Accumulation Pattern of Flavonoids during Fruit Development of Lonicera maackii Determined by Metabolomics
Abstract
:1. Introduction
2. Results and Discussion
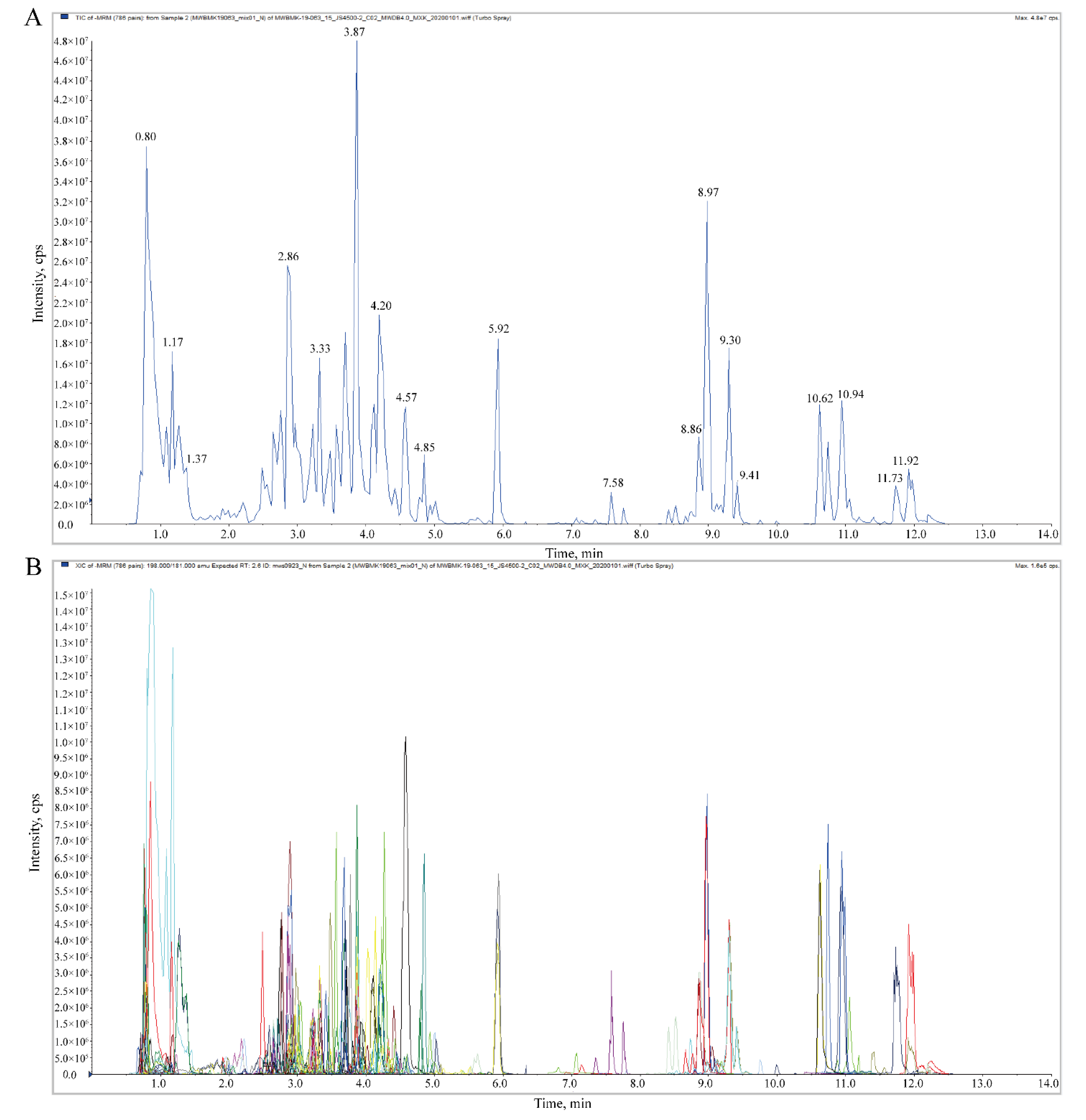
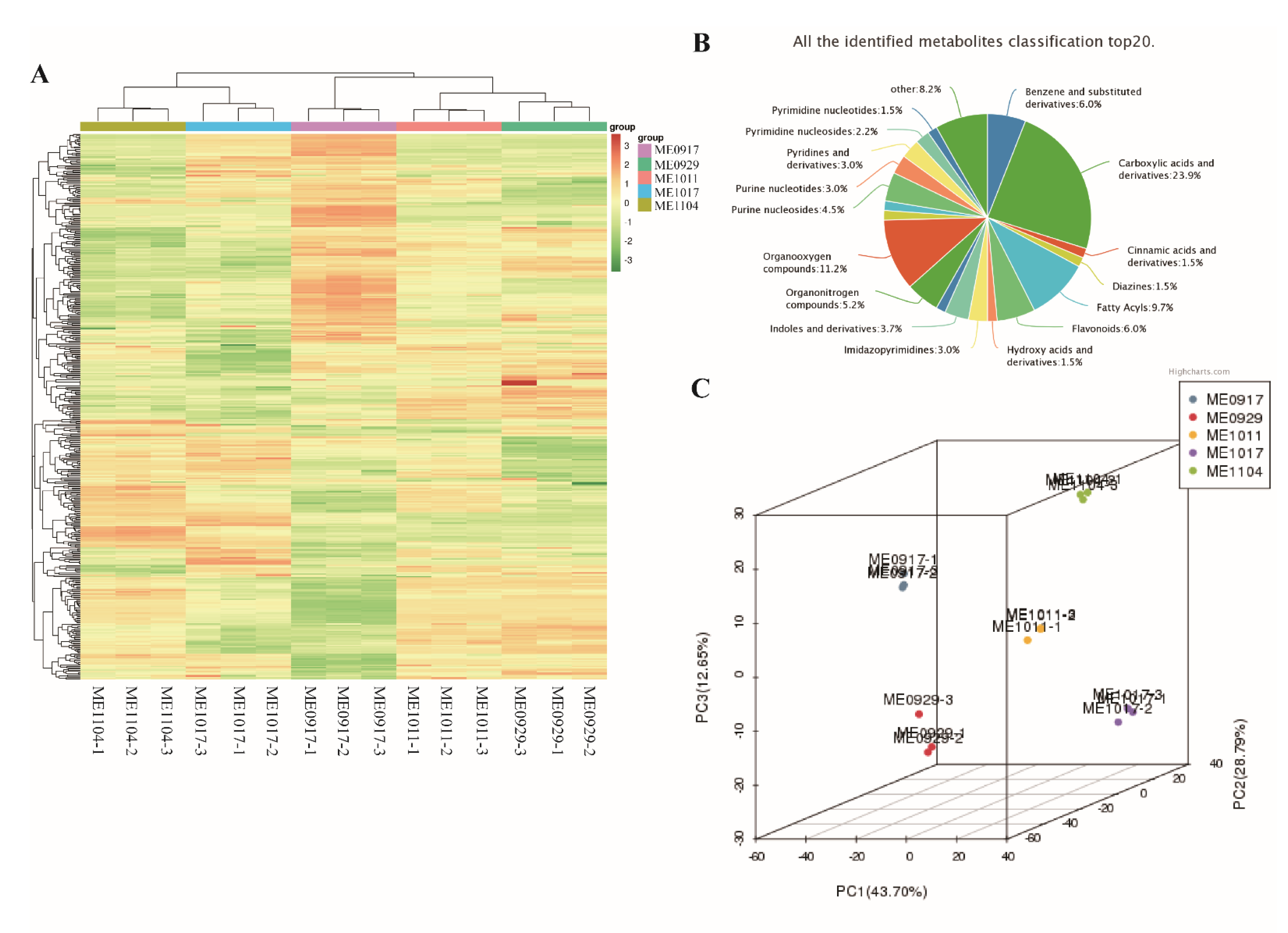
2.1. Overview of Metabolomics Analysis during L. maackii Fruit Development
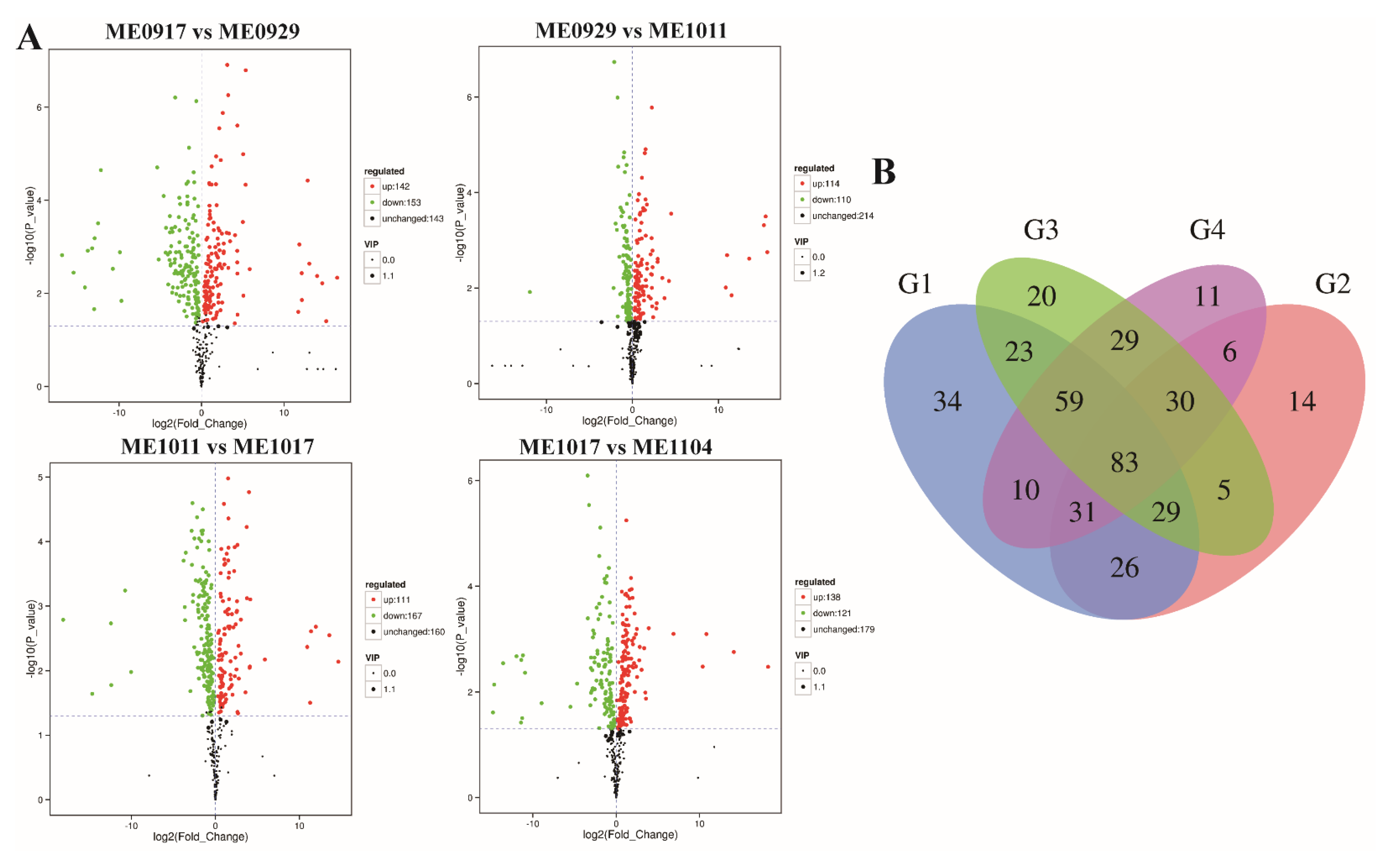
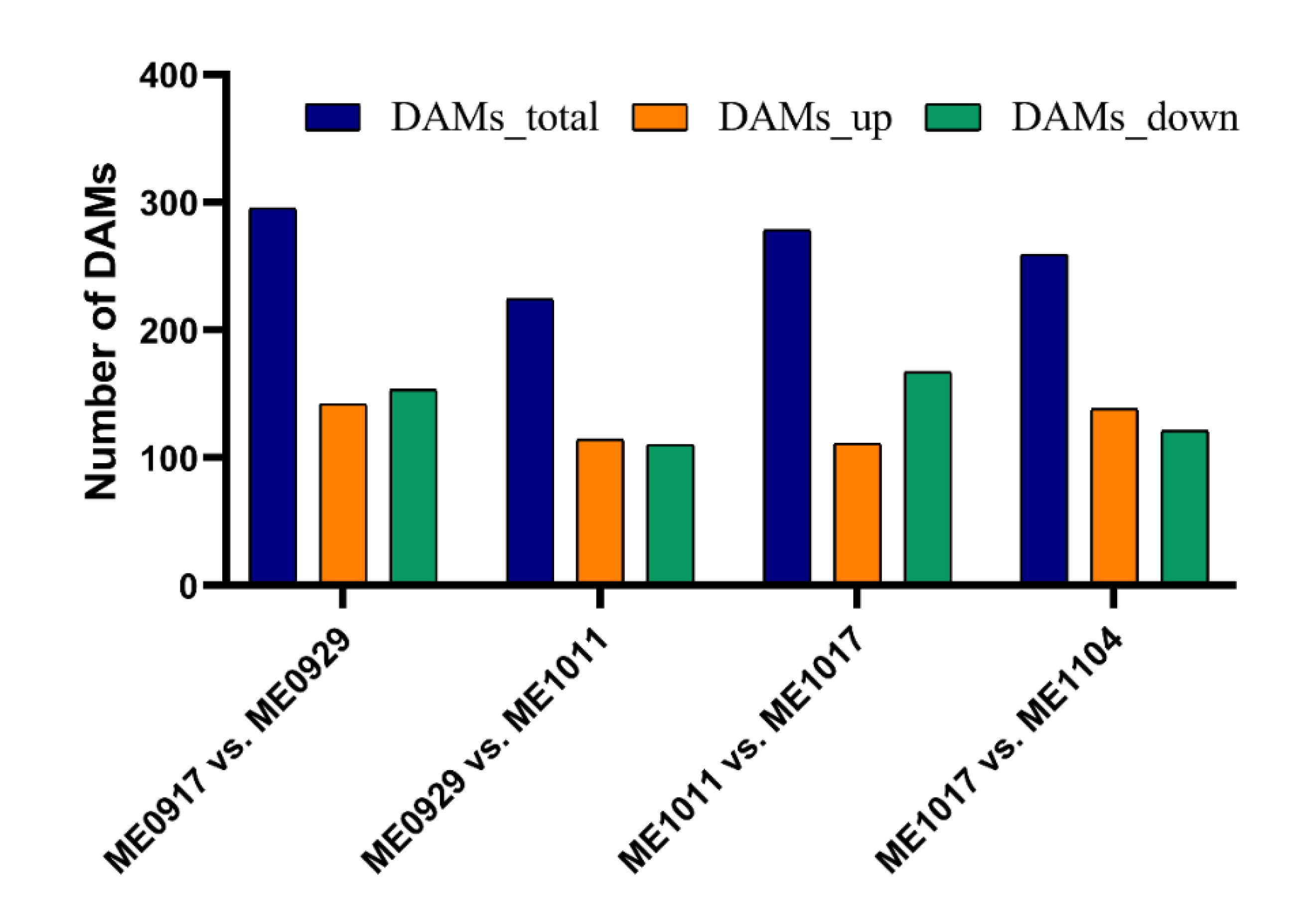
2.2. Analysis of Differential Metabolites during Fruit Development
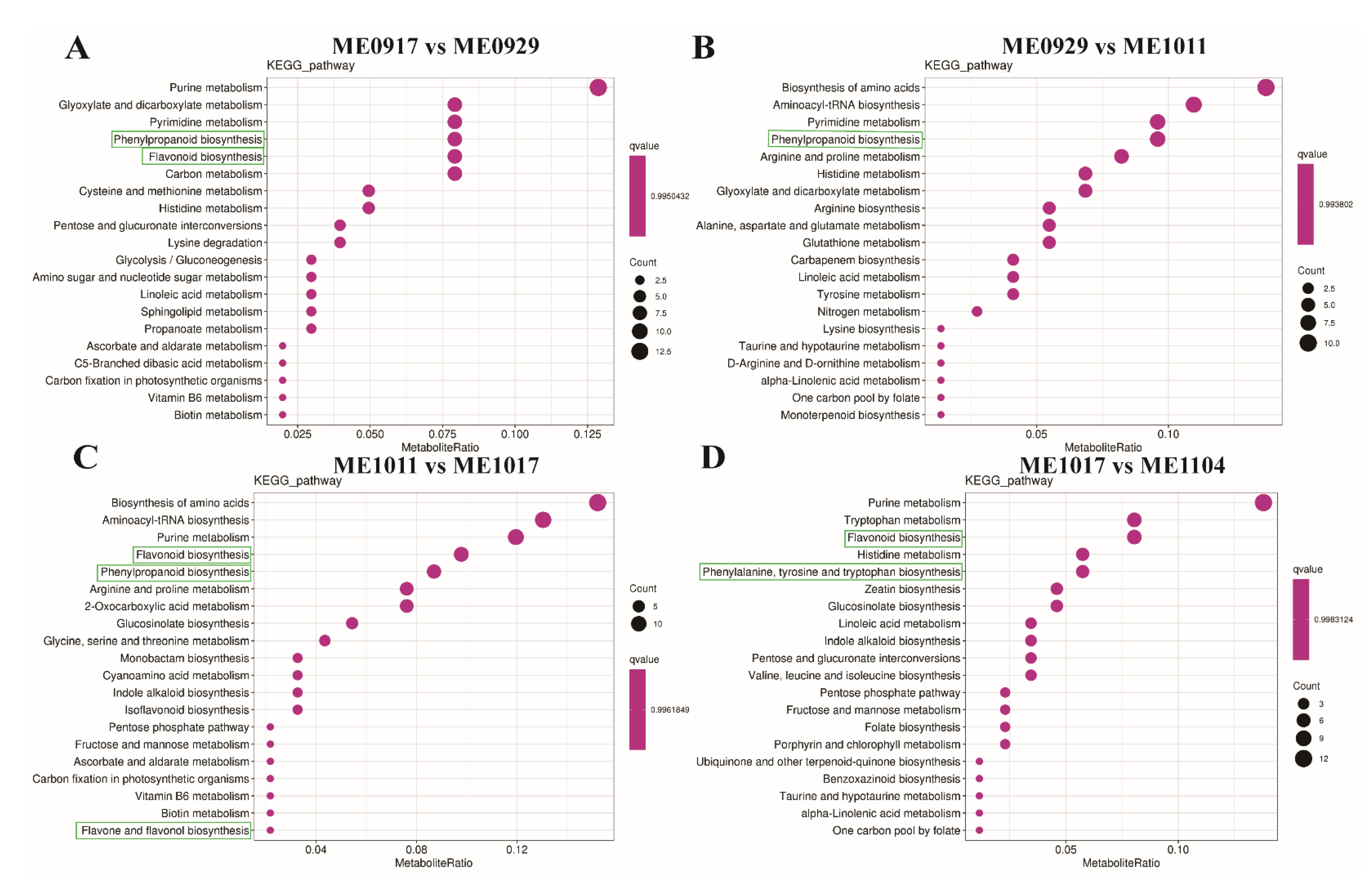
2.3. KEGG Annotation and Enrichment Analysis of Differential Metabolites
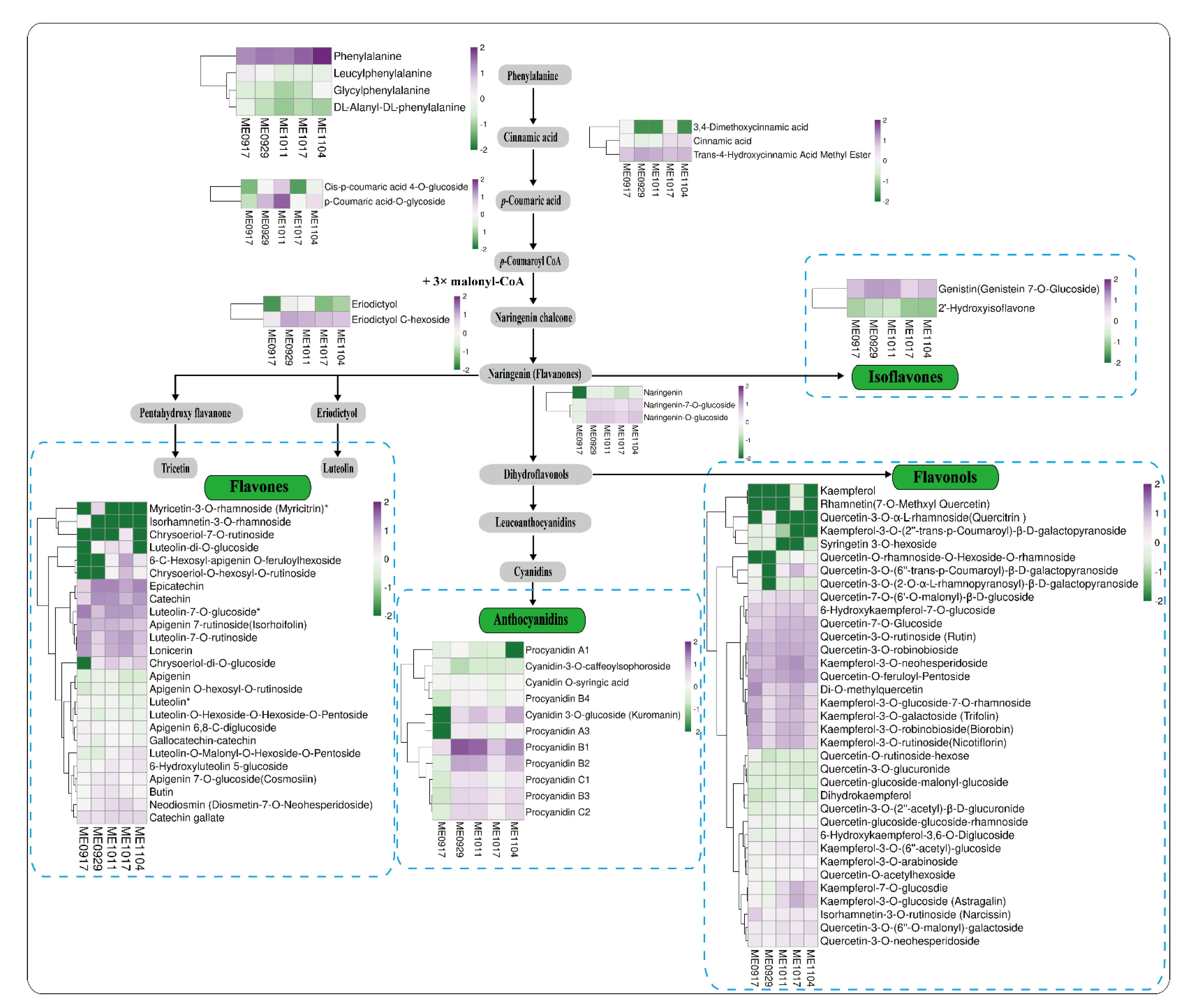
2.4. Accumulation Profiles of Flavonoids during Fruit Development
3. Materials and Methods
3.1. Plant Materials
3.2. Sample Extraction and UPLC–MS/MS System-Based Metabolomics Analysis
3.2.1. Sample Preparation and Extraction
3.2.2. Acquisition Conditions of Chromatography-Mass Spectrometry
3.3. Metabolomics Data Analysis
3.3.1. LC-MS Processing Data
3.3.2. Principal Component Analysis
3.3.3. Clustering Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Sample Availability
References
- Luken, J.O.; Tholemeier, T.C.; Kuddes, L.M.; Kunkel, B. Performance, plasticity, and acclimation of the nonindigenous shrub Lonicera maackii (Caprifoliaceae) in contrasting light environments. Can. J. Bot. 1995, 73, 1953–1961. [Google Scholar] [CrossRef]
- Cipollini, D.; Stevenson, R.; Enright, S.; Eyles, A.; Bonello, P. Phenolic metabolites in leaves of the invasive shrub, Lonicera maackii, and their potential phytotoxic and anti-herbivore effects. J. Chem. Ecol. 2008, 34, 144–152. [Google Scholar] [CrossRef] [PubMed]
- Yong, J.; Lu, C.; Huang, S. Chemical constituents of Lonicera maackii. Chem. Nat. Compd. 2014, 50, 945–947. [Google Scholar] [CrossRef]
- Deardorff, J.; Gorchov, D. Beavers cut, but do not prefer, an invasive shrub, Amur honeysuckle (Lonicera maackii). Biol. Invasions 2021, 23. [Google Scholar] [CrossRef]
- Mandli, J.T.; Lee, X.; Bron, G.M.; Paskewitz, S.M. Integrated tick management in south central wisconsin: Impact of invasive vegetation removal and host-targeted acaricides on the density of questing Ixodes scapularis (acari: Ixodidae) nymphs. J. Med. Entomol. 2021, tjab131. [Google Scholar] [CrossRef]
- Jia, G.; Wang, H.; Yu, P.; Li, P. The complete chloroplast genome of the Lonicera maackii (Caprifoliaceae), an ornamental plant. Mitochondrial DNA B 2020, 5, 560–561. [Google Scholar] [CrossRef] [Green Version]
