Compounds from the Medicines for Malaria Venture Box Inhibit In Vitro Growth of Babesia divergens, a Blood-Borne Parasite of Veterinary and Zoonotic Importance
Abstract
1. Introduction
2. Results and Discussion
3. Materials and Methods
3.1. Chemical Reagents
3.2. In Vitro Cultivation of B. divergens Parasite
3.3. Malaria Box Compound Screening against B. divergens
3.4. Viability Test
3.5. Structural Similarity Measurements
3.6. In Vitro Drug Combination Test
3.7. Statistical Analyses
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Suarez, C.E.; Alzan, H.F.; Silva, M.G.; Rathinasamy, V.; Poole, W.A.; Cooke, B.M. Unravelling the cellular and molecular pathogenesis of bovine babesiosis: Is the sky the limit? Int. J. Parasitol. 2019, 49, 183–197. [Google Scholar] [CrossRef]
- Rizk, M.A.; El-Sayed, S.A.E.; El-Khodery, S.; Yokoyama, N.; Igarashi, I. Discovering the in vitro potent inhibitors against Babesia and Theileria parasites by repurposing the Malaria Box: A review. Vet. Parasitol 2019, 274, 108895. [Google Scholar] [CrossRef]
- Rizk, M.A.; AbouLaila, M.; El-Sayed, S.A.E.; Guswanto, A.; Yokoyama, N.; Igarashi, I. Inhibitory effects of fluoroquinolone antibiotics on Babesia divergens and Babesia microti, blood parasites of veterinary and zoonotic importance. Infect. Drug Resist. 2018, 11, 1605–1615. [Google Scholar] [CrossRef] [PubMed]
- Rizk, M.A.; El-Sayed, S.A.E.; Nassif, M.; Mosqueda, J.; Xuan, X.; Igarashi, I. Assay methods for in vitro and in vivo anti-Babesia drug efficacy testing: Current progress, outlook, and challenges. Vet. Parasitol 2020, 279, 109013. [Google Scholar] [CrossRef]
- Zintl, A.; Mulcahy, G.; Skerrett, H.E.; Taylor, S.M.; Gray, J.S. Babesia divergens, a bovine blood parasite of veterinary and zoonotic importance. Clin. Microbiol Rev. 2003, 16, 622–636. [Google Scholar] [CrossRef] [PubMed]
