First Immunoassay for Measuring Isoaspartate in Human Serum Albumin
Abstract
:1. Introduction
2. Results
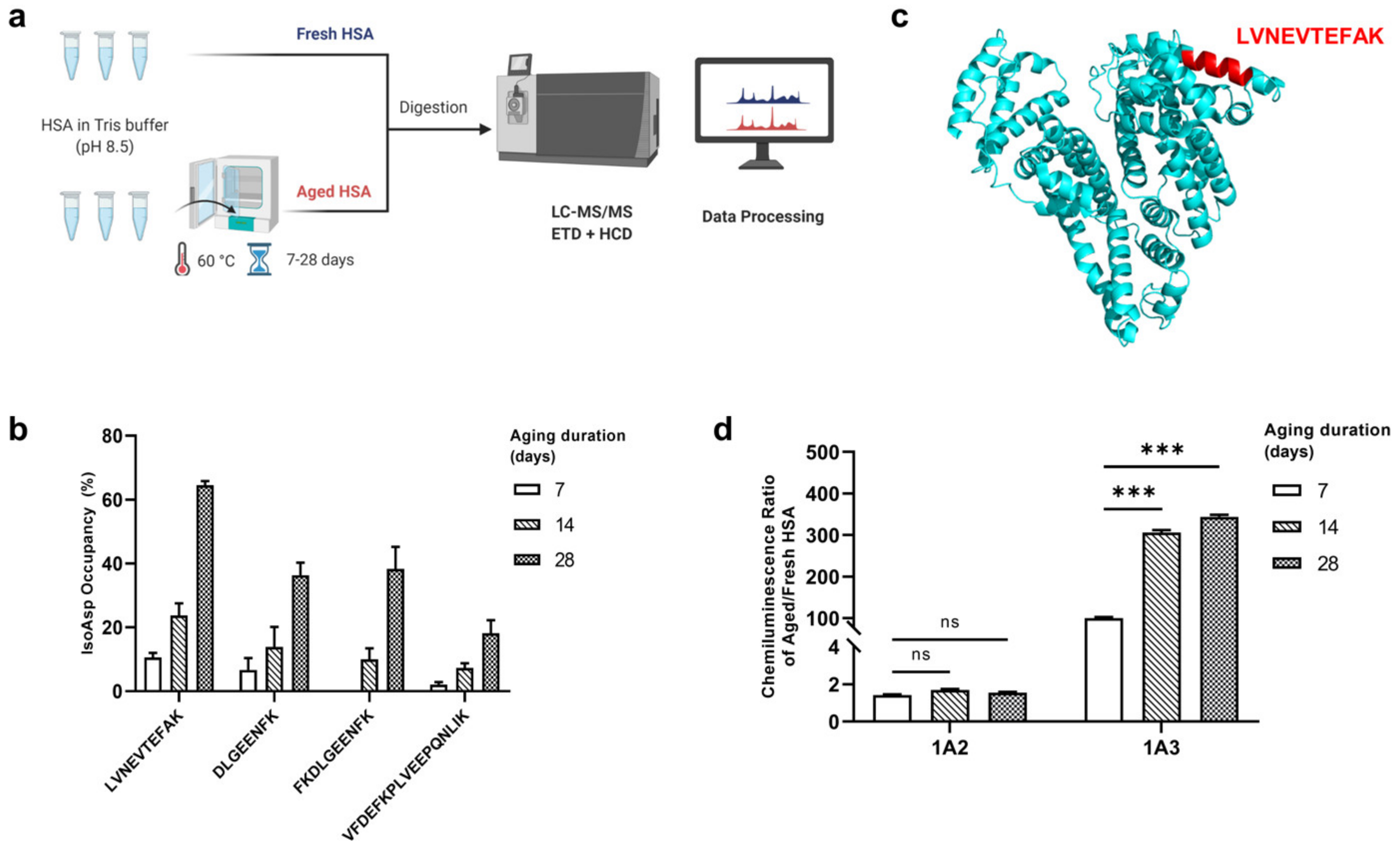
2.1. Identification of the “IsoAsp Meter” Peptide in HSA
2.2. Comparison Results of Different ELISAs
2.3. Indirect ELISA of Deamidated HSA
2.4. Sequence Analysis of mAb Variable Region
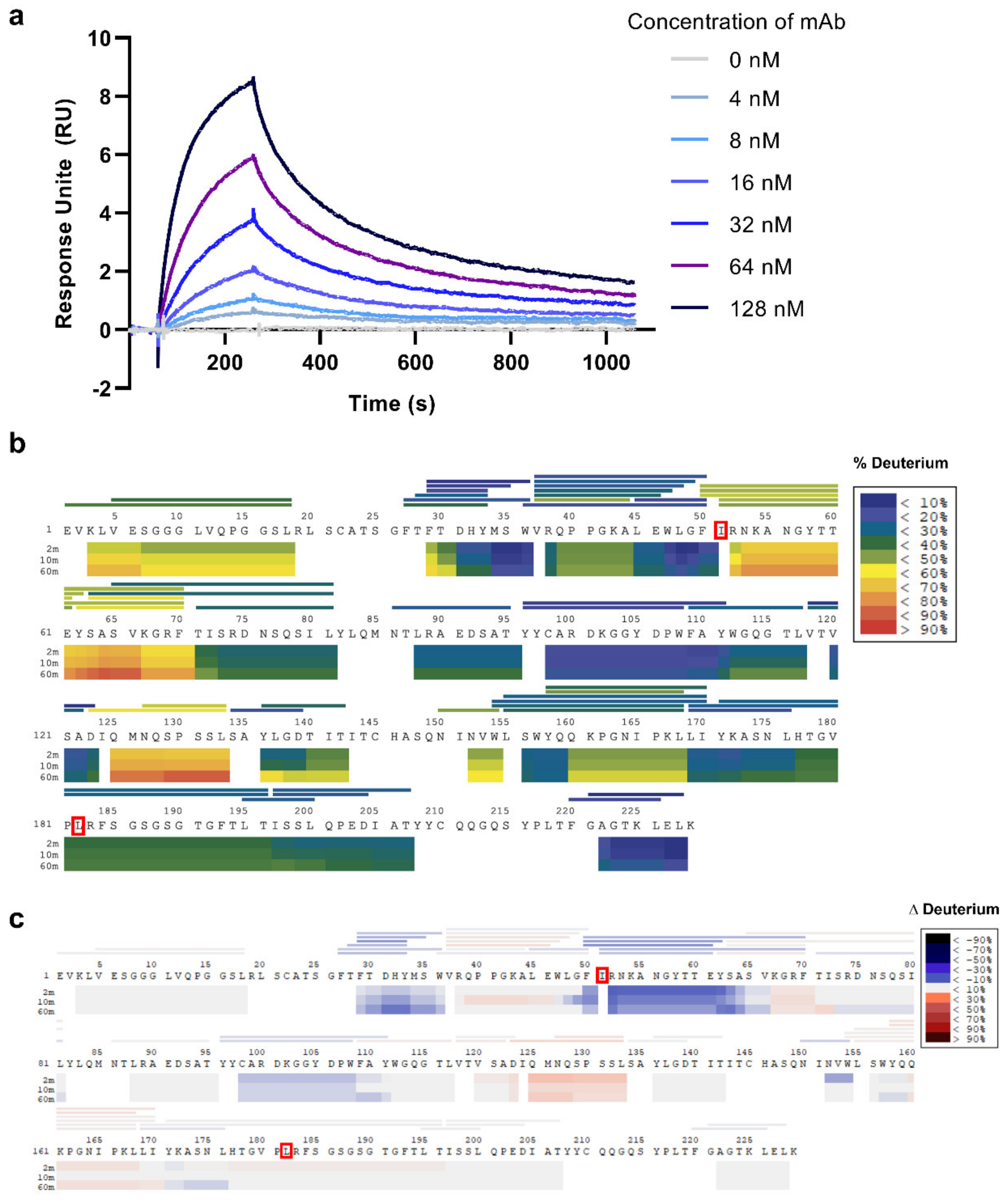
2.5. Kinetic Analyses
2.6. Paratope Mapping
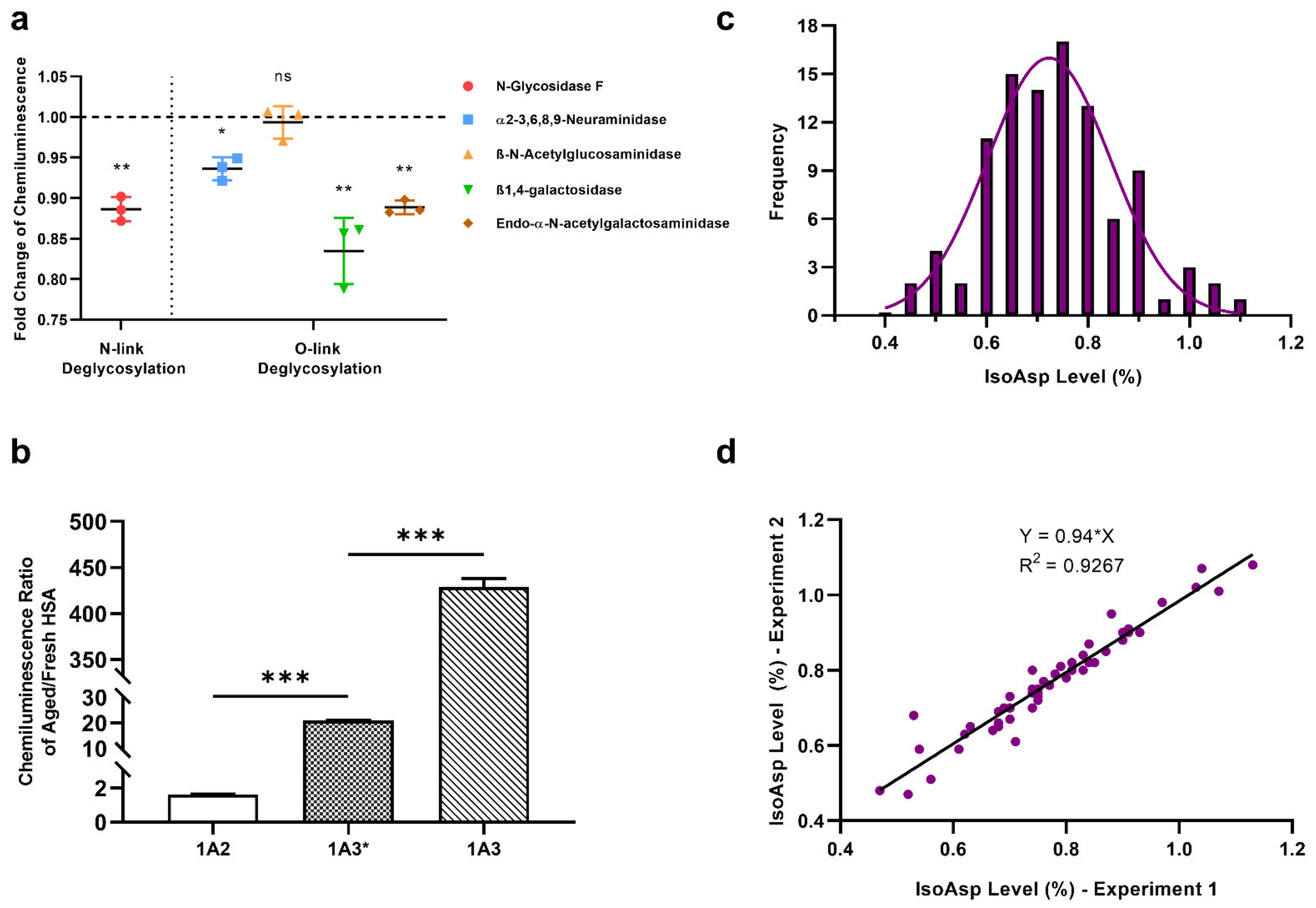
2.7. Glycosylation Effect on 1A3 Specificity
2.8. Effect of the Ig isotype
2.9. Determination of IsoAsp Levels in Healthy Plasma
3. Discussion
4. Materials and Methods
4.1. Artificial Deamidation of Proteins
4.2. Generation of mAbs against the “IsoAsp Meter” in HSA
4.3. Indirect ELISA
4.4. DNA Sequence Analysis of mAbs Variable Regions
4.5. De Novo Protein Sequencing of 1A3
4.6. Surface Plasmon Resonance (SPR)
4.7. Hydrogen-Deuterium Exchange Mass Spectrometry (HDX-MS)
4.8. Deglycosylation of 1A3 mAb
4.9. Production of 1A3* mAb
4.10. ELISA of Blood Plasma from Healthy Donors
5. Conclusions
6. Patents
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Geiger, T.; Clarke, S. Deamidation, Isomerization, and Racemization at Asparaginyl and Aspartyl Residues in Peptides—Succinimide-Linked Reactions That Contribute to Protein-Degradation. J. Biol. Chem. 1987, 262, 785–794. [Google Scholar] [CrossRef]
- Gupta, R.; Srivastava, O.P. Deamidation affects structural and functional properties of human alpha A-crystallin and its oligomerization with alpha B-crystallin. J. Biol. Chem. 2004, 279, 44258–44269. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peters, B.; Trout, B.L. Asparagine deamidation: pH-dependent mechanism from density functional theory. Biochemistry 2006, 45, 5384–5392. [Google Scholar] [PubMed]