- Todd, F.H.; John, L.V. Landscape structure and spread of the exotic shrub Lonicera maackii (Amur honeysuckle) in southwestern Ohio forests. Am. Midl. Nat. 1998, 139, 383–390. [Google Scholar]
- Wang, Y. Extraction of polysaccharides from Lonicera maackii andits anti-oxidation. Guizhou Agric. Sci. 2012, 040, 92–93. [Google Scholar]
- Song, W.; Zhang, X. Quercetin content of Lonicera maackii before and after frost. Guangdong Chem. Ind. 2021, 166–167, 171. [Google Scholar]
- Ovesna, Z.; Vachalkova, A.; Horvathova, K.; Neoplasma, D.T.J. Pentacyclic triterpenoic acids: New chemoprotective compounds. Minireview. Neoplasma 2004, 51, 327–333. [Google Scholar]
- Gu, Z.; Men, S.; Zhu, J.; Hao, Q.; Tong, N.; Liu, Z.A.; Zhang, H.; Shu, Q.; Wang, L. Chalcone synthase is ubiquitinated and degraded via interactions with a RING-H2 protein in petals of Paeonia ‘He Xie’. J. Exp. Bot. 2019, 70, 4749–4762. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Yang, P.; Yang, Q.; Gong, X.; Ma, H.; Dang, K.; Chen, G.; Gao, X.; Feng, B. Analysis of flavonoid metabolites in buckwheat leaves using UPLC-ESI-MS/MS. Molecules 2019, 24, 1310. [Google Scholar] [CrossRef] [Green Version]
- Tsimogiannis, D.; Samiotaki, M.; Panayotou, G.; Oreopoulou, V. Characterization of flavonoid subgroups and hydroxy substitution by HPLC-MS/MS. Molecules 2007, 12, 593–606. [Google Scholar] [CrossRef] [PubMed]
- Sen, S.; Chakraborty, R. The role of antioxidants in human health. ACS Sym. Ser. 2011, 1083, 1–37. [Google Scholar]
- Gligor, O.; Mocan, A.; Moldovan, C.; Locatelli, M.; Crișan, G.; Ferreira, I.C.F.R. Enzyme-assisted extractions of polyphenols—A comprehensive review. Trends Food Sci. Tech. 2019, 88, 302–315. [Google Scholar] [CrossRef]
- Hou, D.-X. Potential mechanisms of cancer chemoprevention by anthocyanins. Curr. Mol. Med. 2003, 3, 149–159. [Google Scholar] [CrossRef]
- Wang, Z.; Yu, Q.; Wanxia, S.; El Mohtar, C.; Zhao, X.; Gmitter, F. Functional study of CHS gene family members in citrus revealed a novel CHS gene affecting the production of flavonoids. BMC Plant Biol. 2018, 18, 189. [Google Scholar] [CrossRef]
- Batra, P.; Sharma, A. Anti-cancer potential of flavonoids: Recent trends and future perspectives. 3 Biotech 2013, 3, 439–459. [Google Scholar] [CrossRef] [Green Version]
- Yang, L.; Zheng, Z.-S.; Cheng, F.; Ruan, X.; Jiang, D.-A.; Cunde, P.; Wang, Q. Seasonal dynamics of metabolites in needles of Taxus wallichiana var. mairei. Molecules 2016, 21, 1403. [Google Scholar] [CrossRef] [Green Version]
