1. Introduction
Drug discovery is a prolonged and complex process that culminates in the manufacture of new drugs. The biomolecular target is selected, and high-performance screening procedures are executed to identify bioactive chemicals for defined aims in traditional drug research and development. It is costly and time-consuming to produce high-performing research testing [
1]. In truth, the chances of success are slim; approximately 1 out of every 5000 drug candidates is expected to be accepted and widely used at some point [
2]. Increased computer capabilities, on the other hand, have enabled the screening of millions of chemical compounds at a reasonable speed and cost. The virtual screening methodology is a computerized method for scanning large libraries of small compounds for the most likely structures with the goal of developing medication [
3,
4,
5]. Virtual screening (VS) is used in the early stages of drug development to identify the most promising lead compounds from large chemical libraries. The development of medications has been sped up in recent years thanks to virtual screening (VS). Virtual screening is divided into two types: structure-based virtual screening (SBVS) and ligand-based virtual screening (LBVS) [
6]. The SBVS approaches strategies to look for indirect chemicals that are suitable for the binding site of the biological target. The molecular docking technique lies at the heart of SBVS approaches [
7]. On the other hand, the LBVS approach is used constantly for the prediction of molecular properties and for measuring molecular similarity because the method to represent the molecules is easy and accurate. The necessity of applications of similarity searching comes from the importance of lead optimization in drug discovery programs, in which close neighbors are looking into an initial lead compound to find decent compounds [
8,
9,
10].
Modern deep learning (DL) approaches have recently been presented in a variety of fields, and they have progressed in recent years, creating a new door for researchers. The hit of DL techniques benefits from the speedy growth of the DL algorithms and the progression of high-performance computing techniques. Moreover, DL techniques have slight generalization errors, which allow them to achieve credible results on certain benchmarks or competitive tests [
11,
12]. In addition, the Siamese network is frequently employed to solve image and text similarity problems. It has been utilized for more complex data samples, particularly heterogeneous data samples with a variety of dimensionality and type properties [
13,
14]. However, some studies reported that pruning the parameters makes the model of deep learning smaller in size, more memory-efficient, more power-efficient, and faster at inference. The whole idea of model pruning is to reduce the number of parameters without much loss in the accuracy of the model, which means cutting down parts of the network that contribute less or nothing to the network during inference [
15,
16,
17].
Various techniques have been utilized in order to augment the retrieval effectiveness of similarity search algorithms. The use of 2D similarity algorithms has gained popularity. Estimating molecular similarity is based on the assumption that structurally similar molecules seem to be more likely to have similar characteristics than structurally different ones. Therefore, the objective of similarity searching is to identify molecules that are similar in structure to the consumer’s reference structures [
18,
19]. A number of coefficient techniques can be used to quantify the similarity/difference between molecule pairs. Many other studies tested the outcomes of various similarity coefficients, showing that the Tanimoto coefficient performed better than the others. As a result, in cheminformatics, the Tanimoto coefficient has become the most often used measure of chemical compound similarity [
20,
21,
22]. Some experiments attempted to combine techniques from other fields. Such that they adapted the techniques from text information retrieval to employ in the cheminformatic domain to improve the similitude of molecular searching [
23]. For example, the Bayesian inference network was based on text retrieval domains, it has been adapted and used in the similarity of molecular searches in virtual screening, and outperforms the Tanimoto technique [
24,
25]. Furthermore, reweighting approaches were employed to model document retrieval in the text field and modified in the cheminformatic area in the retrieval model [
25,
26,
27]. Mohammed Al Dabagh (2017) improved the molecular similarity searching and molecular ranking of chemical compounds in LBVS using quantum mechanics physics theory principles [
28]. Mubarak Hussien (2017) constructed a new similarity measure using current similarity measures by reweighting several bit-strings. Furthermore, the author offered ranking strategies for the development of a replacement ranking approach [
29]. Deep belief networks (DBN) were used by Nasser, Majed, and colleagues (2021) to reweight molecular data wherein many descriptors were used, each reflecting separate relevant aspects, and combining all new features from all descriptors to create a new descriptor for similarity searches [
30,
31].
