Transporter Protein-Guided Genome Mining for Head-to-Tail Cyclized Bacteriocins
Abstract
:1. Introduction
2. Results and Discussion
2.1. Identification of Putative Head-to-Tail Cyclized Bacteriocins
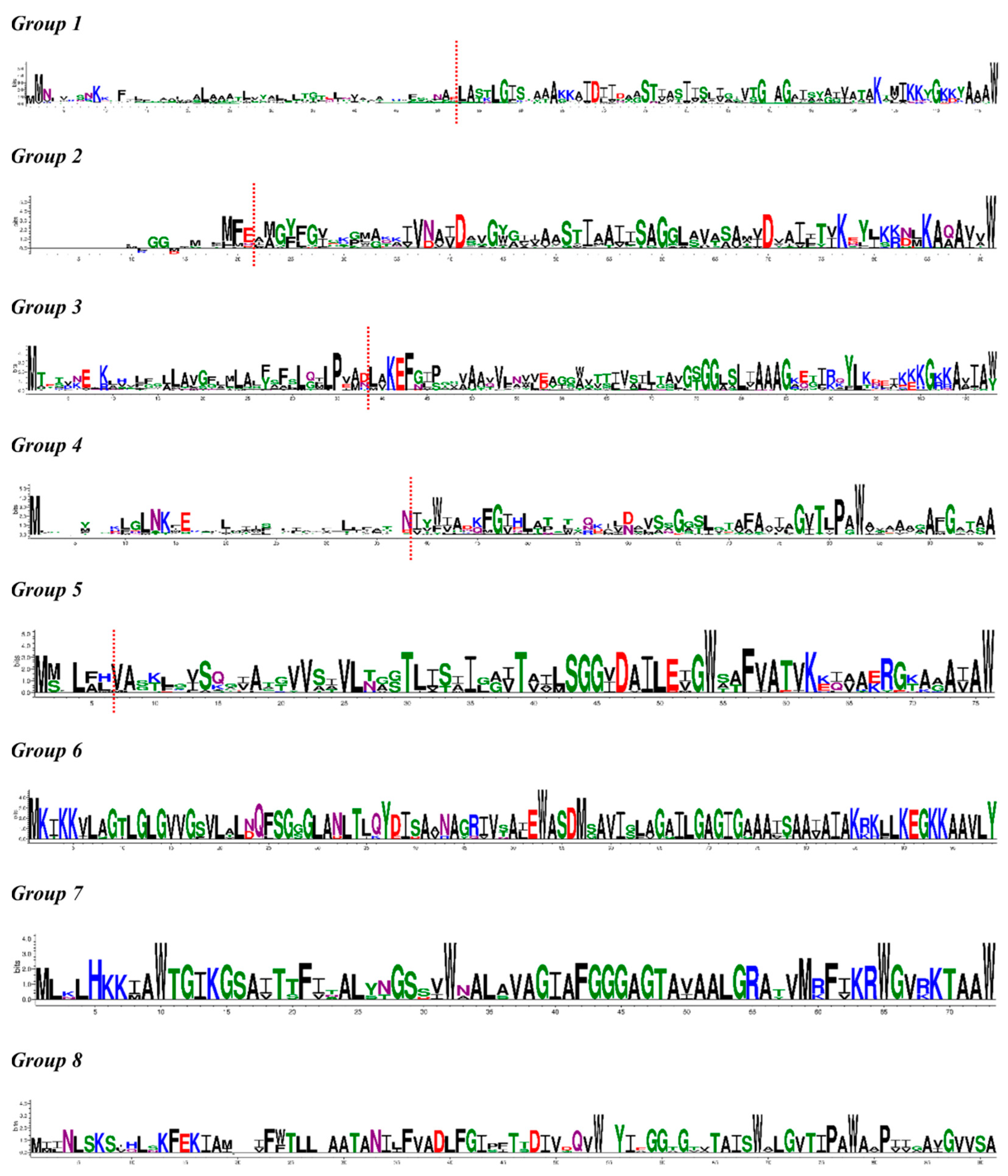
2.2. Amino Acid Sequence Diversity of Identified Precursor Peptides
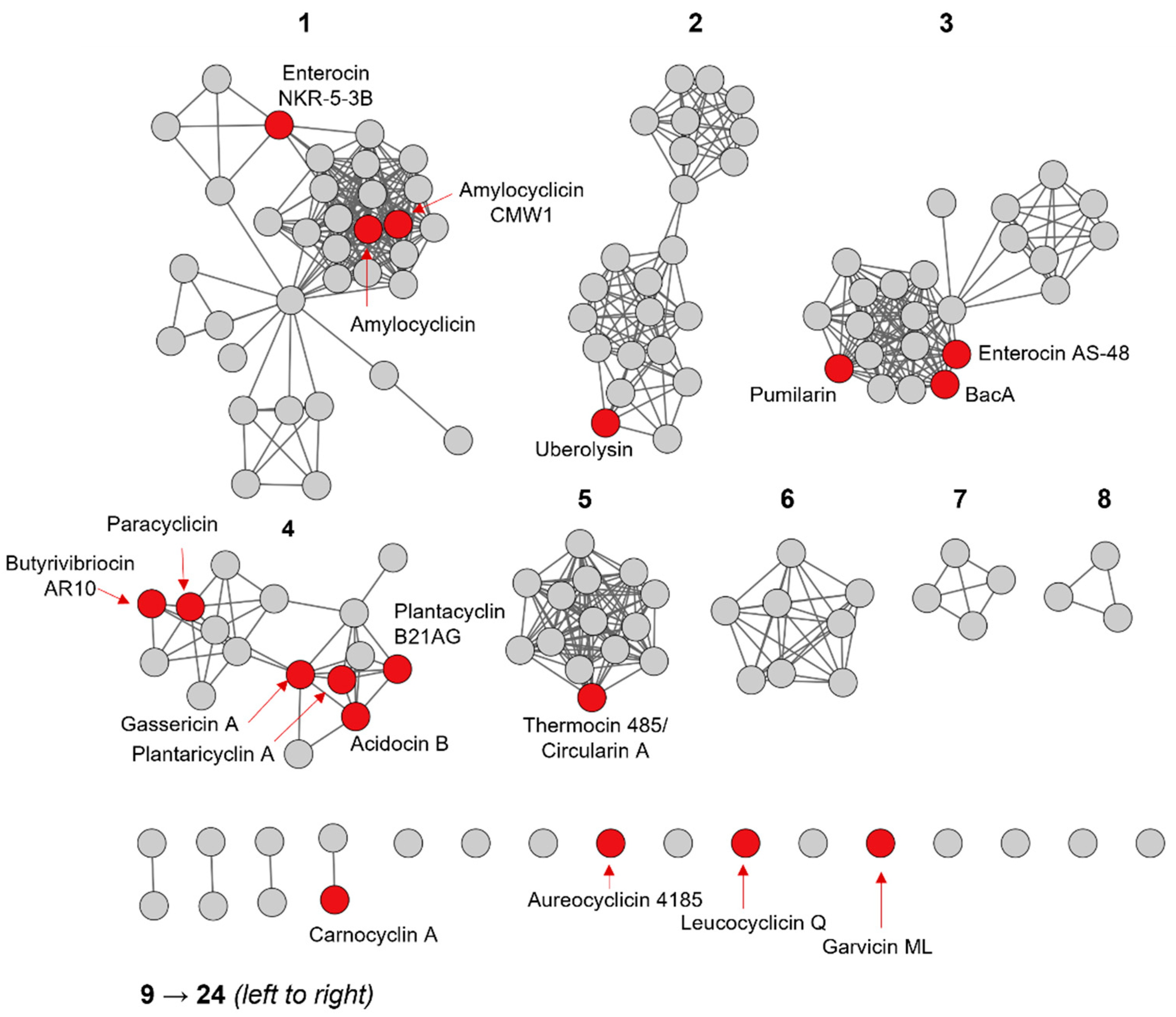
2.3. Primary Sequence Analysis of Identified Groups in the Sequence Similarity Network
2.4. Identification of the Leader Peptide Cleavage Sites
2.5. Biosynthetic Gene Cluster Analysis
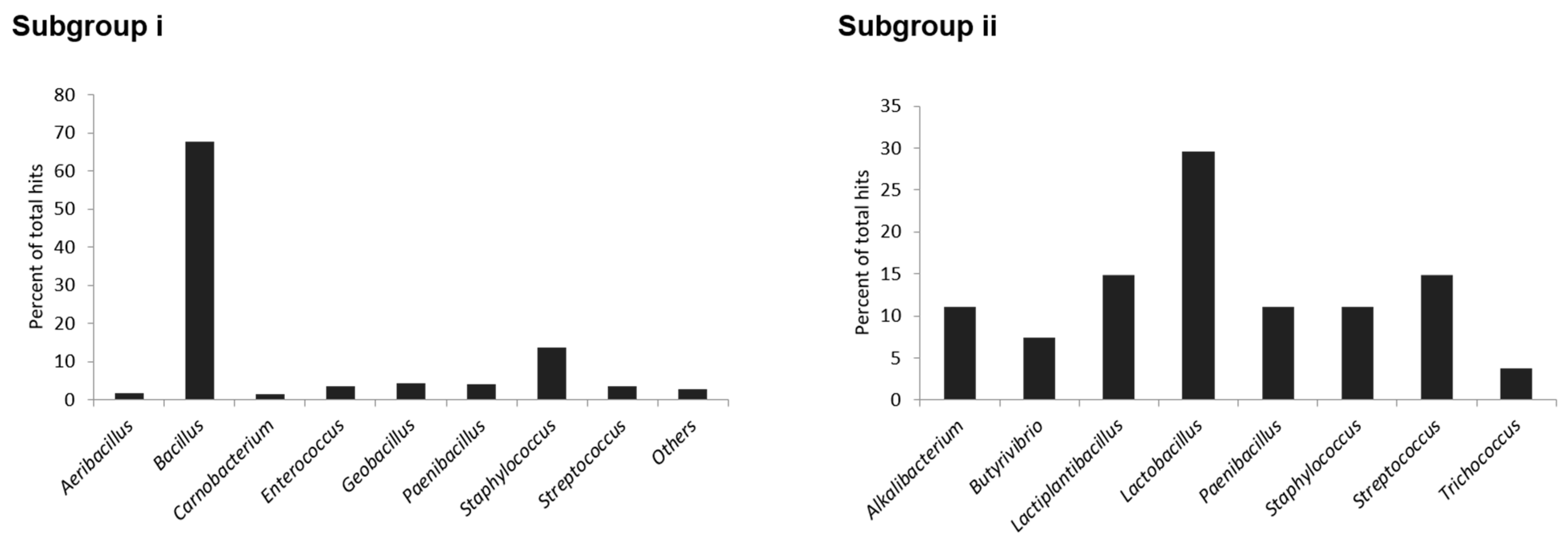
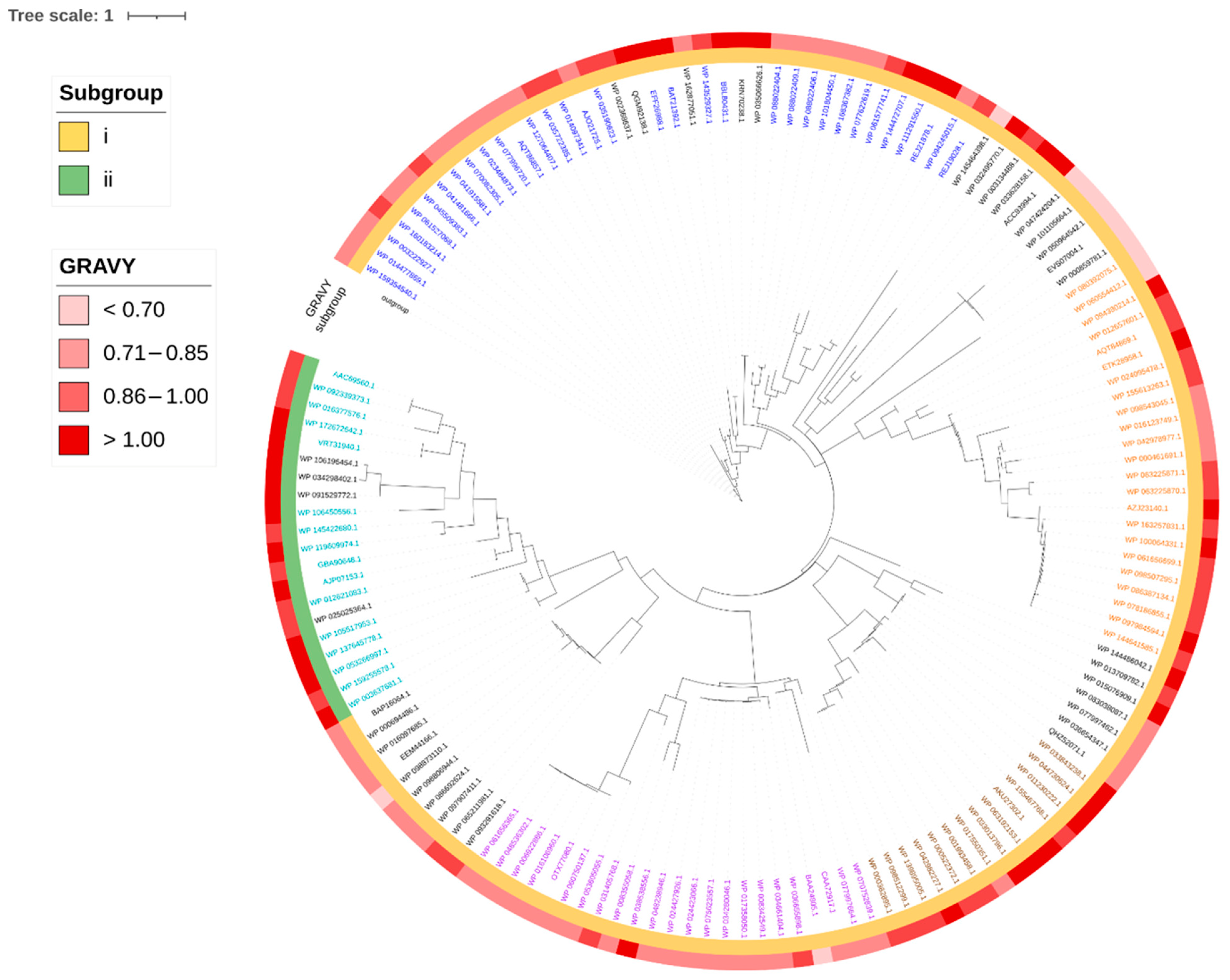
2.6. Phylogenetic Distribution of Identified Head-to-Tail Cyclized Bacteriocins
3. Materials and Methods
3.1. Genome Mining for Head-to-Tail Cyclized Bacteriocins
3.2. Precursor Peptide Sequence Similarity Analysis
3.3. Generation of Precursor Peptide Sequence Logos
3.4. Phylogenetic Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- O’Connor, P.M.; Kuniyoshi, T.M.; Oliveira, R.P.; Hill, C.; Ross, R.P.; Cotter, P.D. Antimicrobials for food and feed; a bacteriocin perspective. Curr. Opin. Biotechnol. 2020, 61, 160–167. [Google Scholar] [CrossRef]
- Johnson, E.M.; Jung, D.Y.; Jin, D.Y.; Jayabalan, D.R.; Yang, D.S.H.; Suh, J.W. Bacteriocins as food preservatives: Challenges and emerging horizons. Crit. Rev. Food Sci. Nutr. 2018, 58, 2743–2767. [Google Scholar] [CrossRef]
- Ben Lagha, A.; Haas, B.; Gottschalk, M.; Grenier, D. Antimicrobial potential of bacteriocins in poultry and swine production. Vet. Res. 2017, 48, 22. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hegarty, J.W.; Guinane, C.M.; Ross, R.P.; Hill, C.; Cotter, P.D. Bacteriocin production: A relatively unharnessed probiotic trait. F1000Research 2016, 5, 2587. [Google Scholar] [CrossRef]
- Bäuerl, C.; Umu, Ö.C.O.; Hernandez, P.E.; Diep, D.B.; Pérez-Martínez, G. A method to assess bacteriocin effects on the gut microbiota of mice. J. Vis. Exp. 2017, 125, 56053. [Google Scholar] [CrossRef] [PubMed]
- Van Belkum, M.J.; Martin-Visscher, L.A.; Vederas, J.C. Structure and genetics of circular bacteriocins. Trends Microbiol. 2011, 19, 411–418. [Google Scholar] [CrossRef] [PubMed]
- Perez, R.H.; Zendo, T.; Sonomoto, K. Circular and leaderless bacteriocins: Biosynthesis, mode of action, applications, and prospects. Front. Microbiol. 2018, 9, 2085. [Google Scholar] [CrossRef]