- Yu, X.; Yu, W.; Zhang, X.; Wang, Y.; Wang, S.; Zhai, H. Simultaneous determination of flavonoids and anthraquinones in honey by using SPE-CE-LIF. Anal. Biochem. 2021, 631, 114373. [Google Scholar] [CrossRef]
- Bovy, A.; Schijlen, E.; Hall, R.D. Metabolic engineering of flavonoids in tomato (Solanum lycopersicum): The potential for metabolomics. Metabolomics 2007, 3, 399. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Treutter, D. Significance of flavonoids in plant resistance: A review. Environ. Chem. Lett. 2006, 4, 147–157. [Google Scholar] [CrossRef]
- Muhlemann, J.; Younts, T.; Muday, G. Flavonols control pollen tube growth and integrity by regulating ROS homeostasis during high-temperature stress. Proc. Natl. Acad. Sci. USA 2018, 115, E11188–E11197. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Winkel-Shirley, B. Flavonoid biosynthesis. A colorful model for genetics, biochemistry, cell biology, and biotechnology. Plant Physiol. 2001, 126, 485–493. [Google Scholar] [CrossRef] [Green Version]
- Qu, T.; Gao, Y.; Li, A.; Li, Z.; Qin, X. Systems biology analysis of the effect and mechanism of total flavonoids of Astragali Radix against cyclophosphamide-induced leucopenia in mice. J. Pharmaceut. Biomed. 2021, 205, 114357. [Google Scholar] [CrossRef] [PubMed]
- Feng, X.; Yu, Q.; Li, B.; Kan, J. Comparative analysis of carotenoids and metabolite characteristics in discolored red pepper and normal red pepper based on non-targeted metabolomics. LWT 2021, 153, 112398. [Google Scholar] [CrossRef]
- Hu, S.-S.; Cao, W.; Dai, H.-B.; Da, J.-H.; Ye, L.-H.; Cao, J.; Li, X.-Y. Ionic-liquid-micelle-functionalized mesoporous Fe3O4 microspheres for ultraperformance liquid chromatography determination of anthraquinones in dietary supplements. J. Agr. Food Chem. 2014, 62, 8822–8829. [Google Scholar] [CrossRef]
- Wang, G.-Y.; Qi, H.-Y.; Shi, Y.-P. Ultrasonic cell grinder extraction of anthraquinones from Radix et Rhizoma Rhei and determination by ultra-performance liquid chromatography. J. Sep. Sci. 2015, 33, 1730–1738. [Google Scholar] [CrossRef]
- Valasi, L.; Kokotou, M.G.; Pappas, C.S. GC-MS, FTIR and Raman spectroscopic analysis of fatty acids of Pistacia vera (Greek variety “Aegina”) oils from two consecutive harvest periods and chemometric differentiation of oils quality. Food Res. Int. 2021, 148, 110590. [Google Scholar] [CrossRef]
- Locatelli, M.; Tammaro, F.; Menghini, L.; Carlucci, G.; Epifano, F.; Genovese, S. Anthraquinone profile and chemical fingerprint of Rhamnus saxatilis L. from Italy. Phytochem. Lett. 2009, 2, 223–226. [Google Scholar] [CrossRef]