On the other hand, many studies have used deep learning methods as prediction or classification models. Some of them have used the DNN model to predict the activities of the selected compounds. Furthermore, other studies have reported that deep learning methods in Siamese architecture as a similarity model produce the best performance in many fields. For example, Jonas et al. (2016) used an LSTM Siamese neural network to calculate the similarity wherein the exponential Manhattan distance was used to measure the similarity between two sentences [
32]. Jun Yu and Mengyan Li et al. (2020) used CNN Siamese architecture to determine whether two people are related, allowing missing persons to be reunited with their kin [
33]. In the drug discovery domain, Devendra Singh Dhami et al. (2020) used images as an input in a Siamese convolutional network architecture to predict drug interactions in the drug discovery area [
34]. Minji Jeon1 et al. (2019) proposed a method for calculating distance, utilizing MLP Siamese neural networks (ReSimNet) in structure-based virtual screen (SBVS) using cosine similarity [
35].
Moreover, some early work in the pruning parameters domain used a gradual pruning scheme based on pruning all the weights in a layer less than some threshold (manually chosen) [
12]. Blundell et al. (2015) introduced Bayes backpropagation for feedforward neural networks. This method gives the uncertainty in their predictions and reduces the model’s parameter count by ordering the weights according to their signal-to-noise-ratio and setting a certain percentage of the weights with the lowest ratios to 0 to prune these weights [
15]. Louizos and Christos et al. (2017) used hierarchical priors to prune nodes instead of individual weights and also used the posterior uncertainties to determine the optimal fixed-point precision to encode the weights [
36]. Chenglong Zhao et al. (2019) proposed a variational Bayesian scheme for pruning convolutional neural networks at the channel level. The variational technique is introduced to estimate the distribution of a newly proposed parameter; based on this, redundant channels can be removed from the model [
37].
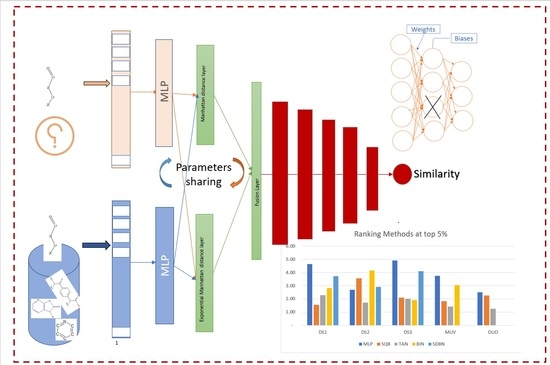
Despite the good performances of the above methods compared to their prior, especially when dealing with molecules that have homogenous active elements structural such as classes of molecules in the MDL Drug Data Report dataset (MDDR_DR2), the performances are not satisfied when dealing with molecules with structurally heterogeneous nature such as classes of molecules in the MDL Drug Data Report dataset (MDDR_DR3, MDDR_DR1) and maximum unbiased validation (MUV) dataset. In this paper, the Siamese multi-layer perceptron model will be used and enhanced in order to achieve the main purpose of this study for improving the performance of similarity searching, especially with molecules that are structurally heterogeneous. The following are the paper’s main contributions:
- (1)
The Siamese multi-layer perceptron will be enhanced by (a) using two distance layers and then a fuse layer that combines the results from two distance layers, with multiple layers added after the fusion layer to improve the similarity recall; (b) pruning nodes in the Siamese similarity model to reduce the number of parameters that contribute less or nothing to the network during inference.
- (2)
In comparison to the benchmark approach and previous studies, the suggested method outperformance in terms of results, especially when dealing with heterogeneous classes of molecules.
4. Results and Discussion
The experimental results from the MDDR-DS1, MDDR-DS2, MDDR-DS3, MUV, and DUD datasets, for the ECFC-4 descriptor, are provided in
Table 6,
Table 7,
Table 8,
Table 9,
Table 10,
Table 11,
Table 12,
Table 13,
Table 14 and
Table 15, respectively, using 1% and 5% cut-offs. These tables show the results of the enhanced Siamese MLP similarity model compared to the benchmark TAN, as well as earlier studies BIN, SQB, and SDBN for MDDR datasets, BIN and SQB for MUV datasets, and SQB for DUD datasets. Every row in the tables displays the recall for the top 1% and 5% of the activity class, with the best recall rate shaded in each row. The mean rows in the tables represent the average for all activity classes, whereas the rows with shaded cells represent the total number of shaded cells for each technique throughout the whole range of activity classes.
