Metabolic Profile of C-Prenyl Coumarins Using Mass Spectrometry-Based Metabolomics
Abstract
:1. Introduction
2. Results
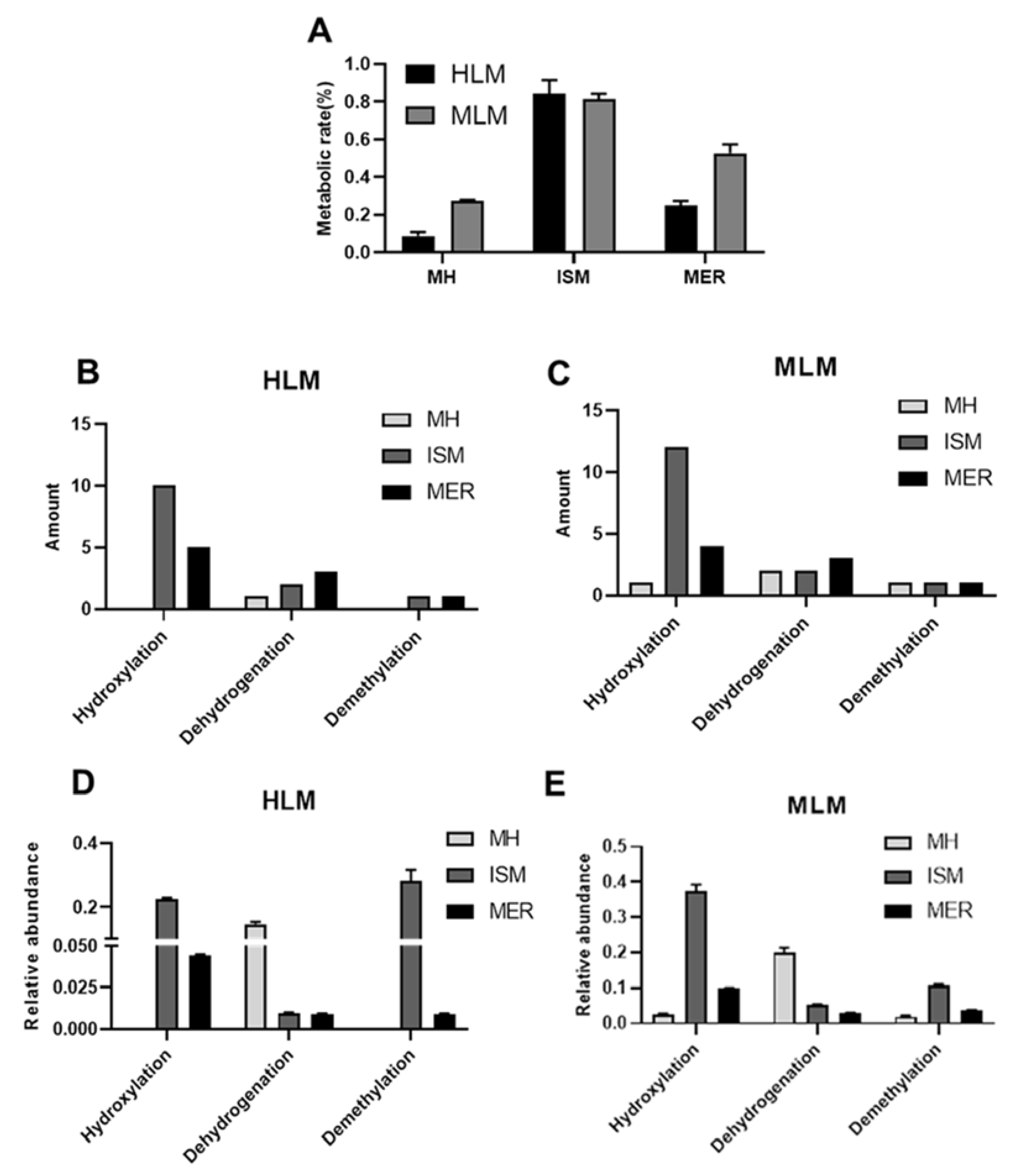