- Van Voorhis, W.C.; Adams, J.H.; Adelfio, R.; Ahyong, V.; Akabas, M.H.; Alano, P.; Alday, A.; Aleman Resto, Y.; Alsibaee, A.; Alzualde, A.; et al. Open Source Drug Discovery with the Malaria Box Compound Collection for Neglected Diseases and Beyond. PLoS Pathog. 2016, 12, e1005763. [Google Scholar] [CrossRef] [PubMed]
- Bennett, T.N.; Paguio, M.; Gligorijevic, B.; Seudieu, C.; Kosar, A.D.; Davidson, E.; Roepe, P.D. Novel, rapid, and inexpensive cell-based quantification of antimalarial drug efficacy. Antimicrob. Agents Chemother. 2004, 48, 1807–1810. [Google Scholar] [CrossRef]
- Spangenberg, T.; Burrows, J.N.; Kowalczyk, P.; McDonald, S.; Wells, T.N.; Willis, P. The open access malaria box: A drug discovery catalyst for neglected diseases. PLoS ONE 2013, 8, e62906. [Google Scholar] [CrossRef] [PubMed]
- Paul, A.S.; Moreira, C.K.; Elsworth, B.; Allred, D.R.; Duraisingh, M.T. Extensive Shared Chemosensitivity between Malaria and Babesiosis Blood-Stage Parasites. Antimicrob. Agents Chemother. 2016, 60, 5059–5063. [Google Scholar] [CrossRef]
- Elabbadi, N.; Ancelin, M.L.; Vial, H.J. Use of radioactive ethanolamine incorporation into phospholipids to assess in vitro antimalarial activity by the semiautomated microdilution technique. Antimicrob. Agents Chemother. 1992, 36, 50–55. [Google Scholar] [CrossRef][Green Version]
- Chulay, J.D.; Haynes, J.D.; Diggs, C.L. Plasmodium falciparum: Assessment of in vitro growth by [3H]hypoxanthine incorporation. Exp. Parasitol. 1983, 55, 138–146. [Google Scholar] [CrossRef]
- Geary, T.G.; Divo, A.A.; Jensen, J.B. An in vitro assay system for the identification of potential antimalarial drugs. J. Parasitol. 1983, 69, 577–583. [Google Scholar] [CrossRef] [PubMed]
- Sherman, I.W. Transport of amino acids and nucleic acid precursors in malarial parasites. Bull. World Health Organ. 1977, 55, 211–225. [Google Scholar]