- Załuski, D.; Olech, M.; Kuźniewski, R.; Verpoorte, R.; Nowak, R.; Smolarz, H.D. LC-ESI-MS/MS profiling of phenolics from Eleutherococcus spp. inflorescences, structure-activity relationship as antioxidants, inhibitors of hyaluronidase and acetylcholinesterase. Saudi Pharm. J. 2017, 25, 734–743. [Google Scholar] [CrossRef] [Green Version]
- Johnson, R.; Lunte, C. A capillary electrophoresis electrospray ionization-mass spectrometry method using a borate background electrolyte for the fingerprinting analysis of flavonoids in Ginkgo biloba herbal supplements. Anal. Methods 2016, 8, 3325. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ji, Y.; Li, Y.; Ren, B.; Liu, X.; Li, Y.; Soar, J. Nitrogen-doped graphene-ionic liquid-glassy carbon microsphere paste electrode for ultra-sensitive determination of quercetin. Microchem. J. 2020, 155, 104689. [Google Scholar] [CrossRef]
- Qu, T.; Wang, E.; Li, A.; Du, G.; Li, Z.; Qin, X.-M. NMR based metabolomic approach revealed cyclophosphamide-induced systematic alterations in a rat model. RSC Adv. 2016, 6, 111020–111030. [Google Scholar] [CrossRef]
- Ouakhssase, A.; Fatini, N.; Addi, E. A facile extraction method followed by UPLC-MS/MS for the analysis of aflatoxins and ochratoxin A in raw coffee beans. Food Addit. Contam. A 2021, 38, 1–10. [Google Scholar] [CrossRef]
- Zhao, X.; Zhang, S.; Liu, D.; Yang, M.; Wei, J. Analysis of flavonoids in Dalbergia odorifera by ultra-performance liquid chromatography with tandem mass spectrometry. Molecules 2020, 25, 389. [Google Scholar] [CrossRef] [Green Version]
- Rivera-Mondragón, A.; Tuenter, E.; Ortiz, O.; Sakavitsi, M.E.; Nikou, T.; Halabalaki, M.; Caballero-George, C.; Apers, S.; Pieters, L.; Foubert, K. UPLC-MS/MS-based molecular networking and NMR structural determination for the untargeted phytochemical characterization of the fruit of Crescentia cujete (Bignoniaceae). Phytochemistry 2020, 177, 112438. [Google Scholar] [CrossRef]
- Peng, F.; Yin, H.; Du, B.; Niu, K.; Ren, X.; Yang, Y. Anti-fatigue activity of purified flavonoids prepared from chestnut (Castanea mollissima) flower. J. Funct. Foods 2021, 79, 104365. [Google Scholar] [CrossRef]
- Yang, Y.; Jin, X.; Jiao, X.; Li, J.; Liang, L.; Ma, Y.; Liu, R.; Li, Z. Advances in pharmacological actions and mechanisms of flavonoids from traditional chinese medicine in treating chronic obstructive pulmonary disease. Evid. Based Complement. Alternat. Med. 2020, 2020, 8871105. [Google Scholar] [CrossRef]
