In Silico Structure-Guided Optimization and Molecular Simulation Studies of 3-Phenoxy-4-(3-trifluoromethylphenyl)pyridazines as Potent Phytoene Desaturase Inhibitors
Abstract
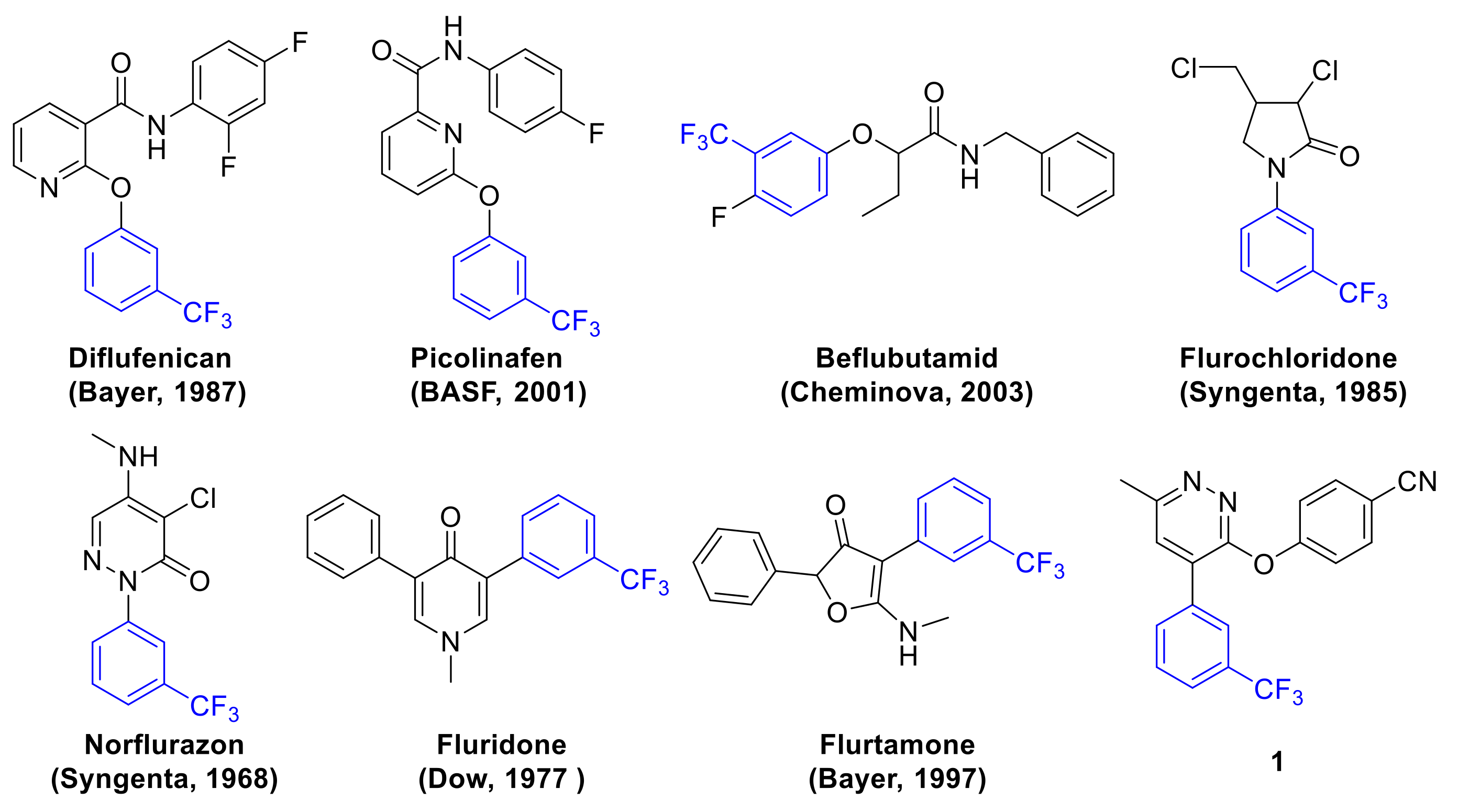
:1. Introduction
2. Results
2.1. Design of Compounds 2–5
2.2. Chemistry
2.3. Herbicidal Activity and SAR
2.4. Crop Selectivity
2.5. Synechococcus PDS Binding Affinity
2.6. Molecular Simulation Studies
3. Discussion
4. Materials and Methods
4.1. Synthetic Chemistry
4.1.1. Preparation of the Intermediates 7–12
4.1.2. General Route to Prepare Compounds 2–5
4.2. Protein Overexpression and Purification
4.3. Surface Plasmon Resonance Assay
4.4. Herbicidal Activity Assay
4.5. Crop Selectivity
4.6. Mocelcuar Simulation Studies
4.7. clogP Calculation
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Sample Availability
References
- Abouziena, H.F.; Haggag, W.M. Weed Control in Clean Agriculture: A Review. Planta Daninha 2016, 34, 377–392. [Google Scholar] [CrossRef]
- Liu, J.; Hua, R.; Lv, P.; Tang, J.; Wang, Y.; Cao, H.; Wu, X.; Li, Q.X. Novel hydrolytic de-methylthiolation of the s-triazine herb-icide prometryn by Leucobacter sp. JW-1. Sci. Total Environ. 2017, 579, 115–123. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Wang, B.; Xi, Z. Development of protoporphyrinogen IX oxidase inhibitors for sustainable agriculture. In Crop Protection Products for Sustainable Agriculture; American Chemical Society: Colombia, WA, USA, 2021; Volume 1390, pp. 11–41. [Google Scholar]
- Lin, H.Y.; Chen, X.; Dong, J.; Yang, J.F.; Xiao, H.; Ye, Y.; Li, L.H.; Zhan, C.G.; Yang, W.C.; Yang, G.F. Rational redesign of en-zyme via the combination of quantum mechanics/molecular mechanics, molecular dynamics, and structural biology study. J. Am. Chem. Soc. 2021, 143, 15674–15687. [Google Scholar] [CrossRef]