- Robinson, N.E.; Robinson, A.B. Molecular clocks. Proc. Natl. Acad. Sci. USA 2001, 98, 944–949. [Google Scholar] [CrossRef] [Green Version]
- Goldberg, A.L. Protein degradation and protection against misfolded or damaged proteins. Nature 2003, 426, 895–899. [Google Scholar] [CrossRef]
- Bohme, L.; Bar, J.W.; Hoffmann, T.; Manhart, S.; Ludwig, H.H.; Rosche, F.; Demuth, H.U. Isoaspartate residues dramatically influence substrate recognition and turnover by proteases. Biol. Chem. 2008, 389, 1043–1053. [Google Scholar]
- Doyle, H.A.; Gee, R.J.; Mamula, M.J. Altered immunogenicity of isoaspartate containing proteins. Autoimmunity 2007, 40, 131–137. [Google Scholar] [CrossRef]
- Yang, H.; Lyutvinskiy, Y.; Soininen, H.; Zubarev, R.A. Alzheimer’s disease and mild cognitive impairment are associated with elevated levels of isoaspartyl residues in blood plasma proteins. J. Alzheimers Dis. 2011, 27, 113–118. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chatterjee, T.; Das, G.; Chatterjee, B.K.; Dhar, J.; Ghosh, S.; Chakrabarti, P. The role of isoaspartate in fibrillation and its prevention by Protein-L-isoaspartyl methyltransferase. Biochim. Biophys. Acta Gen. Subj. 2020, 1864, 129500. [Google Scholar] [CrossRef]
- Wilmarth, P.A.; Tanner, S.; Dasari, S.; Nagalla, S.R.; Riviere, M.A.; Bafna, V.; Pevzner, P.A.; David, L.L. Age-related changes in human crystallins determined from comparative analysis of post-translational modifications in young and aged lens: Does deamidation contribute to crystallin insolubility? J. Proteome Res. 2006, 5, 2554–2566. [Google Scholar]
- Hardy, J.A.; Higgins, G.A. Alzheimer’s disease: The amyloid cascade hypothesis. Science 1992, 256, 184–185. [Google Scholar] [CrossRef] [PubMed]
- Roher, A.E.; Lowenson, J.D.; Clarke, S.; Wolkow, C.; Wang, R.; Cotter, R.J.; Reardon, I.M.; Zurcherneely, H.A.; Heinrikson, R.L.; Ball, M.J.; et al. Structural Alterations in the Peptide Backbone of Beta-Amyloid Core Protein May Account for Its Deposition and Stability in Alzheimers-Disease. J. Biol. Chem. 1993, 268, 3072–3083. [Google Scholar] [CrossRef]