- Ghidoli, M.; Colombo, F.; Sangiorgio, S.; Landoni, M.; Giupponi, L.; Nielsen, E.; Pilu, R. Food containing bioactive flavonoids and other phenolic or sulfur phytochemicals with antiviral effect: Can we design a promising diet against COVID-19? Front. Nutr. 2021, 8, 661331. [Google Scholar] [CrossRef]
- Dai, F.; Luo, G.; Wang, Z.; Kuang, Z.; Li, Z.; Huang, J.; Tang, C. Possible involvement of flavonoids in response of mulberry (Morus alba L.) to infection with Ralstonia solanacearum (Smith 1896) Yabuuchi et al., 1996. Eur. J. Hortic. Sci. 2019, 84, 161–170. [Google Scholar] [CrossRef]
- Zhang, L.; Zhang, Z.; Fang, S.; Liu, Y.; Shang, X. Integrative analysis of metabolome and transcriptome reveals molecular regulatory mechanism of flavonoid biosynthesis in Cyclocarya paliurus under salt stress. Ind. Crop. Prod. 2021, 170, 113823. [Google Scholar] [CrossRef]
- Jiang, C.-K.; Ma, J.-Q.; Liu, Y.-F.; Chen, J.-D.; Ni, D.-J.; Chen, L. Identification and distribution of a single nucleotide polymorphism responsible for the catechin content in tea plants. Hortic. Res. Engl. 2020, 7, 24. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kuo, M.-Y.; Liao, M.-F.; Chen, F.-L.; Li, Y.-C.; Yang, M.-L.; Lin, R.-H.; Kuan, Y.-H. Luteolin attenuates the pulmonary inflammatory response involves abilities of antioxidation and inhibition of MAPK and NFκB pathways in mice with endotoxin-induced acute lung injury. Food Chem. Toxicol. 2011, 49, 2660–2666. [Google Scholar] [CrossRef] [PubMed]
- Han, X.; Sun, S.; Sun, Y.; Song, Q.; Zhu, J.; Song, N.; Sun, T.; Xia, M.; Ding, J.; Lu, M.; et al. Small molecule-driven NLRP3 inflammation inhibition via interplay between ubiquitination and autophagy: Implications for Parkinson disease. Autophagy 2019, 15, 1–22. [Google Scholar] [CrossRef]
- BinMowyna, M.; Alfaris, N. Kaempferol suppresses acetaminophen-induced liver damage by upregulation/activation of SIRT1. Pharm. Biol. 2021, 59, 146–156. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Han, J.; Jinjin, D.; Fan, X.; Cai, Y.; Li, J.; Wang, T.; Zhou, J.; Shang, J. Metabolomics characterizes the effects and mechanisms of quercetin in nonalcoholic fatty liver disease development. Int. J. Mol. Sci. 2019, 20, 1220. [Google Scholar] [CrossRef] [Green Version]
- Pan, R.Y.; Ma, J.; Kong, X.X.; Wang, X.F.; Li, S.S.; Qi, X.L.; Yan, Y.H.; Cheng, J.; Liu, Q.; Jin, W.; et al. Sodium rutin ameliorates Alzheimer’s disease-like pathology by enhancing microglial amyloid-β clearance. Sci. Adv. 2019, 5, eaau6328. [Google Scholar] [CrossRef] [Green Version]