When comparing the MDDR-DS1 recall results for the top 1% and 5% in
Table 6 and
Table 7, the suggested enhanced Siamese MLP technique was clearly superior to the benchmark TAN method and prior studies BIN, SQB, and SDBN in terms of the mean and number of shaded cells. The suggested technique has the highest mean value (30.69) in
Table 6, followed by SDBN, BIN, SQB, and lastly, TAN methods. In the suggested approach, the shaded cells have a value of (7). The suggested approach has the highest mean value (50.463) in
Table 7, followed by SDBN, BIN, SQB, and lastly TAN methods. In the suggested technique, shaded cells have a value of (9).
Furthermore, the MDDR-DS2 recall values obtained at the top 1%, as shown in
Table 8, demonstrate that the suggested Siamese MLP technique outperforms the benchmark TAN method. In view of the number of shaded cells, the MLP approach produced the best retrieval recall results, and the suggested method’s mean value is extremely close to that of prior research. However, by comparison, The MDDR-DS2 recall values obtained at the top 5% in
Table 9 clearly shows that the suggested Siamese MLP approach outperforms the benchmark TAN method only. In terms of the mean and number of shaded cells, the BIN approach produced the best retrieval recall results. Next, the second is SQB, SDBN, and finally, TAN in view of the mean value.
In addition, the MDDR-DS3 recall values recorded at the top 1% and 5% in
Table 10 and
Table 11, respectively, show that the proposed enhanced Siamese MLP method is obviously superior to the benchmark TAN method and methods from other studies. Likewise, in
Table 10, the proposed method gave the best retrieval recall results in view of the mean and number of shaded cells, compared to prior studies and benchmark TAN, followed by SDBN, BIN, SQB, and finally, TAN methods. By comparison, in
Table 11, the suggested enhanced Siamese MLP method was obviously superior to the benchmark TAN method and other studies. The second one is SDBN, followed by TAN, BIN, and SQB.
Moreover, the MUV dataset recall values recorded at the top 1% and 5% in
Table 12 and
Table 13, respectively, show that the proposed enhanced Siamese MLP method is obviously superior to the benchmark TAN method and other studies. Likewise, in
Table 12, the proposed method gave the best retrieval recall results in view of the mean and number of shaded cells, compared to the TAN method and methods from other studies, followed by BIN, SQB, and finally, TAN methods. However, by comparison, in
Table 13, the proposed enhanced Siamese MLP method was obviously superior to the benchmark TAN method and methods from other studies. Next, the second one is BIN, followed by SQB and TAN.
Moreover, the DUD dataset recall values recorded at the top 1% and 5% in
Table 14 and
Table 15, respectively, show that the proposed enhanced Siamese MLP method is obviously superior to the benchmark TAN method and methods from other studies. Likewise, in
Table 14, the proposed method gave the best retrieval recall results in view of the mean and number of shaded cells, compared to the previous study and benchmark TAN. Furthermore, in
Table 15, the proposed enhanced Siamese MLP method was obviously superior to the benchmark TAN method and the previous study SQB.
The experimental results for pruning nodes on MDDR-DS1, MDDR-DS2, MDDR-DS3, MUV, and DUD datasets are shown in
Figure 4,
Figure 5,
Figure 6,
Figure 7,
Figure 8,
Figure 9,
Figure 10,
Figure 11,
Figure 12 and
Figure 13, respectively. In these figures, the x axis represents the pruning ratio starting from 0% and ending with 90%, 95%, 95%, 70%, and 98% in DS1, DS2, DS3, MUV, and DUD datasets, respectively. They y axis represents the level of retrieval recall values for each class in the dataset. The classes of molecules are represented as color lines. The tables that contain on pruning ratio of recall values for each dataset are available as
Supplementary Materials.
Figure 4 shows the level of the retrieval recall values at different pruning ratios for each class at the top 1% in DS1. We note that the recall values of most classes remain the same until they reach 80% of the pruning ratio, while some classes increased slightly, such as class 7, and decreased by a little in others, such as classes 2 and 9. Furthermore,
Figure 5 shows the level of retrieval recall values at different pruning ratios for each class at the top 5% in DS1. The recall values of most classes remain the same values until they reach 80% of the pruning ratio, while some classes decreased, such as classes 2 and 10.