- Duke, S.O. The history and current status of glyphosate. Pest Manag. Sci. 2018, 74, 1027–1034. [Google Scholar] [CrossRef]
- Zhao, L.-X.; Jiang, M.-J.; Hu, J.-J.; Zou, Y.-L.; Cheng, Y.; Ren, T.; Gao, S.; Fu, Y.; Ye, F. Design, Synthesis, and Herbicidal Activity of Novel Diphenyl Ether Derivatives Containing Fast Degrading Tetrahydrophthalimide. J. Agric. Food Chem. 2020, 68, 3729–3741. [Google Scholar] [CrossRef]
- Zuo, Y.; Wu, Q.; Su, S.-W.; Niu, C.-W.; Xi, Z.; Yang, G.-F. Synthesis, Herbicidal Activity, and QSAR of Novel N-Benzothiazolyl-pyrimidine-2,4-diones as Protoporphyrinogen Oxidase Inhibitors. J. Agric. Food Chem. 2016, 64, 552–562. [Google Scholar] [CrossRef]
- Yang, J.; Guan, A.; Wu, Q.; Cui, D.; Liu, C. Design, synthesis and herbicidal evaluation of novel uracil derivatives containing an isoxazoline moiety. Pest Manag. Sci. 2020, 76, 3395–3402. [Google Scholar] [CrossRef]
- Xu, H.; Zou, X.M.; Zhu, Y.Q.; Liu, B.; Tao, H.L.; Hu, X.H.; Song, H.B.; Hu, F.Z.; Wang, Y.; Yang, H.Z. Synthesis and herbicidal activity of novel α,α,α-trifluoro-m-tolyl pyridazinone derivatives. Pest Manag. Sci. 2006, 62, 522–530. [Google Scholar]
- Matthias, W.; Gerhard, H. Herbicides with Bleaching Properties. In Modern Crop Protection Compounds; John Wiley & Sons Ltd.: Chichester, UK, 2019; pp. 213–302. [Google Scholar]
- Zhang, H.; Wang, J.; Ji, Z.; Sun, X.; Tian, Q.; Wei, S.; Ji, Z. Discovery, SAR, and putative mode of action of N-benzyl-2-methoxybenzamides as potential bleaching herbicides. Pest Manag. Sci. 2021, 77, 2804–2811. [Google Scholar] [CrossRef] [PubMed]
- Heap, I. The International Herbicide-Resistant Weed Database. 2020. Available online: www.weedscience.org (accessed on 1 November 2021).