- Acedo, J.Z.; Chiorean, S.; Vederas, J.C.; van Belkum, M.J. The expanding structural variety among bacteriocins from Gram-positive bacteria. FEMS Microbiol. Rev. 2018, 42, 805–828. [Google Scholar] [CrossRef]
- Towle, K.M.; Vederas, J.C. Structural features of many circular and leaderless bacteriocins are similar to those in saposins and saposin-like peptides. MedChemComm 2017, 8, 276–285. [Google Scholar] [CrossRef] [Green Version]
- Acedo, J.Z.; van Belkum, M.J.; Lohans, C.T.; Towle, K.M.; Miskolzie, M.; Vederas, J.C. Nuclear magnetic resonance solution structures of lacticin Q and aureocin A53 reveal a structural motif conserved among leaderless bacteriocins with broad-spectrum activity. Biochemistry 2016, 55, 733–742. [Google Scholar] [CrossRef]
- Martin-Visscher, L.A.; Yoganathan, S.; Sit, C.S.; Lohans, C.T.; Vederas, J.C. The activity of bacteriocins from Carnobacterium maltaromaticum UAL307 against Gram-negative bacteria in combination with EDTA treatment. FEMS Microbiol. Lett. 2011, 317, 152–159. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sawa, N.; Zendo, T.; Kiyofuji, J.; Fujita, K.; Himeno, K.; Nakayama, J.; Sonomoto, K. Identification and characterization of lactocyclicin Q, a novel cyclic bacteriocin produced by Lactococcus sp. strain QU 12. Appl. Environ. Microbiol. 2009, 75, 1552–1558. [Google Scholar] [CrossRef] [Green Version]
- Masuda, Y.; Ono, H.; Kitagawa, H.; Ito, H.; Mu, F.; Sawa, N.; Zendo, T.; Sonomoto, K. Identification and characterization of leucocyclicin Q, a novel cyclic bacteriocin produced by Leuconostoc mesenteroides TK41401. Appl. Environ. Microbiol. 2011, 77, 8164–8170. [Google Scholar] [CrossRef] [Green Version]
- Gálvez, A.; Maqueda, M.; Martínez-Bueno, M.; Valdivia, E. Bactericidal and bacteriolytic action of peptide antibiotic AS-48 against Gram-positive and Gram-negative bacteria and other organisms. Res. Microbiol. 1989, 140, 57–68. [Google Scholar] [CrossRef]
- Gálvez, A.; Maqueda, M.; Martínez-Bueno, M.; Valdivia, E. Permeation of bacterial cells, permeation of cytoplasmic, and artificial membrane vesicles, and channel formation on lipid bilayers by peptide antibiotic AS-48. J. Bacteriol. 1991, 173, 886–892. [Google Scholar] [CrossRef] [Green Version]