- Peng, X.; Liu, H.; Chen, P.; Tang, F.; Hu, Y.; Wang, F.; Pi, Z.; Zhao, M.; Chen, N.; Chen, H.; et al. A chromosome-scale genome Aassembly of paper mulberry (Broussonetia papyrifera) provides new insights into its forage and papermaking usage. Mol. Plant 2019, 12, 661–677. [Google Scholar] [CrossRef]
- Chen, W.; Gong, L.; Guo, Z.; Wang, W.; Zhang, H.; Liu, X.; Yu, S.; Xiong, L.; Luo, J. A novel integrated method for large-scale detection, identification, and quantification of widely targeted metabolites: Application in the study of rice metabolomics. Mol. Plant 2013, 6, 1769–1780. [Google Scholar] [CrossRef] [Green Version]
- Ogata, H.; Goto, S.; Sato, K.; Fujibuchi, W.; Bono, H.; Kanehisa, M. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 1999, 27, 29–34. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, Y.; Zhang, R.; Song, Y.; He, J.; Sun, J.; Bai, J.; An, Z.; Dong, L.; Zhan, Q.; Abliz, Z. RRLC-MS/MS-based metabonomics combined with in-depth analysis of metabolic correlation network: Finding potential biomarkers for breast cancer. Analyst 2009, 134, 2003–2011. [Google Scholar] [CrossRef] [PubMed]
- Fraga, C.G.; Clowers, B.H.; Moore, R.J.; Zink, E. Signature-discovery approach for sample matching of a nerve-agent precursor using liquid chromatography-mass spectrometry, XCMS, and chemometrics. Anal. Chem. 2010, 82, 4165–4173. [Google Scholar] [CrossRef]
| Metabolite Name | T1 vs. T2 | T2 vs. T3 | T3 vs. T4 | T4 vs. T5 | ||||
|---|---|---|---|---|---|---|---|---|
| Flavone | Log2FC | VIP | Log2FC | VIP | Log2FC | VIP | Log2FC | VIP |
| Isorhamnetin-3-O-rhamnoside | −12.97 | 1.14 | - | - | - | - | - | - |
| Myricetin-3-O-rhamnoside | - | - | - | - | - | - | - | - |
| Luteolin | - | - | - | - | −0.88 | 1.10 | - | - |
| Luteolin-7-O-glucoside | −3.56 | 1.12 | 2.26 | 1.25 | - | - | −1.13 | 1.14 |
| 6-Hydroxyluteolin 5-glucoside | - | - | 1.89 | 1.22 | −0.95 | 1.06 | 1.42 | 1.18 |
| Luteolin-di-O-glucoside | - | - | 2.13 | 1.14 | - | - | −14.79 | 1.15 |
| Luteolin-O-Malonyl-O-Hexoside-O-Pentoside | - | - | 3.72 | 1.20 | - | - | - | - |
| Luteolin-O-Hexoside-O-Hexoside-O-Pentoside | −1.57 | 1.09 | 1.56 | 1.22 | −0.85 | 1.06 | - | - |
| Apigenin | - | - | - | - | −2.14 | 1.17 | - | - |
| Apigenin 7-O-glucoside | 1.50 | 1.13 | - | - | −1.48 | 1.18 | 0.86 | 1.09 |
| Apigenin 7-rutinoside | −0.80 | 1.09 | - | - | - | - | - | - |
| Apigenin 6,8-C-diglucoside | - | - | - | - | - | - | −1.30 | 1.19 |