2.1. In Vitro Metabolism of MH, ISM, and MER by HLM and MLM
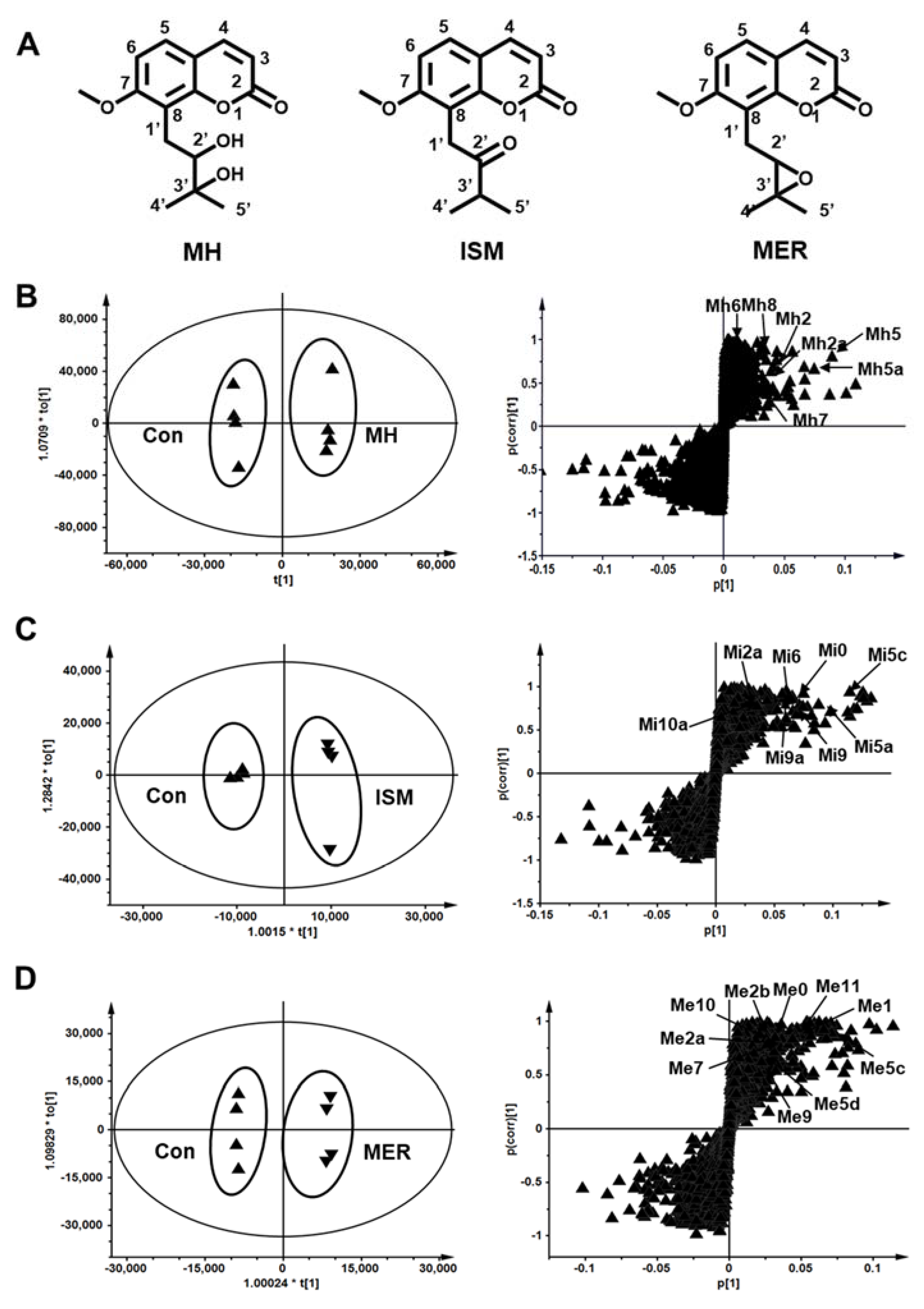
2.2. Metabolomic Profiling of MH, ISM, and MER Metabolites in Mice