- Gong, X.; Martin-Visscher, L.A.; Nahirney, D.; Vederas, J.C.; Duszyk, M. The circular bacteriocin, carnocyclin A, forms anion-selective channels in lipid bilayers. Biochim. Biophys. Acta 2009, 1788, 1797–1803. [Google Scholar] [CrossRef] [Green Version]
- Kawai, Y.; Ishii, Y.; Arakawa, K.; Uemura, K.; Saitoh, B.; Nishimura, J.; Kitazawa, H.; Yamazaki, Y.; Tateno, Y.; Itoh, T.; et al. Structural and functional differences in two cyclic bacteriocins with the same sequences produced by lactobacilli. Appl. Environ. Microbiol. 2004, 70, 2906–2911. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sánchez-Barrena, M.J.; Martínez-Ripoll, M.; Gálvez, A.; Valdivia, E.; Maqueda, M.; Cruz, V.; Albert, A. Structure of bacteriocin AS-48: From soluble state to membrane bound state. J. Mol. Biol. 2003, 334, 541–549. [Google Scholar] [CrossRef]
- Cruz, V.L.; Ramos, J.; Melo, M.N.; Martinez-Salazar, J. Bacteriocin AS-48 binding to model membranes and pore formation as revealed by coarse-grained simulations. Biochim. Biophys. Acta 2013, 1828, 2524–2531. [Google Scholar] [CrossRef]
- Martin-Visscher, L.A.; Gong, X.; Duszyk, M.; Vederas, J.C. The three-dimensional structure of carnocyclin A reveals that many circular bacteriocins share a common structural motif. J. Biol. Chem. 2009, 284, 28674–28681. [Google Scholar] [CrossRef] [Green Version]
- Gabrielsen, C.; Brede, D.A.; Hernández, P.E.; Nes, I.F.; Diep, D.B. The maltose ABC transporter in Lactococcus lactis facilitates high-level sensitivity to the circular bacteriocin garvicin ML. Antimicrob. Agents Chemother 2012, 56, 2908–2915. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Trivella, D.B.B.; de Felicio, R. The tripod for bacterial natural product discovery: Genome mining, silent pathway induction, and mass spectrometry-based molecular networking. mSystems 2018, 3. [Google Scholar] [CrossRef] [Green Version]
- O’Neill, E.C.; Schorn, M.; Larson, C.B.; Millán-Aguiñaga, N. Targeted antibiotic discovery through biosynthesis-associated resistance determinants: Target directed genome mining. Crit. Rev. Microbiol. 2019, 45, 255–277. [Google Scholar] [CrossRef]
- Sekurova, O.N.; Schneider, O.; Zotchev, S.B. Novel bioactive natural products from bacteria via bioprospecting, genome mining and metabolic engineering. Microb. Biotechnol. 2019, 12, 828–844. [Google Scholar] [CrossRef] [Green Version]