- Finder, V.H.; Glockshuber, R. Amyloid-beta aggregation. Neurodegener. Dis. 2007, 4, 13–27. [Google Scholar] [CrossRef] [PubMed]
- Selkoe, D.J.; Hardy, J. The amyloid hypothesis of Alzheimer’s disease at 25 years. EMBO Mol. Med. 2016, 8, 595–608. [Google Scholar] [CrossRef] [PubMed]
- Fukuda, H.; Shimizu, T.; Nakajima, M.; Mori, H.; Shirasawa, T. Synthesis, aggregation, and neurotoxicity of the Alzheimer’s A beta 1-42 amyloid peptide and its isoaspartyl isomers. Bioorg. Med. Chem. Lett. 1999, 9, 953–956. [Google Scholar] [CrossRef]
- Sugiki, T.; Utsunomiya-Tate, N. Site-specific aspartic acid isomerization regulates self-assembly and neurotoxicity of amyloid-beta. Biochem. Biophys. Res. Commun. 2013, 441, 493–498. [Google Scholar] [CrossRef]
- Kozin, S.A.; Cheglakov, I.B.; Ovsepyan, A.A.; Telegin, G.B.; Tsvetkov, P.O.; Lisitsa, A.V.; Makarov, A.A. Peripherally Applied Synthetic Peptide isoAsp7-A beta(1-42) Triggers Cerebral beta-Amyloidosis. Neurotox. Res. 2013, 24, 370–376. [Google Scholar]
- Johnson, B.A.; Shirokawa, J.M.; Geddes, J.W.; Choi, B.H.; Kim, R.C.; Aswad, D.W. Protein L-Isoaspartyl Methyltransferase in Postmortem Brains of Aged Humans. Neurobiol. Aging 1991, 12, 19–24. [Google Scholar] [CrossRef]
- Gnoth, K.; Piechotta, A.; Kleinschmidt, M.; Konrath, S.; Schenk, M.; Taudte, N.; Ramsbeck, D.; Rieckmann, V.; Geissler, S.; Eichentopf, R.; et al. Targeting isoaspartate-modified Abeta rescues behavioral deficits in transgenic mice with Alzheimer’s disease-like pathology. Alzheimers Res. Ther. 2020, 12, 149. [Google Scholar]
- Blennow, K. A Review of Fluid Biomarkers for Alzheimer’s Disease: Moving from CSF to Blood. Neurol. Ther. 2017, 6 (Suppl. 1), 15–24. [Google Scholar] [CrossRef]