- Irvin, A.D.; Young, E.R. Further studies on the uptake of tritiated nucleic acid precursors by Babesia spp. of cattle and mice. Int. J. Parasitol. 1979, 9, 109–114. [Google Scholar] [CrossRef]
- Hassan, H.F. Purine-metabolising enzymes in Babesia divergens. Zeit. Parasitenkund. 1987, 73, 121–125. [Google Scholar]
- Lobo, C.A. Babesia divergens and Plasmodium falciparum use common receptors, glycophorins A and B, to invade the human red blood cell. Infect. Immun. 2005, 73, 649–651. [Google Scholar] [CrossRef]
- Bork, S.; Yokoyama, N.; Ikehara, Y.; Kumar, S.; Sugimoto, C.; Igarashi, I. Growth-inhibitory effect of heparin on Babesia parasites. Antimicrob. Agents Chemother. 2004, 48, 236–241. [Google Scholar] [CrossRef]
- Baum, J.; Chen, L.; Healer, J.; Lopaticki, S.; Boyle, M.; Triglia, T.; Ehlgen, F.; Ralph, S.A.; Beeson, J.G.; Cowman, A.F. Reticulocyte-binding protein homologue 5-an essential adhesin involved in invasion of human erythrocytes by Plasmodium falciparum. Int J. Parasitol. 2009, 39, 371–380. [Google Scholar] [CrossRef]
- Boyle, M.J.; Richards, J.S.; Gilson, P.R.; Chai, W.; Beeson, J.G. Interactions with heparin-like molecules during erythrocyte invasion by Plasmodium falciparum merozoites. Blood 2010, 115, 4559–4568. [Google Scholar] [CrossRef]
- Kobayashi, K.; Kato, K.; Sugi, T.; Takemae, H.; Pandey, K.; Gong, H.; Tohya, Y.; Akashi, H. Plasmodium falciparum BAEBL binds to heparan sulfate proteoglycans on the human erythrocyte surface. J. Biol. Chem. 2010, 285, 1716–1725. [Google Scholar] [CrossRef]
- Kobayashi, K.; Takano, R.; Takemae, H.; Sugi, T.; Ishiwa, A.; Gong, H.; Recuenco, F.C.; Iwanaga, T.; Horimoto, T.; Akashi, H.; et al. Analyses of interactions between heparin and the apical surface proteins of Plasmodium falciparum. Sci. Rep. 2013, 3, 3178. [Google Scholar] [CrossRef] [PubMed]