- Poorinmohammad, N.; Bagheban-Shemirani, R.; Hamedi, J. Genome mining for ribosomally synthesised and post-translationally modified peptides (RiPPs) reveals undiscovered bioactive potentials of actinobacteria. Antonie Van Leeuwenhoek 2019, 112, 1477–1499. [Google Scholar] [CrossRef]
- Huo, L.; Zhao, X.; Acedo, J.Z.; Estrada, P.; Nair, S.K.; van der Donk, W.A. Characterization of a dehydratase and methyltransferase in the biosynthesis of a ribosomally-synthesized and post-translationally modified peptide in Lachnospiraceae. Chembiochem 2019, 21, 190–199. [Google Scholar] [CrossRef]
- Acedo, J.Z.; Bothwell, I.R.; An, L.; Trouth, A.; Frazier, C.; van der Donk, W.A. O-Methyltransferase-mediated incorporation of a β-amino acid in lanthipeptides. J. Am. Chem. Soc. 2019, 141, 16790–16801. [Google Scholar] [CrossRef] [PubMed]
- Van Heel, A.J.; de Jong, A.; Montalbán-López, M.; Kok, J.; Kuipers, O.P. BAGEL3: Automated identification of genes encoding bacteriocins and (non-)bactericidal posttranslationally modified peptides. Nucleic Acids Res. 2013, 41, W448–W453. [Google Scholar] [CrossRef] [PubMed]
- Van Heel, A.J.; Montalbán-López, M.; Quentin, O.; Kuipers, O.P. Genome-guided identification of novel head-to-tail cyclized antimicrobial peptides, exemplified by the discovery of pumilarin. Microb. Genom. 2017, 3. [Google Scholar] [CrossRef] [Green Version]
- Xin, B.; Liu, H.; Zheng, J.; Xie, C.; Gao, Y.; Dai, D.; Peng, D.; Ruan, L.; Chen, H.; Sun, M. In silico analysis highlights the diversity and novelty of circular bacteriocins in sequenced microbial genomes. mSystems 2020, 5, e00047-20. [Google Scholar] [CrossRef]
- Tietz, J.I.; Schwalen, C.J.; Patel, P.S.; Maxson, T.; Blair, P.M.; Tai, H.C.; Zakai, U.I.; Mitchell, D.A. A new genome-mining tool redefines the lasso peptide biosynthetic landscape. Nat. Chem. Biol. 2017, 13, 470–478. [Google Scholar] [CrossRef] [PubMed]
- Vezina, B.; Rehm, B.H.A.; Smith, A.T. Bioinformatic prospecting and phylogenetic analysis reveals 94 undescribed circular bacteriocins and key motifs. BMC Microbiol. 2020, 20, 77. [Google Scholar] [CrossRef]
- Cochrane, S.A.; Vederas, J.C. Lipopeptides from Bacillus and Paenibacillus spp.: A gold mine of antibiotic candidates. Med. Res. Rev. 2016, 36, 4–31. [Google Scholar] [CrossRef]