Figure 6 shows the level of retrieval recall values at different pruning ratios for each class at the top 1% in DS2. We note that the recall values of most classes remain the same until they reach 90% of the pruning ratio, while some classes increased slightly, such as classes 5 and 8, and decreased by a little in others, such as classes 1, 3, and 4. Furthermore,
Figure 7 shows the level of retrieval recall values at different pruning ratios for each class at the top 5% in DS2. The recall values of most classes remain the same until they reach 90% of the pruning ratio, except for class 6, which remains until more than 95% of the pruning ratio, while some classes decreased by a little, such as class 4, or increased slightly, such as class 5.
Figure 8 shows the level of retrieval recall values at different pruning ratios for each class at the top 1% in DS3. We note that the recall values of most classes remain the same until they reach 80% of the pruning ratio, while some classes increased slightly, such as class 4, a more than 95% pruning ratio, and decreased by a little in others, such as classes 1, 7, and 8. Furthermore,
Figure 9 shows the retrieval recall values at different pruning ratios for each class at the top 5% in DS3. The recall values of most classes remain the same until they reach 80% of the pruning ratio, except for class 4, which increased until more than 95%, while some classes decreased by a little, such as classes 1, 3, 7, and 8.
Figure 10 shows the level of retrieval recall values at different pruning ratios for each class at the top 1% in MUV. We note that the recall values of most classes remained the same until they reached 60% of the pruning ratio, while some classes increased slightly, such as classes 3 and 10, and decreased by a little in others, such as class 8. Moreover,
Figure 11 shows the retrieval recall values at different pruning ratios for each class at the top 5% in MUV. The recall values of most classes remained the same until they reached 60% of the pruning ratio, while some classes increased slightly, such as classes 1, 3 and 5, and decreased by a little in others, such as classes 3 and 4.
Furthermore,
Figure 12 shows the level of retrieval recall values at different pruning ratios for each class at the top 1% in DUD. We note that the recall values of most classes remained the same until they reached 80% of the pruning ratio, while some classes increased slightly, such as classes 4,6, and 9, and decreased by a little in others, such as classes 3, 8, and 12. Moreover,
Figure 13 shows the retrieval recall values at different pruning ratios for each class at the top 5% in DUD. The recall values of most classes remained the same until they reached 80% of the pruning ratio, while some classes increased slightly, such as classes 11 and 12, and decreased by a little in others, such as classes 3 and 10.
Moreover, the Kendall W concordance test has been used;
Table 16 shows the ranking of the enhanced Siamese multilayer perceptron method based on previous studies of TAN, BIN, SQB, and SDBN using Kendall W test results for MDDR-DS1, MDDR-DS2, MDDR-DS3, MUV, and DUD at the top 1% and 5%. The first method is the benchmark method, which is the Tanimoto coefficient TAN; the second method is Bayesian inference [
24]; the third method is quantum similarity search SQB-Complex [
28]; the last method is multi-descriptor-based on Stack of deep belief networks SDBN [
31]. The results of the Kendall W test of the top 1% for all used datasets show that the values of associated probability (
p) are less than 0.05. This indicates that the enhanced Siamese multilayer perceptron method is significant in the top 1% for all cases. As a result, the overall ranking of all methods indicates that the enhanced Siamese multilayer perceptron method is superior to previous studies and benchmark TAN. The overall ranking for methods showed that MLP has the top ranks among other methods. This is the same as with the results of the Kendall W test of the top 5%; the results show that the values of associated probability (p) are less than 0.05. This indicates that the enhanced Siamese multilayer perceptron method is significant in the top 5%. As a result, the overall ranking of all methods indicates that the enhanced Siamese multilayer perceptron method is superior to previous studies for all datasets and the overall ranking for the method showed that Siamese multilayer perceptron method has the top ranks among other methods, except in DR2, in which the BIN method was better than MLP.
Figure 14 and
Figure 15 show the ranking of the enhanced Siamese multilayer perceptron method based on TAN, BIN, SQB, and SDBN using the Kendall W test results for DS1, DS2,DS3, MUV, and DUD at the top 1% and 5%.