- Boyom, F.F.; Fokou, P.V.; Tchokouaha, L.R.; Spangenberg, T.; Mfopa, A.N.; Kouipou, R.M.; Mbouna, C.J.; Donfack, V.F.; Zollo, P.H. Repurposing the open access malaria box to discover potent inhibitors of Toxoplasma gondii and Entamoeba histolytica. Antimicrob. Agents Chemother. 2014, 58, 5848–5854. [Google Scholar] [CrossRef] [PubMed]
- Ingram-Sieber, K.; Cowan, N.; Panic, G.; Vargas, M.; Mansour, N.R.; Bickle, Q.D.; Wells, T.N.; Spangenberg, T.; Keiser, J. Orally active antischistosomal early leads identified from the open access malaria box. PLoS Negl. Trop Dis. 2014, 8, e2610. [Google Scholar] [CrossRef]
- Bessoff, K.; Spangenberg, T.; Foderaro, J.E.; Jumani, R.S.; Ward, G.E.; Huston, C.D. Identification of Cryptosporidium parvum active chemical series by Repurposing the open access malaria box. Antimicrob. Agents Chemother. 2014, 58, 2731–2739. [Google Scholar] [CrossRef] [PubMed]
- Rizk, M.A.; El-Sayed, S.A.; Terkawi, M.A.; Youssef, M.A.; El Said el Sel, S.; Elsayed, G.; El-Khodery, S.; El-Ashker, M.; Elsify, A.; Omar, M.; et al. Optimization of a Fluorescence-Based Assay for Large-Scale Drug Screening against Babesia and Theileria Parasites. PLoS ONE 2015, 10, e0125276. [Google Scholar] [CrossRef]
- Rizk, M.A.; El-Sayed, S.A.; AbouLaila, M.; Tuvshintulga, B.; Yokoyama, N.; Igarashi, I. Large-scale drug screening against Babesia divergens parasite using a fluorescence-based high-throughput screening assay. Vet. Parasitol. 2016, 227, 93–97. [Google Scholar] [CrossRef]
- Lau, A.O.; Pedroni, M.J.; Bhanot, P. Target specific-trisubstituted pyrrole inhibits Babesia bovis erythrocytic growth. Exp. Parasitol. 2013, 133, 365–368. [Google Scholar] [CrossRef]
- El-Sayed, S.A.E.; Rizk, M.A.; Terkawi, M.A.; Yokoyama, N.; Igarashi, I. Molecular identification and antigenic characterization of Babesia divergens Erythrocyte Binding Protein (BdEBP) as a potential vaccine candidate. Parasitol. Int. 2017, 66, 721–726. [Google Scholar] [CrossRef]
