C.; Donnelly, M.K.; Bergstrom, G.C.; Walker, L.P.; Gibson, D.M. An optimized microplate assay system for quantitative evaluation of plant cell wall-degrading enzyme activity of fungal culture extracts. Biotechnol. Bioeng. 2009, 102, 1033–1044. [Google Scholar] [CrossRef]
- Miller, G.L. Use of dinitrosalicylic acid reagent for determination of reducing sugar. Anal. Chem. 1959, 31, 426–428. [Google Scholar] [CrossRef]
- Chang, V.S.; Holtzapple, M.T. Fundamental factors affecting biomass enzymatic reactivity. Appl. Biochem. Biotechnol. Part A Enzym. Eng. Biotechnol. 2000, 38, 5–37. [Google Scholar] [CrossRef]
- Hames, B.; Ruiz, R.; Scarlata, C.; Sluiter, A.; Sluiter, J.; Templeton, D. Preparation of samples for compositional analysis. Lab. Anal. Proced. 2008, 1617, 65–71. [Google Scholar]
- Sluiter, A.; Hames, B.; Ruiz, R.; Scarlata, C.; Sluiter, J.; Templeton, D.; Crocker, D. Determination of Structural Carbohydrates and Lignin in Biomass, Laboratory Analytical Procedure; National Renewable Energy Laboratory: Golden, CO, USA, 2011. Available online: https://www.nrel.gov/docs/gen/fy13/42618.pdf (accessed on 8 November 2021).
- Zhang, M.; Zheng, R.; Chen, J.; Huang, H. Investigation on the determination of lignocellulosics components by NREL method. Chin. J. Anal. Lab. 2010, 29, 15–18. [Google Scholar]
- Carpita, N.C.; Defernez, M.; Findlay, K.; Wells, B.; Shoue, D.A.; Catchpole, G.; Wilson, R.H.; McCann, M.C. Cell wall architecture of the elongating maize coleoptile. Plant Physiol. 2001, 127, 551–565. [Google Scholar] [CrossRef]
- Zheng, Y.; Cosgrove, D.J.; Ning, G. High-resolution field emission scanning electron microscopy (FESEM) imaging of cellulose microfibril organization in plant primary cell walls. Microsc. Microanal. 2017, 23, 1048–1054. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Liang, X.; Gleason, M.L.; Zhang, R.; Sun, G. Comparative genomics of Botryosphaeria dothidea and B kuwatsukai, causal agents of apple ring rot, reveals both species expansion of pathogenicity-related genes and variations in virulence gene content during speciation. IMA Fungus 2018, 9, 243–257. [Google Scholar] [CrossRef] [PubMed]
- Zaccaron, A.Z.; Bluhm, B.H. The genome sequence of Bipolaris cookei reveals mechanisms of pathogenesis underlying target leaf spot of sorghum. Sci. Rep. 2017, 7, 1–15. [Google Scholar] [CrossRef]
- Baroncelli, R.; Amby, D.B.; Zapparata, A.; Sarrocco, S.; Vannacci, G.; Le Floch, G.; Harrison, R.J.; Holub, E.; Sukno, S.A.; Sreenivasaprasad, S.; et al. Gene family expansions and contractions are associated with host range in plant pathogens of the genus Colletotrichum. BMC Genom. 2016, 17, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Lin, L.; Feilong, G.; Zhichao, Z.; Lijun, P.; Yu, H.; Xiuliang, Z.; Jinfeng, Y.; Wenwu, Y.; Zengyan, Z. High-quality genome assembly-based and functional analyses reveal the pathogenesis mechanisms and evolutionary landscape of wheat sharp eyespot Rhizoctonia cerealis. Res. Sq. 2021. [Google Scholar] [CrossRef]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I.; Bork, P. Interactive tree of life (iTOL) v5: An online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021, 49, W293–W296. [Google Scholar] [CrossRef] [PubMed]
- Metsalu, T.; Vilo, J. ClustVis: A web tool for visualizing clustering of multivariate data using principal component analysis and heatmap. Nucleic Acids Res. 2015, 43, W566–W570. [Google Scholar] [CrossRef] [PubMed]

| Pathogenic Species | Phylum | Host | Disease | Reference |
|---|---|---|---|---|
| Bipolaris sorokiniana | Ascomycota | Cereals (e.g., wheat) and grasses (e.g., switchgrass). | Causes disease on the root, leaf and stem, and head tissue. | [25,26] |
| Fusarium graminearum | Ascomycota | Cereals (e.g., wheat) and grasses (e.g., switchgrass) | Causes Fusarium head blight and Gibberella ear rot and stalk rot. | [28,29,30] |
| Gaeumannomyces graminis | Ascomycota | Cereals (e.g., wheat) and grasses. | Colonizes the root and crown tissue, causing Turfgrass disease. | [27] |
| Rhizoctonia cerealis | Basidiomycota | Cereals (e.g., wheat) and grasses (e.g., switchgrass) | Causes sharp eyespot and root rot. | [31,32] |
| Sclerotinia sclerotiorum | Ascomycota | Dicotyledonous herbaceous species (e.g., rapeseed, soybean) | Causes Sclerotinia head rot, Sclerotinia stalk rot, and Sclerotinia wilt. | [24] |
| Valsa mali var. mali | Ascomycota | Apple tree | Preferentially infects apple trees, causing canker diseases. | [20,38] |
| Botryosphaeria dothidea | Ascomycota | Trees and shrubs (e.g., apple and other fruit trees) | Disease symptoms are associated with twig, branch and stem cankers, tip and branch dieback, fruit rot, etc. | [21,22] |
| Colletotrichum gloeosporioides (teleomorph: Glomerella cingulata) | Ascomycota | Trees (e.g., apple tree), cereals and grasses, legumes, vegetables | Causes anthracnose disease | [23] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dou, Y.; Yang, Y.; Mund, N.K.; Wei, Y.; Liu, Y.; Wei, L.; Wang, Y.; Du, P.; Zhou, Y.; Liesche, J.; et al. Comparative Analysis of Herbaceous and Woody Cell Wall Digestibility by Pathogenic Fungi. Molecules 2021, 26, 7220. https://doi.org/10.3390/molecules26237220
Dou Y, Yang Y, Mund NK, Wei Y, Liu Y, Wei L, Wang Y, Du P, Zhou Y, Liesche J, et al. Comparative Analysis of Herbaceous and Woody Cell Wall Digestibility by Pathogenic Fungi. Molecules. 2021; 26(23):7220. https://doi.org/10.3390/molecules26237220
Chicago/Turabian StyleDou, Yanhua, Yan Yang, Nitesh Kumar Mund, Yanping Wei, Yisong Liu, Linfang Wei, Yifan Wang, Panpan Du, Yunheng Zhou, Johannes Liesche, and et al. 2021. "Comparative Analysis of Herbaceous and Woody Cell Wall Digestibility by Pathogenic Fungi" Molecules 26, no. 23: 7220. https://doi.org/10.3390/molecules26237220
APA StyleDou, Y., Yang, Y., Mund, N. K., Wei, Y., Liu, Y., Wei, L., Wang, Y., Du, P., Zhou, Y., Liesche, J., Huang, L., Fang, H., Zhao, C., Li, J., Wei, Y., & Chen, S. (2021). Comparative Analysis of Herbaceous and Woody Cell Wall Digestibility by Pathogenic Fungi. Molecules, 26(23), 7220. https://doi.org/10.3390/molecules26237220