- Paraskevaidi, M.; Morais, C.L.M.; Lima, K.M.G.; Snowden, J.S.; Saxon, J.A.; Richardson, A.M.T.; Jones, M.; Mann, D.M.A.; Allsop, D.; Martin-Hirsch, P.L.; et al. Differential diagnosis of Alzheimer’s disease using spectrochemical analysis of blood. Proc. Natl. Acad. Sci. USA 2017, 114, E7929–E7938. [Google Scholar] [CrossRef] [Green Version]
- Jack, C.R., Jr.; Bennett, D.A.; Blennow, K.; Carrillo, M.C.; Dunn, B.; Haeberlein, S.B.; Holtzman, D.M.; Jagust, W.; Jessen, F.; Karlawish, J.; et al. Contributors, NIA-AA Research Framework: Toward a biological definition of Alzheimer’s disease. Alzheimers Dement 2018, 14, 535–562. [Google Scholar] [CrossRef] [PubMed]
- Zou, K.; Abdullah, M.; Michikawa, M. Current Biomarkers for Alzheimer’s Disease: From CSF to Blood. J. Pers. Med. 2020, 10, 85. [Google Scholar] [CrossRef] [PubMed]
- Maes, M.; DeVos, N.; Wauters, A.; Demedts, P.; Maurits, V.; Neels, H.; Bosmans, E.; Altamura, C.; Lin, A.; Song, C.; et al. Inflammatory markers in younger vs elderly normal volunteers and in patients with Alzheimer’s disease. J. Psychiatr. Res. 1999, 33, 397–405. [Google Scholar] [CrossRef]
- Kim, T.S.; Pae, C.U.; Yoon, S.J.; Jang, W.Y.; Lee, N.J.; Kim, J.J.; Lee, S.J.; Lee, C.; Paik, I.H.; Lee, C.U. Decreased plasma antioxidants in patients with Alzheimer’s disease. Int. J. Geriatr. Psychiatry 2006, 21, 344–348. [Google Scholar]
- Kim, J.W.; Byun, M.S.; Lee, J.H.; Yi, D.; Jeon, S.Y.; Sohn, B.K.; Lee, J.Y.; Shin, S.A.; Kim, Y.K.; Kang, K.M.; et al. Serum albumin and beta-amyloid deposition in the human brain. Neurology 2020, 95, E815–E826. [Google Scholar] [CrossRef] [PubMed]
- Taneja, V. Sex Hormones Determine Immune Response. Front. Immunol. 2018, 9, 1931. [Google Scholar] [CrossRef]