- Rizk, M.A.; El-Sayed, S.A.E.; AbouLaila, M.; Yokoyama, N.; Igarashi, I. Evaluation of the inhibitory effect of N-acetyl-L-cysteine on Babesia and Theileria parasites. Exp. Parasitol. 2017, 179, 43–48. [Google Scholar] [CrossRef]
- O’Boyle, N.M.; Sayle, R.A. Comparing structural fingerprints using a literature based similarity benchmark. J. Cheminformatics 2016, 8, 2206–2219. [Google Scholar] [CrossRef]
- Backman, T.W.; Cao, Y.; Girke, T. ChemMine tools: An online service for analyzing and clustering small molecules. Nucleic Acids Res. 2011, 39, W486–W491. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.; Charisi, A.; Cheng, L.C.; Jiang, T.; Girke, T. ChemmineR: A compound mining framework for R. Bioinformatics 2008, 24, 1733–1744. [Google Scholar] [CrossRef] [PubMed]
- El-Sayed, S.A.E.; Rizk, M.A.; Yokoyama, N.; Igarashi, I. Evaluation of the in vitro and in vivo inhibitory effect of thymoquinone on piroplasm parasites. Parasit. Vectors 2019, 12, 37. [Google Scholar] [CrossRef] [PubMed]
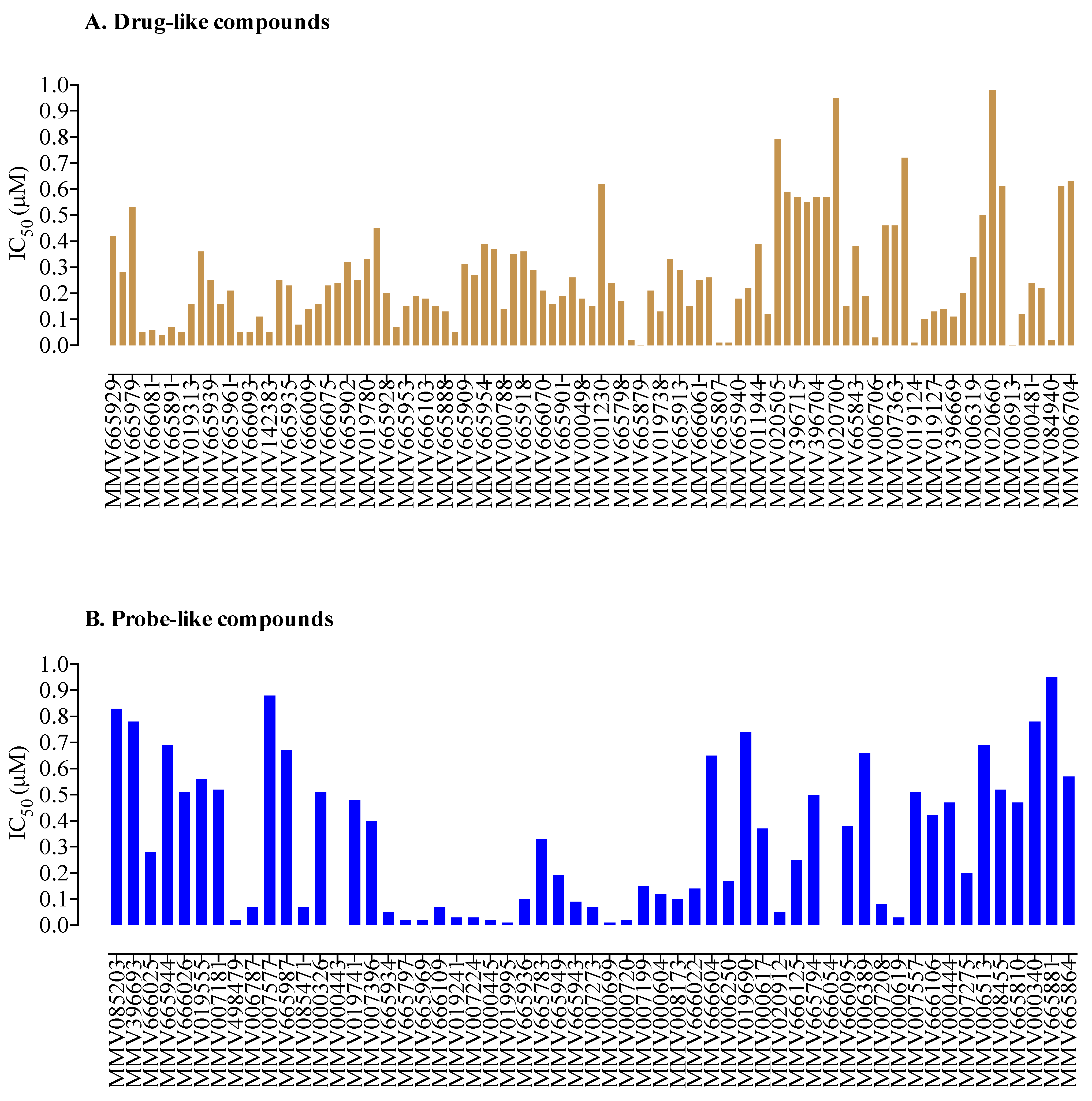
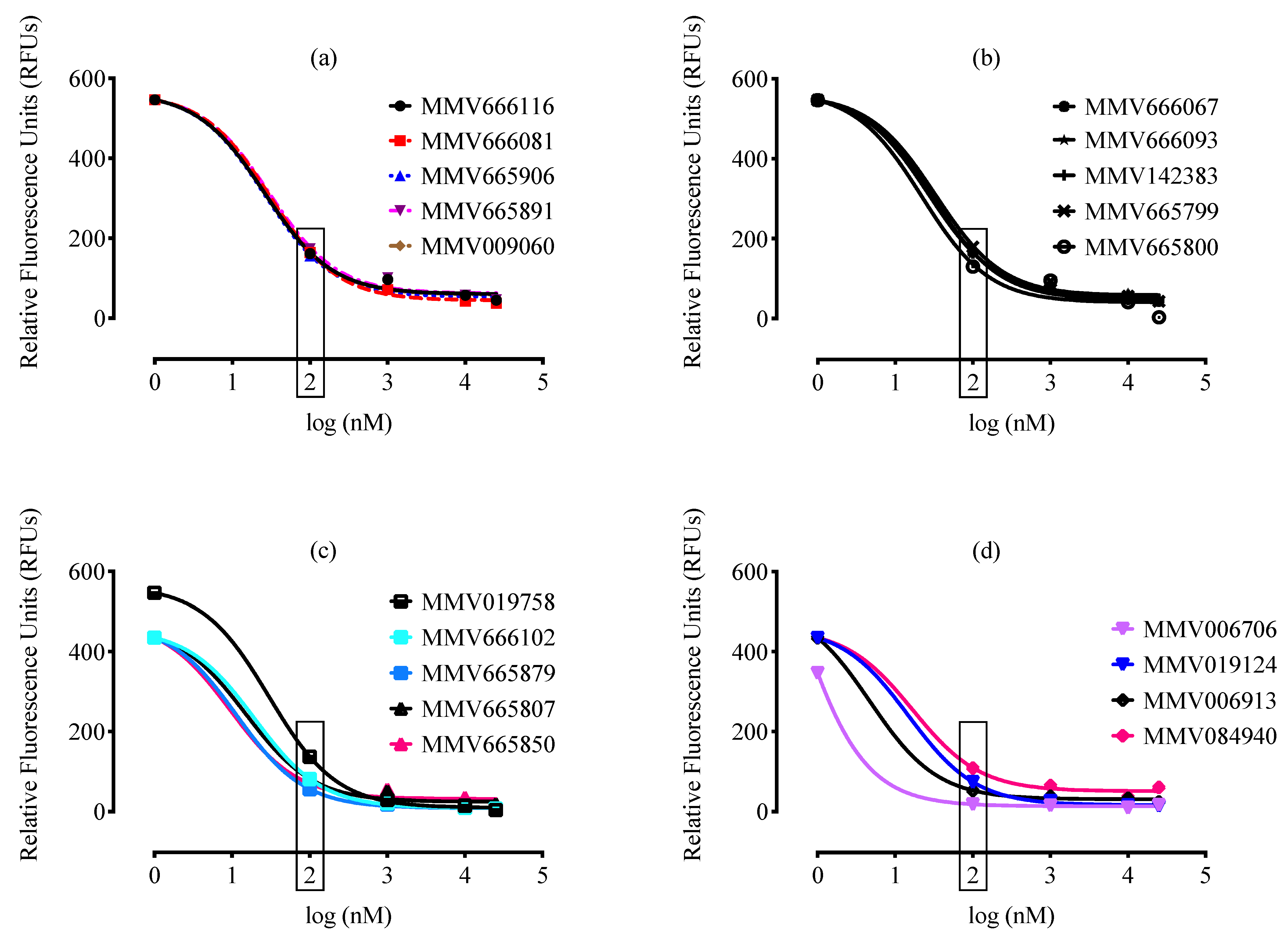
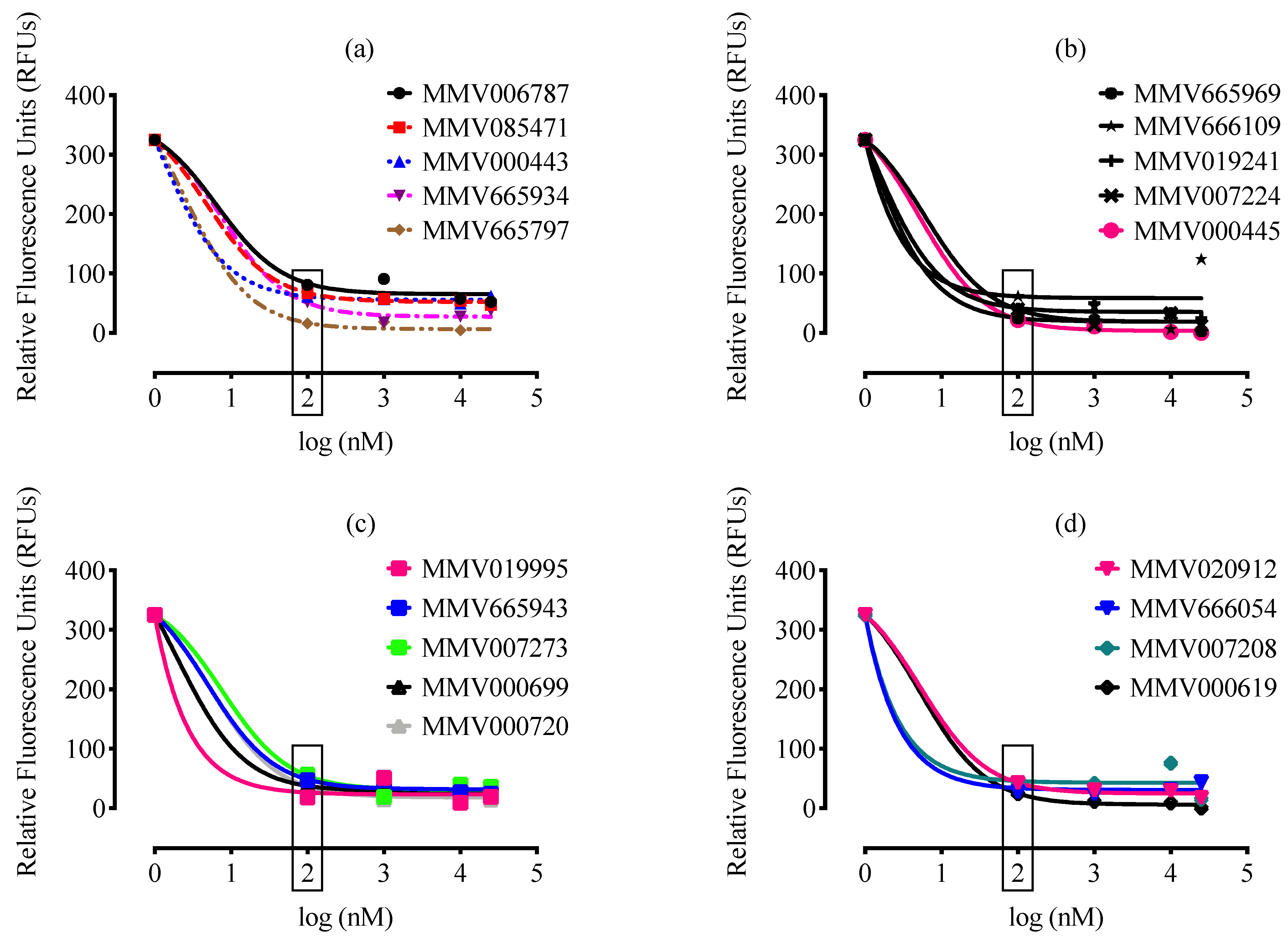


| Compound ID a | Set * | MW * (g/mol) | IC50 (nM) * P. falciparum | IC50 (nM) b B. divergens | CC50 (nM) c Against MRC-5 | SI d |
|---|---|---|---|---|---|---|
| MMV665807 | Drug-like | 315.67 | ND | 10 | 240 | 24 |
| MMV665850 | Drug-like | 273.71 | ND | 10 | 15878 | 1587 |
| MMV019124 | Drug-like | 373.40 | 1089.5 | 10 | 32000 | 3200 |
| MMV006913 | Drug-like | 199.24 | 1290 | 1 | 32000 | 32000 |
| MMV019995 | Probe-like | 462.42 | 563 | 10 | 11956 | 1195 |
| MMV000699 | Probe-like | 559.62 | 454 | 10 | 32000 | 3200 |
| MMV666054 | Probe-like | 487.76 | 513 | 1 | 7785 | 7785 |
| Drug Combination | C a | FICD1 | FICD2 | ΣFIC | Degree of Interaction b |
|---|---|---|---|---|---|