| Apigenin O-hexosyl-O-rutinoside | 2.30 | 1.07 | - | - | −1.33 | 1.17 | - | - |
| 6-C-Hexosyl-apigenin O-feruloylhexoside | - | - | 13.58 | 1.26 | 5.90 | 1.17 | −4.69 | 1.19 |
| Butin | 1.83 | 1.13 | 0.45 | 1.18 | −1.30 | 1.18 | −0.70 | 1.18 |
| Chrysoeriol-7-O-rutinoside | - | - | - | - | 14.64 | 1.17 | −14.64 | 1.19 |
| Chrysoeriol-di-O-glucoside | 14.65 | 1.13 | 2.41 | 1.17 | −1.53 | 1.04 | 1.48 | 1.16 |
| Chrysoeriol-O-hexosyl-O-rutinoside | - | - | - | - | - | - | - | - |
| Luteolin-7-O-rutinoside | −3.27 | 1.14 | 2.30 | 1.20 | 0.79 | 1.09 | −2.07 | 1.19 |
| Diosmetin-7-O-Neohesperidoside | 1.95 | 1.14 | 1.13 | 1.24 | 1.06 | 1.16 | −2.60 | 1.20 |
| Epicatechin | 4.34 | 1.14 | - | - | −1.83 | 1.18 | 1.77 | 1.20 |
| Catechin | 4.33 | 1.14 | −0.33 | 1.10 | −2.05 | 1.17 | 1.91 | 1.19 |
| Lonicerin | −3.14 | 1.14 | 2.27 | 1.23 | 0.72 | 1.14 | −2.34 | 1.20 |
| Catechin gallate | - | - | 1.28 | 1.19 | - | - | - | - |
| Gallocatechin-catechin | −2.70 | 1.14 | 2.26 | 1.26 | - | - | −1.30 | 1.16 |
| Flavonol | Log2FC | VIP | Log2FC | VIP | Log2FC | VIP | Log2FC | VIP |
| Kaempferol | - | - | - | - | 0.59 | 1.15 | −11.28 | 1.13 |
| Dihydrokaempferol | 0.76 | 1.06 | 1.68 | 1.26 | - | - | −2.92 | 1.18 |
| Kaempferol-3-O-arabinoside | - | - | - | - | 0.44 | 1.14 | −1.13 | 1.20 |
| Kaempferol-7-O-glucosdie | −1.97 | 1.12 | 4.51 | 1.26 | 2.58 | 1.18 | −1.40 | 1.20 |
| Kaempferol-3-O-glucoside | −1.66 | 1.11 | 4.24 | 1.25 | 2.63 | 1.18 | −1.34 | 1.19 |
| Kaempferol-3-O-galactoside | −3.72 | 1.14 | 2.16 | 1.25 | - | - | −1.25 | 1.17 |
| 6-Hydroxykaempferol-7-O-glucoside | −0.36 | 1.04 | - | - | - | - | - | - |
| Kaempferol-3-O-(6″-acetyl)-glucoside | 1.69 | 1.07 | - | - | - | - | 0.80 | 1.11 |
| Kaempferol-3-O-(2″-trans-p-Coumaroyl)-β-d-galactopyranoside | - | - | - | - | - | - | - | - |
| Kaempferol-3-O-robinobioside | −2.90 | 1.13 | 2.33 | 1.26 | - | - | −1.75 | 1.19 |
| Kaempferol-3-O-rutinoside | −2.81 | 1.14 | 2.23 | 1.26 | - | - | −1.80 | 1.18 |
| Kaempferol-3-O-neohesperidoside | - | - | 1.53 | 1.26 | 0.71 | 1.18 | −1.91 | 1.20 |
| Kaempferol-3-O-glucoside-7-O-rhamnoside | −3.20 | 1.14 | 2.35 | 1.25 | 0.81 | 1.12 | −2.14 | 1.18 |
| 6-Hydroxykaempferol-3,6-O-Diglucoside | - | - | 2.82 | 1.21 | - | - | 1.19 | 1.17 |
| 7-O-Methxyl Quercetin | - | - | - | - | 11.96 | 1.18 | −11.96 | 1.20 |
| Di-O-methylquercetin | −3.84 | 1.14 | 0.33 | 1.19 | 1.58 | 1.18 | −2.63 | 1.20 |
| Quercetin-3-O-α-l-rhamnoside | - | - | - | - | - | - | - | - |
| Quercetin-7-O-Glucoside | - | - | 1.01 | 1.22 | - | - | 0.16 | 1.05 |