- Xu, H.; Hu, X.-H.; Zou, X.-M.; Liu, B.; Zhu, Y.-Q.; Wang, Y.; Hu, F.-Z.; Yang, H.-Z. Synthesis and Herbicidal Activities of Novel 3-N-Substituted Amino-6-methyl-4-(3-trifluoromethylphenyl)pyridazine Derivatives. J. Agric. Food Chem. 2008, 56, 6567–6572. [Google Scholar] [CrossRef]
- Brausemann, A.; Gemmecker, S.; Koschmieder, J.; Ghisla, S.; Beyer, P.; Einsle, O. Structure of Phytoene Desaturase Provides Insights into Herbicide Binding and Reaction Mechanisms Involved in Carotene Desaturation. Structure 2017, 25, 1222–1232.e3. [Google Scholar] [CrossRef]
- Xiong, L.; Li, H.; Jiang, L.-N.; Ge, J.-M.; Yang, W.-C.; Zhu, X.L.; Yang, G.-F. Structure-Based Discovery of Potential Fungicides as Succinate Ubiquinone Oxidoreductase Inhibitors. J. Agric. Food Chem. 2017, 65, 1021–1029. [Google Scholar] [CrossRef]
- Morris, G.M.; Huey, R.; Lindstrom, W.; Sanner, M.F.; Belew, R.K.; Goodsell, D.S.; Olson, A.J. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J. Comput. Chem. 2009, 30, 2785–2791. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, H.; Zhu, Y.-Q.; Zou, X.-M.; Liu, B.; Wang, Y.; Hu, F.-Z.; Yang, H.-Z. Synthesis and herbicidal activities of novel 3-(substituted benzyloxy or phenoxy)-6-methyl-4-(3-trifluoromethylphenyl)pyridazine derivatives. Pest Manag. Sci. 2011, 68, 276–284. [Google Scholar] [CrossRef]
- Zhang, K.; Sun, W.; Huang, L.; Zhu, K.; Pei, F.; Zhu, L.; Wang, Q.; Lu, Y.; Zhang, H.; Jin, H.; et al. Identifying glyceraldehyde 3-phosphate dehydrogenase as a cyclic adenosine diphosphoribose binding protein by photoaffinity potein-liand labeling approach. J. Am. Chem. Soc. 2017, 139, 156–170. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.-W.; Yu, S.-Y.; Pang, Z.-L.; Ma, D.-J.; Liang, L.; Wang, X.; Wei, T.; Yang, H.-Z.; Ma, Y.-Q.; Xi, Z. Discovery of a Broad-Spectrum Fluorogenic Agonist for Strigolactone Receptors through a Computational Approach. J. Agric. Food Chem. 2021, 69, 10486–10495. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.; Hu, X.; Zhu, Y.; Zou, X.; Liu, B.; Hu, F.; Yang, H. Synthesis and herbicidal activities of novel 4-(3-trifluoromethylphenyl)-2H-pyridazin-3-one derivatives. Sci. China Ser. B Chem. 2010, 53, 157–166. [Google Scholar] [CrossRef]
- Fraser, P.D.; Linden, H.; Sandmann, G. Purification and reactivation of recombinant Synechococcus phytoene desaturase from an overexpressing strain of Escherichia coli. Biochem. J. 1993, 291, 687–692. [Google Scholar] [CrossRef] [Green Version]