- Zhao, M.; Guo, C. Multipronged Regulatory Functions of Serum Albumin in Early Stages of Amyloid-beta Aggregation. ACS Chem. Neurosci. 2021, 12, 2409–2420. [Google Scholar] [CrossRef]
- Qin, Z.X.; Kaufman, R.S.; Khoury, R.N.; Khoury, M.K.; Aswad, D.W. Isoaspartate Accumulation in Mouse Brain Is Associated with Altered Patterns of Protein Phosphorylation and Acetylation, Some of Which Are Highly Sex-Dependent. PLoS ONE 2013, 8, e80758. [Google Scholar] [CrossRef] [Green Version]
- Qin, Z.X.; Dimitrijevic, A.; Aswad, D.W. Accelerated protein damage in brains of PIMT +/− mice; a possible model for the variability of cognitive decline in human aging. Neurobiol. Aging 2015, 36, 1029–1036. [Google Scholar] [CrossRef] [Green Version]
- World Health Organization. Global Status Report on Blood Safety and Availability 2016; World Health Organization: Geneva, Switzerland, 2017. [Google Scholar]
- Cournoyer, J.J.; Pittman, J.L.; Ivleva, V.B.; Fallows, E.; Waskell, L.; Costello, C.E.; O’Connor, P.B. Deamidation: Differentiation of aspartyl from isoaspartyl products in peptides by electron capture dissociation. Protein Sci. 2005, 14, 452–463. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lyutvinskiy, Y.; Yang, H.; Rutishauser, D.; Zubarev, R.A. In silico instrumental response correction improves precision of label-free proteomics and accuracy of proteomics-based predictive models. Mol. Cell. Proteom. 2013, 12, 2324–2331. [Google Scholar] [CrossRef] [PubMed] [Green Version]

| Indirect ELISA | Indirect Sandwich ELISA | ||||
|---|---|---|---|---|---|
| Plate | White Polystyrene | Copper Coated | White Polystyrene | White Polystyrene | Copper-coated |
| Blocking buffer | 10% Milk in PBST | ||||
| Capture antibody | N/A | 15C7 anti-HSA mAb | 1A3 mAb | 1A3 mAb | |
| Primary/Detection antibody | 1A3 mAb | 1A3 mAb | 1A2 mAb | 1E1 anti-HSA mAb | |
| Secondary antibody | Peroxidase AffiniPure Polyclonal Goat Anti-Mouse IgG + IgM (H + L) | ||||