| MMV006913 + DA | 0.75 + 0.75 | 0.41 | 0.46 | 0.87 | Additive |
| 0.75 + 0.50 | 0.47 | 0.39 | 0.86 | Additive | |
| 0.50 + 0.75 | 0.74 | 0.93 | 1.67 | Indifference | |
| 0.50 + 0.50 | 0.88 | 1.03 | 1.91 | Indifference | |
| MMV666054 + DA | 0.75 + 0.75 | 1.6 | 0.22 | 1.82 | Indifference |
| 0.75 + 0.50 | 1.01 | 0.1 | 1.11 | Indifference | |
| 0.50 + 0.75 | 1.29 | 0.05 | 1.34 | Indifference | |
| 0.50 + 0.50 | 2.51 | 0.51 | 3.02 | Antagonism |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rizk, M.A.; El-Sayed, S.A.E.-S.; Alkhoudary, M.S.; Alsharif, K.F.; Abdel-Daim, M.M.; Igarashi, I. Compounds from the Medicines for Malaria Venture Box Inhibit In Vitro Growth of Babesia divergens, a Blood-Borne Parasite of Veterinary and Zoonotic Importance. Molecules 2021, 26, 7118. https://doi.org/10.3390/molecules26237118
Rizk MA, El-Sayed SAE-S, Alkhoudary MS, Alsharif KF, Abdel-Daim MM, Igarashi I. Compounds from the Medicines for Malaria Venture Box Inhibit In Vitro Growth of Babesia divergens, a Blood-Borne Parasite of Veterinary and Zoonotic Importance. Molecules. 2021; 26(23):7118. https://doi.org/10.3390/molecules26237118
Chicago/Turabian StyleRizk, Mohamed Abdo, Shimaa Abd El-Salam El-Sayed, Mahmoud S. Alkhoudary, Khalaf F. Alsharif, Mohamed M. Abdel-Daim, and Ikuo Igarashi. 2021. "Compounds from the Medicines for Malaria Venture Box Inhibit In Vitro Growth of Babesia divergens, a Blood-Borne Parasite of Veterinary and Zoonotic Importance" Molecules 26, no. 23: 7118. https://doi.org/10.3390/molecules26237118
APA StyleRizk, M. A., El-Sayed, S. A. E.-S., Alkhoudary, M. S., Alsharif, K. F., Abdel-Daim, M. M., & Igarashi, I. (2021). Compounds from the Medicines for Malaria Venture Box Inhibit In Vitro Growth of Babesia divergens, a Blood-Borne Parasite of Veterinary and Zoonotic Importance. Molecules, 26(23), 7118. https://doi.org/10.3390/molecules26237118