- Pang, Z.; Zhang, X.; Ma, F.; Liu, J.; Zhang, H.; Wang, J.; Wen, X.; Xi, Z. Comparative studies of potential binding pocket resi-dues reveal the molecular basis of ShHTL receptors in the perception of GR24 in Striga. J. Agric. Food Chem. 2020, 68, 12729–12737. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.-W.; Zhang, R.-B.; Ismail, I.; Xue, Z.-Y.; Liang, L.; Yu, S.-Y.; Wen, X.; Xi, Z. Design, Herbicidal Activity, and QSAR Analysis of Cycloalka[d]quinazoline-2,4-dione–Benzoxazinones as Protoporphyrinogen IX Oxidase Inhibitors. J. Agric. Food Chem. 2019, 67, 9254–9264. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.-W.; Li, Q.; Wen, K.; Ismail, I.; Liu, D.-D.; Niu, C.-W.; Wen, X.; Yang, G.-F.; Xi, Z. Synthesis and Herbicidal Activity of Pyrido[2,3-d]pyrimidine-2,4-dione–Benzoxazinone Hybrids as Protoporphyrinogen Oxidase Inhibitors. J. Agric. Food Chem. 2017, 65, 5278–5286. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.-W.; Zhang, R.-B.; Yu, S.-Y.; Liang, L.; Ismail, I.; Li, Y.-H.; Xu, H.; Wen, X.; Xi, Z. Discovery of Novel N-Isoxazolinylphenyltriazinones as Promising Protoporphyrinogen IX Oxidase Inhibitors. J. Agric. Food Chem. 2019, 67, 12382–12392. [Google Scholar] [CrossRef] [PubMed]
- Li, K.-J.; Qu, R.-Y.; Liu, Y.-C.; Yang, J.-F.; Devendar, P.; Chen, Q.; Niu, C.-W.; Xi, Z.; Yang, G.-F. Design, Synthesis, and Herbicidal Activity of Pyrimidine–Biphenyl Hybrids as Novel Acetohydroxyacid Synthase Inhibitors. J. Agric. Food Chem. 2018, 66, 3773–3782. [Google Scholar] [CrossRef]
- Case, D.A.; Babin, V.; Berryman, J.T.; Betz, R.M.; Cai, Q.; Cerutti, D.S.; Cheatham, T.E.; Darden, T.; Duke, R.E.; Gohlke, H. AMBER 14; University of California: San Francisco, CA, USA, 2014. [Google Scholar]
- Yang, J.-F.; Yin, C.-Y.; Wang, D.; Jia, C.-Y.; Hao, G.-F.; Yang, G.-F. Molecular Determinants Elucidate the Selectivity in Abscisic Acid Receptor and HAB1 Protein Interactions. Front. Chem. 2020, 8, 425. [Google Scholar] [CrossRef] [PubMed]
- Liang, L.; Yu, S.; Li, Q.; Wang, X.; Wang, D.; Xi, Z. Design, synthesis, and molecular simulation studies of N-phenyltetrahydroquinazolinones as protoporphyrinogen IX oxidase inhibitors. Bioorg. Med. Chem. 2021, 39, 116165. [Google Scholar] [CrossRef]
- Zhang, R.B.; Yu, S.Y.; Liang, L.; Ismail, I.; Wang, D.W.; Li, Y.H.; Xu, H.; Wen, X.; Xi, Z. Design, synthesis, and molecular mech-anism studies of N-phenylisoxazoline-thiadiazolo[3,4-a]pyridazine hybrids as protoporphyrinogen IX oxidase inhibitors. J. Agric. Food Chem. 2020, 68, 13672–13684. [Google Scholar] [CrossRef]