- Daas, M.S.; Rosana, A.R.R.; Acedo, J.Z.; Nateche, F.; Kebbouche-Gana, S.; Vederas, J.C.; Case, R. Draft genome sequences of Bacillus cereus E41 and Bacillus anthracis F34 isolated from Algerian salt lakes. Genome Announc. 2017, 5, e00383-17. [Google Scholar] [CrossRef] [Green Version]
- Daas, M.S.; Acedo, J.Z.; Rosana, A.R.R.; Orata, F.D.; Reiz, B.; Zheng, J.; Natache, F.; Case, R.J.; Kebbouche-Gana, S.; Vederas, J.C. Bacillus amyloliquefaciens ssp. plantarum F11 isolated from Algerian salty lake as a source of biosurfactants and bioactive lipopeptides. FEMS Microbiol. Lett. 2018, 365. [Google Scholar] [CrossRef]
- Daas, M.S.; Rosana, A.R.R.; Acedo, J.Z.; Douzane, M.; Nateche, F.; Kebbouche-Gana, S.; Vederas, J.C. Draft genome sequence of Bacillus paralicheniformis F47, isolated from an Algerian salty lake. Genome Announc. 2018, 6, e00190-18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Daas, M.S.; Rosana, A.R.R.; Acedo, J.Z.; Douzane, M.; Nateche, F.; Kebbouche-Gana, S.; Vederas, J.C. Insights into the draft genome sequence of bioactives-producing Bacillus thuringiensis DNG9 isolated from Algerian soil-oil slough. Stand Genom. Sci. 2018, 13, 25. [Google Scholar] [CrossRef] [Green Version]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]
- Bogaardt, C.; Tonder, A.J.; Brueggemann, A.B. Genomic analyses of pneumococci reveal a wide diversity of bacteriocins–including pneumocyclicin, a novel circular bacteriocin. BMC Genom. 2015, 16, 554. [Google Scholar] [CrossRef] [Green Version]
- Wang, C.Y.; Patel, N.; Wholey, W.Y.; Dawid, S. ABC transporter content diversity in Streptococcus pneumoniae impacts competence regulation and bacteriocin production. Proc. Natl. Acad. Sci. USA 2018, 115, E5776–E5785. [Google Scholar] [CrossRef] [Green Version]
- Sievers, F.; Higgins, D.G. Clustal Omega. Curr. Protoc. Bioinform. 2014, 48, 3.13.1–3.13.16. [Google Scholar] [CrossRef] [PubMed]
- Crooks, G.E.; Hon, G.; Chandonia, J.M.; Brenner, S.E. WebLogo: A sequence logo generator. Genome Res. 2004, 14, 1188–1190. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kourie, J.I.; Shorthouse, A.A. Properties of cytotoxic peptide-formed ion channels. Am. J. Physiol. Cell Physiol. 2000, 278, C1063–C1087. [Google Scholar] [CrossRef]