| Detection | Chemiluminescence | Absorbance | Chemiluminescence | Chemiluminescence | Absorbance |
| S/N at 6% isoAsp | 39.9 | 12.1 | 0.8 | 0.6 | 0.8 |
| p value | <10−5 | <10−5 | 7.0 × 10−3 | <10−5 | <10−5 |
| S/N at 0.6% isoAsp | 4.3 | 3.6 | N/A | ||
| No. | Poition of Glycans | ∆M (kD) |
|---|---|---|
| a | HexNAc[4]Hex[4] | +1.461 |
| b | HexNAc[4]Hex[3] | +1.298 |
| c | dHex[1]HexNAc[3]Hex[3] | +1.241 |
| d | dHex[1]HexNAc[4]Hex[6] | +1.931 |
| e | dHex[1]HexNAc[4]Hex[4] | +1.607 |
| f | dHex[1]HexNAc[4]Hex[3] | +1.445 |
| g | HexNAc[2]Hex[5] | +1.216 |
| h | HexNAc[4]Hex[5]NeuAc[1] | +1.914 |
| Antibody-Antigen | ka (M−1 s−1) | kd (s−1) | KD (M) |
|---|---|---|---|
| 1A3-aHSA | 2.6 ± 0.5 × 10−5 | 5.3 ± 0.4 × 10−3 | 2.1 ± 0.3 × 10−8 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, J.; Lundström, S.L.; Seelow, S.; Rodin, S.; Meng, Z.; Astorga-Wells, J.; Jia, Q.; Zubarev, R.A. First Immunoassay for Measuring Isoaspartate in Human Serum Albumin. Molecules 2021, 26, 6709. https://doi.org/10.3390/molecules26216709
Wang J, Lundström SL, Seelow S, Rodin S, Meng Z, Astorga-Wells J, Jia Q, Zubarev RA. First Immunoassay for Measuring Isoaspartate in Human Serum Albumin. Molecules. 2021; 26(21):6709. https://doi.org/10.3390/molecules26216709
Chicago/Turabian StyleWang, Jijing, Susanna L. Lundström, Sven Seelow, Sergey Rodin, Zhaowei Meng, Juan Astorga-Wells, Qinyu Jia, and Roman A. Zubarev. 2021. "First Immunoassay for Measuring Isoaspartate in Human Serum Albumin" Molecules 26, no. 21: 6709. https://doi.org/10.3390/molecules26216709
APA StyleWang, J., Lundström, S. L., Seelow, S., Rodin, S., Meng, Z., Astorga-Wells, J., Jia, Q., & Zubarev, R. A. (2021). First Immunoassay for Measuring Isoaspartate in Human Serum Albumin. Molecules, 26(21), 6709. https://doi.org/10.3390/molecules26216709