- Wang, D.W.; Liang, L.; Xue, Z.Y.; Yu, S.Y.; Zhang, R.B.; Wang, X.; Xu, H.; Wen, X.; Xi, Z. Discovery of N-phenylaminomethylthioacetylpyrimidine-2,4-diones as protoporphyrinogen IX oxidase inhibitors through a reaction in-termediate derivation approach. J. Agric. Food Chem. 2021, 69, 4081–4092. [Google Scholar] [CrossRef] [PubMed]




| Compd | R1 | R2 | Dosage g ai/ha | ECHCG a | DIGSA | SETFA | ABUJU | AMARE | ECLPR | clogP |
|---|---|---|---|---|---|---|---|---|---|---|
| 2a | -CH3 | 4-CF3 | 750 | 8 b | 9 | 8 | 10 | 9 | 8 | 5.16 |
| 600 | 7 | 9 | 7 | 10 | 9 | 8 | ||||
| 300 | 6 | 8 | 6 | 6 | 8 | 7 | ||||
| 150 | 2 | 6 | 2 | 1 | 3 | 3 | ||||
| 2b | -CH3 | 3-F | 750 | 2 | 4 | 4 | 7 | 7 | 2 | 5.32 |
| 2c | -CH3 | 3,4-diF | 750 | 3 | 7 | 5 | 10 | 9 | 5 | 5.44 |
| 2d | -CH3 | 3-F,4-Cl | 750 | 7 | 8 | 8 | 10 | 9 | 7 | 6.08 |
| 600 | 7 | 8 | 7 | 5 | 7 | 3 | ||||
| 2e | -CH3 | 3-F,4-Br | 750 | 5 | 5 | 5 | 10 | 8 | 7 | 6.23 |
| 2f | -CH3 | 3-F,4-CN | 750 | 6 | 6 | 6 | 10 | 8 | 6 | 5.00 |
| 2g | -CH3 | 2-F,4-Cl | 750 | 5 | 5 | 5 | 10 | 8 | 7 | 5.85 |
| 2h | -CH3 | 3,4,5-triF | 750 | 7 | 8 | 8 | 10 | 9 | 7 | 5.53 |
| 600 | 7 | 8 | 7 | 5 | 7 | 3 | ||||
| 3a | -CH2CH3 | 4-CF3 | 750 | 3 | 8 | 5 | 7 | 9 | 5 | 6.70 |
| 3b | -CH2CH3 | 3-F | 750 | 3 | 7 | 8 | 3 | 7 | 3 | 5.85 |
| 3c | -CH2CH3 | 3,4-diF | 750 | 1 | 8 | 8 | 1 | 10 | 1 | 5.97 |
| 600 | 1 | 8 | 7 | 1 | 5 | 1 | ||||
| 3d | -CH2CH3 | 3-F,4-Cl | 750 | 5 | 8 | 7 | 10 | 8 | 5 | 6.61 |
| 600 | 1 | 7 | 1 | 6 | 8 | 1 | ||||
| 3e | -CH2CH3 | 3-F,4-Br | 750 | 1 | 3 | 3 | 1 | 7 | 2 | 6.76 |
| 3f | -CH2CH3 | 3-F,4-CN | 750 | 1 | 2 | 4 | 5 | 7 | 1 | 5.52 |
| 3g | -CH2CH3 | 2-F,4-Cl | 750 | 3 | 3 | 3 | 5 | 6 | 5 | 5.84 |
| 3h | -CH2CH3 | 3,4,5-triF | 750 | 8 | 9 | 9 | 10 | 9 | 8 | 6.06 |
| 600 | 8 | 8 | 7 | 10 | 9 | 6 | ||||
| 300 | 5 | 8 | 6 | 9 | 8 | 1 | ||||
| 3i | -CH2CH3 | 4-CN | 750 | 5 | 5 | 5 | 8 | 6 | 2 | 5.33 |
| 4a |  | 4-CF3 | 750 | 7 | 6 | 7 | 10 | 10 | 7 | 6.66 |
| 4b |  | 3-F | 750 | 1 | 1 | 1 | 1 | 1 | 1 | 5.81 |
| 4c |  | 3,4-diF | 750 | 5 | 8 | 7 | 10 | 6 | 1 | 5.94 |
| 4d |  | 3-F,4-Cl | 750 | 8 | 10 | 8 | 8 | 10 | 7 | 6.58 |
| 600 | 3 | 9 | 4 | 1 | 7 | 1 | ||||
| 4e |  | 3-F,4-Br | 750 | 1 | 1 | 1 | 1 | 1 | 1 | 6.73 |
| 4f |  | 3-F,4-CN | 750 | 1 | 1 | 1 | 1 | 1 | 1 | 5.48 |
| 4g |  | 2-F,4-Cl | 750 | 1 | 1 | 1 | 1 | 1 | 1 | 6.35 |
| 4h |  | 3,4,5-triF | 750 | 7 | 5 | 4 | 10 | 7 | 3 | 6.03 |
| 4i |  | 4-CN | 750 | 2 | 4 | 3 | 3 | 7 | 10 | 5.29 |