- Acedo, J.Z.; Towle, K.M.; Lohans, C.T.; Miskolzie, M.; McKay, R.T.; Doerksen, T.A.; Vederas, J.C.; Martin-Visscher, L.A. Identification and three-dimensional structure of carnobacteriocin XY, a class IIb bacteriocin produced by carnobacteria. FEBS Lett. 2017, 591, 1349–1359. [Google Scholar] [CrossRef]
- Acedo, J.Z.; van Belkum, M.J.; Lohans, C.T.; McKay, R.T.; Miskolzie, M.; Vederas, J.C. Solution structure of acidocin B, a circular bacteriocin produced by Lactobacillus acidophilus M46. Appl. Environ. Microbiol. 2015, 81, 2910–2918. [Google Scholar] [CrossRef] [Green Version]
- Trifinopoulos, J.; Nguyen, L.T.; von Haeseler, A.; Minh, B.Q. W-IQ-TREE: A fast online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Res. 2016, 44, W232–W235. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Letunic, I.; Bork, P. Interactive tree of life (iTOL) v3: An online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 2016, 44, W242–W245. [Google Scholar] [CrossRef]
- Samyn, B.; Martínez-Bueno, M.; Devreese, B.; Maqueda, M.; Gálvez, A.; Valdivia, E.; Coyette, J.; Beeumen, J.V. The cyclic structure of the enterococcal peptide antibiotic AS-48. FEBS Lett. 1994, 352, 87–90. [Google Scholar] [CrossRef] [Green Version]
- Kemperman, R.; Kuipers, A.; Karsens, H.; Nauta, A.; Kuipers, O.; Kok, J. Identification and characterization of two novel clostridial bacteriocins, circularin A and closticin 574. Appl. Environ. Microbiol. 2003, 69, 1589–1597. [Google Scholar] [CrossRef] [Green Version]
- Wirawan, R.E.; Swanson, K.M.; Kleffmann, T.; Jack, R.W.; Tagg, J.R. Uberolysin: A novel cyclic bacteriocin produced by Streptococcus uberis. Microbiology 2007, 153, 1619–1630. [Google Scholar] [CrossRef] [Green Version]
- Martin-Visscher, L.A.; van Belkum, M.J.; Garneau-Tsodikova, S.; Whittal, R.M.; Zheng, J.; McMullen, L.M.; Vederas, J.C. Isolation and characterization of carnocyclin A, a novel circular bacteriocin produced by Carnobacterium maltaromaticum UAL307. Appl. Environ. Microbiol. 2008, 74, 4756–4763. [Google Scholar] [CrossRef] [Green Version]
- Himeno, K.; Rosengren, K.J.; Inoue, T.; Perez, R.H.; Colgrave, M.L.; Lee, H.S.; Chan, L.Y.; Henriques, S.T.; Fujita, K.; Isibashi, N.; et al. Identification, characterization, and three-dimensional structure of the novel circular bacteriocin, enterocin NKR-5-3B, from Enterococcus faecium. Biochemistry 2015, 54, 4863–4876. [Google Scholar] [CrossRef]