| Quercetin-3-O-glucuronide | - | - | - | - | - | - | - | - |
| Quercetin-O-acetylhexoside | - | - | - | - | - | - | 1.42 | 1.15 |
| Quercetin-3-O-(2″-acetyl)-β-d-glucuronide | - | - | - | - | - | - | 0.81 | 1.11 |
| Quercetin-3-O-(6″-O-malonyl)-galactoside | - | - | 1.12 | 1.25 | −0.73 | 1.18 | 1.25 | 1.17 |
| Quercetin-7-O-(6′-O-malonyl)-β-d-glucoside | - | - | 1.27 | 1.16 | −1.17 | 1.17 | 1.20 | 1.19 |
| Quercetin-3-O-(6″-trans-p-Coumaroyl)-β-d-galactopyranoside | −13.27 | 1.14 | 15.28 | 1.26 | 4.16 | 1.18 | −3.40 | 1.20 |
| Quercetin-O-feruloyl-Pentoside | −0.95 | 1.14 | 0.85 | 1.22 | 0.61 | 1.14 | −0.80 | 1.19 |
| Quercetin-3-O-rutinoside (Rutin) | −0.67 | 1.01 | 0.80 | 1.09 | - | - | - | - |
| Quercetin-3-O-robinobioside | −0.72 | 1.07 | 0.68 | 1.19 | - | - | - | - |
| Quercetin-3-O-(2-O-α-l-rhamnopyranosyl)-β-d-galactopyranoside | −13.03 | 1.10 | 11.00 | 1.26 | - | - | - | - |
| Quercetin-3-O-neohesperidoside | −1.04 | 1.06 | 0.88 | 1.13 | 0.72 | 1.10 | −0.76 | 1.15 |
| Quercetin-glucoside-malonyl-glucoside | - | - | - | - | - | - | - | - |
| Quercetin-O-rhamnoside-O-Hexoside-O-rhamnoside | - | - | 11.52 | 1.23 | 2.63 | 1.08 | −2.04 | 1.08 |
| Quercetin-O-rutinoside-hexose | −3.62 | 1.14 | 0.96 | 1.17 | 0.70 | 1.12 | - | - |
| Quercetin-glucoside-glucoside-rhamnoside | −2.70 | 1.14 | 1.14 | 1.07 | - | - | - | - |
| Syringetin 3-O-hexoside | - | - | −11.92 | 1.24 | - | - | - | - |
| Isorhamnetin-3-O-rutinoside | −3.25 | 1.14 | - | - | - | - | - | - |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Qi, Z.; Zhao, R.; Xu, J.; Ge, Y.; Li, R.; Li, R. Accumulation Pattern of Flavonoids during Fruit Development of Lonicera maackii Determined by Metabolomics. Molecules 2021, 26, 6913. https://doi.org/10.3390/molecules26226913
Qi Z, Zhao R, Xu J, Ge Y, Li R, Li R. Accumulation Pattern of Flavonoids during Fruit Development of Lonicera maackii Determined by Metabolomics. Molecules. 2021; 26(22):6913. https://doi.org/10.3390/molecules26226913
Chicago/Turabian StyleQi, Zengxing, Ran Zhao, Jing Xu, Yanrui Ge, Ruofan Li, and Ruili Li. 2021. "Accumulation Pattern of Flavonoids during Fruit Development of Lonicera maackii Determined by Metabolomics" Molecules 26, no. 22: 6913. https://doi.org/10.3390/molecules26226913
APA StyleQi, Z., Zhao, R., Xu, J., Ge, Y., Li, R., & Li, R. (2021). Accumulation Pattern of Flavonoids during Fruit Development of Lonicera maackii Determined by Metabolomics. Molecules, 26(22), 6913. https://doi.org/10.3390/molecules26226913