| 5a | -CH(CH3)2 | 4-CF3 | 750 | 1 | 1 | 1 | 1 | 1 | 1 | 6.09 |
| 5b | -CH(CH3)2 | 3-F | 750 | 1 | 1 | 1 | 1 | 1 | 1 | 5.35 |
| 5c | -CH(CH3)2 | 3,4-diF | 750 | 1 | 1 | 1 | 1 | 1 | 1 | 5.42 |
| 5d | -CH(CH3)2 | 3-F,4-Cl | 750 | 1 | 1 | 1 | 1 | 1 | 1 | 6.06 |
| 5e | -CH(CH3)2 | 3-F,4-Br | 750 | 1 | 1 | 1 | 1 | 1 | 1 | 6.21 |
| 5f | -CH(CH3)2 | 3-F,4-CN | 750 | 1 | 1 | 1 | 1 | 1 | 1 | 4.78 |
| 5g | -CH(CH3)2 | 2-F,4-Cl | 750 | 1 | 1 | 1 | 1 | 1 | 1 | 5.86 |
| 5h | -CH(CH3)2 | 3,4,5-triF | 750 | 1 | 1 | 1 | 1 | 1 | 1 | 5.49 |
| 5i | -CH(CH3)2 | 2-CN | 750 | 1 | 1 | 1 | 1 | 1 | 1 | 6.78 |
| 1 | -CH3 | 4-CN | 750 | 3 | 7 | 5 | 6 | 7 | 5 | 4.80 |
| diflufenican | 750 | 6 | 6 | 6 | 10 | 8 | 6 | 4.23 | ||
| 600 | 2 | 7 | 6 | 5 | 9 | 1 | ||||
| 300 | 1 | 6 | 5 | 2 | 9 | 1 | ||||
| 150 | 1 | 5 | 2 | 1 | 9 | 1 | ||||
| Compd | Dosage g ai/ha | Maize | Rice | Wheat | Rape | Soybean | Cotton | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Post | Pre | Post | Pre | Post | Pre | Post | Pre | Post | Pre | Post | Pre | ||
| 2a | 300 | 40 | 0 | 50 | 0 | 0 | 0 | 40 | 0 | 70 | 0 | 60 | 0 |
| diflufenican | 300 | 5 | 0 | 30 | 0 | 40 | 0 | 40 | 0 | 50 | 0 | 50 | 0 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, L.; Wang, D.; Ma, D.; Zhang, D.; Zhou, N.; Wang, J.; Xu, H.; Xi, Z. In Silico Structure-Guided Optimization and Molecular Simulation Studies of 3-Phenoxy-4-(3-trifluoromethylphenyl)pyridazines as Potent Phytoene Desaturase Inhibitors. Molecules 2021, 26, 6979. https://doi.org/10.3390/molecules26226979
Yang L, Wang D, Ma D, Zhang D, Zhou N, Wang J, Xu H, Xi Z. In Silico Structure-Guided Optimization and Molecular Simulation Studies of 3-Phenoxy-4-(3-trifluoromethylphenyl)pyridazines as Potent Phytoene Desaturase Inhibitors. Molecules. 2021; 26(22):6979. https://doi.org/10.3390/molecules26226979
Chicago/Turabian StyleYang, Lijun, Dawei Wang, Dejun Ma, Di Zhang, Nuo Zhou, Jing Wang, Han Xu, and Zhen Xi. 2021. "In Silico Structure-Guided Optimization and Molecular Simulation Studies of 3-Phenoxy-4-(3-trifluoromethylphenyl)pyridazines as Potent Phytoene Desaturase Inhibitors" Molecules 26, no. 22: 6979. https://doi.org/10.3390/molecules26226979
APA StyleYang, L., Wang, D., Ma, D., Zhang, D., Zhou, N., Wang, J., Xu, H., & Xi, Z. (2021). In Silico Structure-Guided Optimization and Molecular Simulation Studies of 3-Phenoxy-4-(3-trifluoromethylphenyl)pyridazines as Potent Phytoene Desaturase Inhibitors. Molecules, 26(22), 6979. https://doi.org/10.3390/molecules26226979