- Borrero, J.; Brede, D.A.; Skaugen, M.; Diep, D.B.; Herranz, C.; Nes, I.F.; Cintas, L.M.; Hernández, P.E. Characterization of garvicin ML, a novel circular bacteriocin produced by Lactococcus garvieae DCC43, isolated from mallard ducks (Anas platyrhynchos). Appl. Environ. Microbiol. 2011, 77, 369–373. [Google Scholar] [CrossRef] [Green Version]
- Potter, A.; Ceotto, H.; Coelho, M.L.; Guimaráes, A.J.; Bastos, M.C. The gene cluster of aureocyclicin 4185: The first cyclic bacteriocin of Staphylococcus aureus. Microbiology 2014, 160, 917–928. [Google Scholar] [CrossRef] [Green Version]
- Kurata, A.; Yamaguchi, T.; Kira, M.; Kishimoto, N. Characterization and heterologous expression of an antimicrobial peptide from Bacillus amyloliquefaciens CMW1. Biotechnol. Biotechnol. Equip. 2019, 33, 886–893. [Google Scholar] [CrossRef] [Green Version]
- Scholz, R.; Vater, J.; Budiharjo, A.; Wang, Z.; He, Y.; Dietel, K.; Schwecke, T.; Herfort, S.; Lasch, P.; Borriss, R. Amylocyclicin, a novel circular bacteriocin produced by Bacillus amyloliquefaciens FZB42. J. Bacteriol. 2014, 196, 1842–1852. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tomita, H.; Fujimoto, S.; Tanimoto, K.; Ike, Y. Cloning and genetic and sequence analyses of the bacteriocin 21 determinant encoded on the Enterococcus faecalis pheromone-responsive conjugative plasmid pPD1. J. Bacteriol. 1997, 179, 7843–7855. [Google Scholar] [CrossRef] [Green Version]
- Egan, K. Discovery and Evaluation of Novel and Characterised Bacteriocins for Future Applications. Ph.D. Thesis, School of Microbiology, University College Cork, Cork, Ireland, 2018. [Google Scholar]
- Kawai, Y.; Saito, T.; Kitazawa, H.; Itoh, T. Gassericin A; an uncommon cyclic bacteriocin produced by Lactobacillus gasseri LA39 linked at N- and C-terminal ends. Biosci. Biotechnol. Biochem. 1998, 62, 2438–2440. [Google Scholar] [CrossRef] [PubMed]
- Kalmokoff, M.L.; Cyr, T.D.; Hefford, M.A.; Whitford, M.F.; Teather, R.M. Butyrivibriocin AR10, a new cyclic bacteriocin produced by the ruminal anaerobe Butyrivibrio fibrisolvens AR10: Characterization of the gene and peptide. Can. J. Microbiol. 2003, 49, 763–773. [Google Scholar] [CrossRef] [PubMed]
- Collins, F.W.J.; O’Connor, P.M.; O’Sullivan, O.; Gómez-Sala, B.; Rea, M.C.; Hill, C.; Ross, R.P. Bacteriocin gene-trait matching across the complete Lactobacillus pan-genome. Sci. Rep. 2017, 7, 3481. [Google Scholar] [CrossRef] [Green Version]
- Golneshin, A. Characterisation of Bacteriocin Genes and Proteins from Lactobacillus plantarum B21 as Potential New Antimicrobial Agents and Natural Food Preservatives; School of Applied Sciences, RMIT University: Melbourne, Australia, 2014. [Google Scholar]
- Borrero, J.; Kelly, E.; O’Connor, P.M.; Kelleher, P.; Scully, C.; Cotter, P.D.; Mahony, J.; van Sinderen, D. Purification, characterization and heterologous production of plantaricyclin A, a novel circular bacteriocin produced by Lactobacillus plantarum NI326. Appl. Environ. Microbiol. 2017, 84, e01801–e01817. [Google Scholar] [CrossRef] [Green Version]
- Gerlt, J.A.; Bouvier, J.T.; Davidson, D.B.; Imker, H.J.; Sadkhin, B.; Slater, D.R.; Whalen, K.L. Enzyme Function Initiative-Enzyme Similarity Tool (EFI-EST): A web tool for generating protein sequence similarity networks. Biochim. Biophys. Acta 2015, 1854, 1019–1037. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zallot, R.; Oberg, N.; Gerlt, J.A. The EFI web resource for genomic enzymology tools: Leveraging protein, genome, and metagenome databases to discover novel enzymes and metabolic pathways. Biochemistry 2019, 58, 4169–4182. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [PubMed] [Green Version]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Major, D.; Flanzbaum, L.; Lussier, L.; Davies, C.; Caldo, K.M.P.; Acedo, J.Z. Transporter Protein-Guided Genome Mining for Head-to-Tail Cyclized Bacteriocins. Molecules 2021, 26, 7218. https://doi.org/10.3390/molecules26237218
Major D, Flanzbaum L, Lussier L, Davies C, Caldo KMP, Acedo JZ. Transporter Protein-Guided Genome Mining for Head-to-Tail Cyclized Bacteriocins. Molecules. 2021; 26(23):7218. https://doi.org/10.3390/molecules26237218
Chicago/Turabian StyleMajor, Daniel, Lara Flanzbaum, Leah Lussier, Carly Davies, Kristian Mark P. Caldo, and Jeella Z. Acedo. 2021. "Transporter Protein-Guided Genome Mining for Head-to-Tail Cyclized Bacteriocins" Molecules 26, no. 23: 7218. https://doi.org/10.3390/molecules26237218
APA StyleMajor, D., Flanzbaum, L., Lussier, L., Davies, C., Caldo, K. M. P., & Acedo, J. Z. (2021). Transporter Protein-Guided Genome Mining for Head-to-Tail Cyclized Bacteriocins. Molecules, 26(23), 7218. https://doi.org/10.3390/molecules26237218