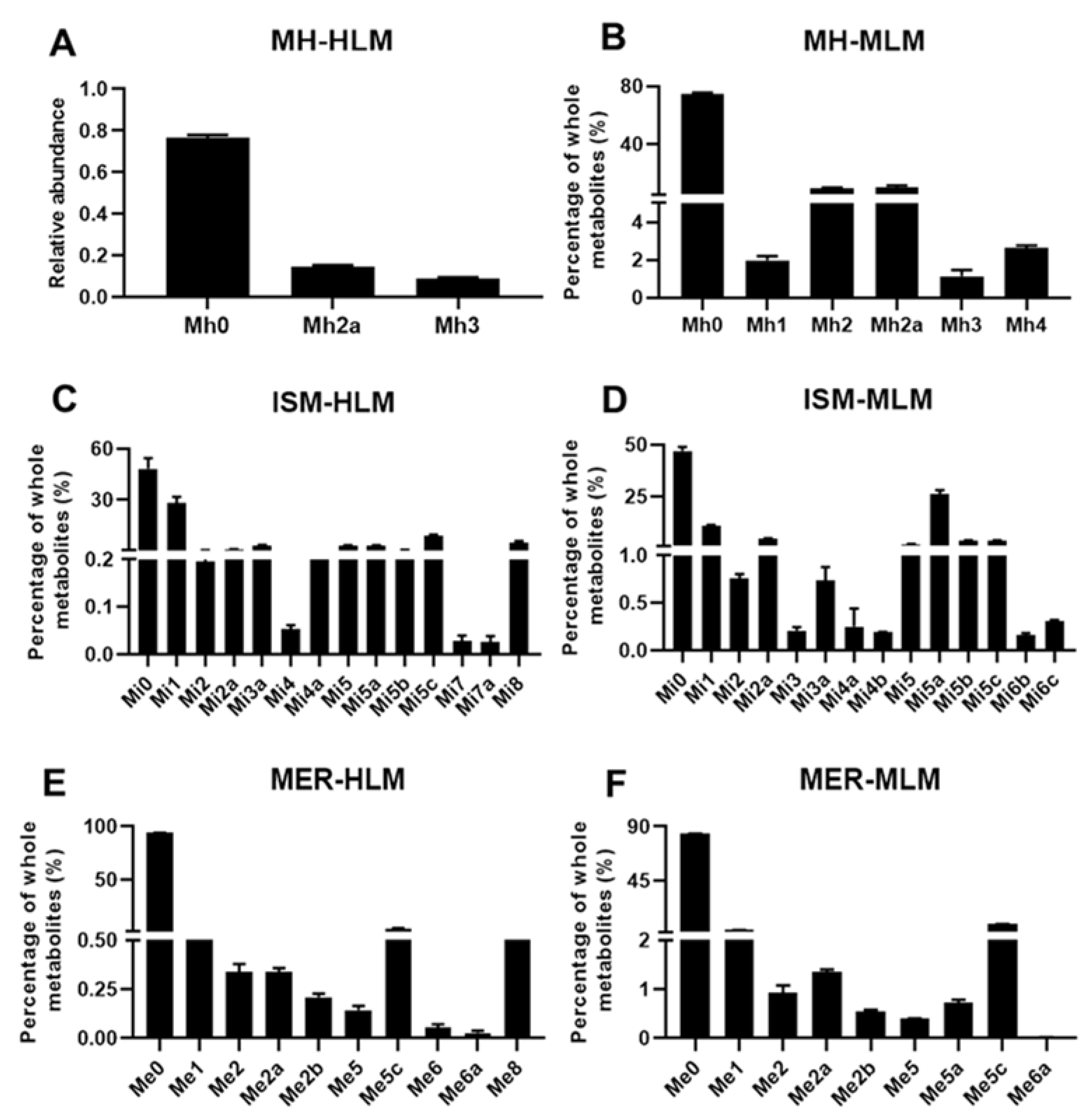
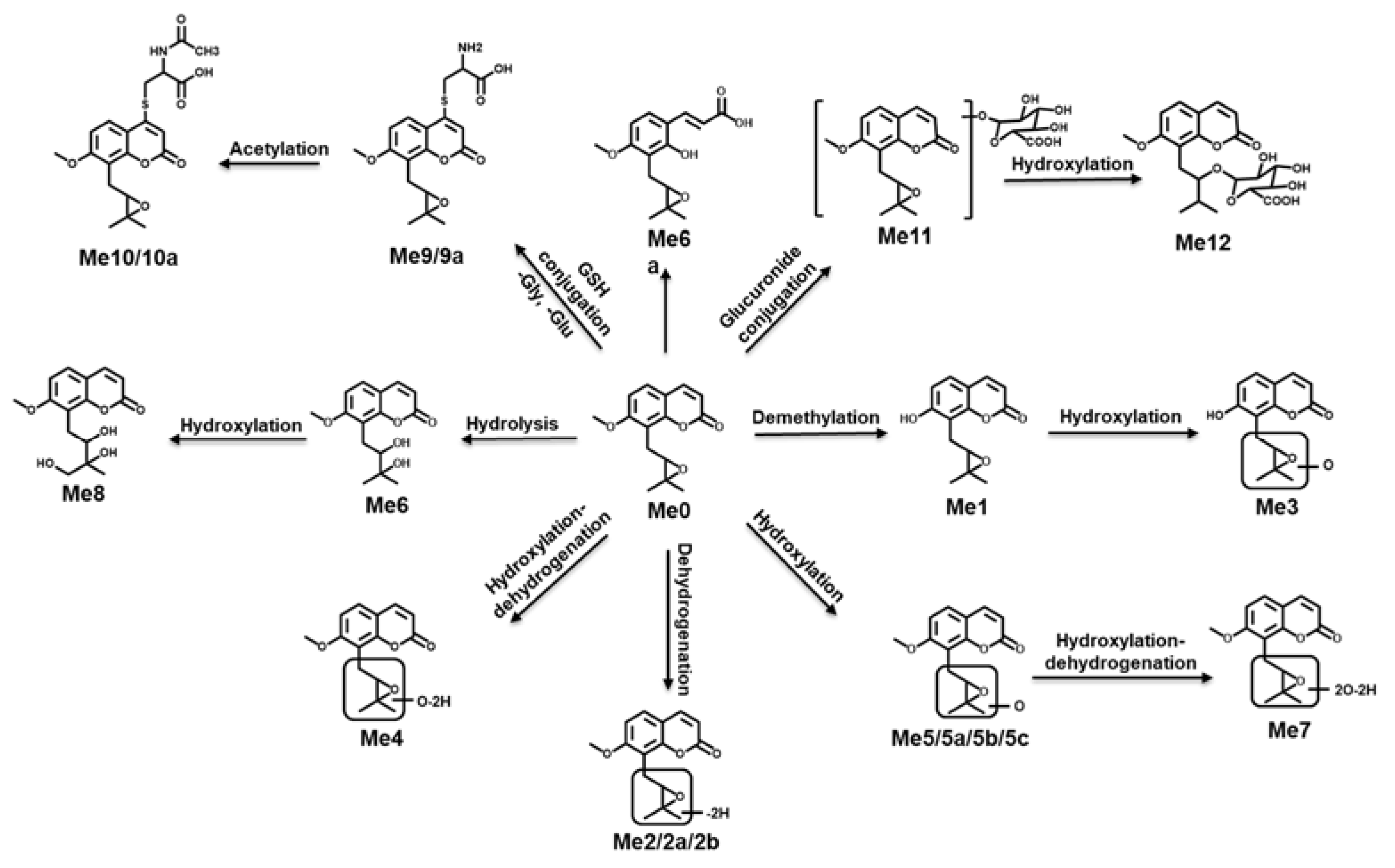
2.3. Structure Elucidation of MH, ISM, and MER Metabolites
2.3.1. Structure Elucidation of MH Metabolites
2.3.2. Structure Elucidation of ISM Metabolites
2.3.3. Structure Elucidation of MER Metabolites
2.4. CYPs Involved in the Formation of MH, ISM, and MER Metabolites
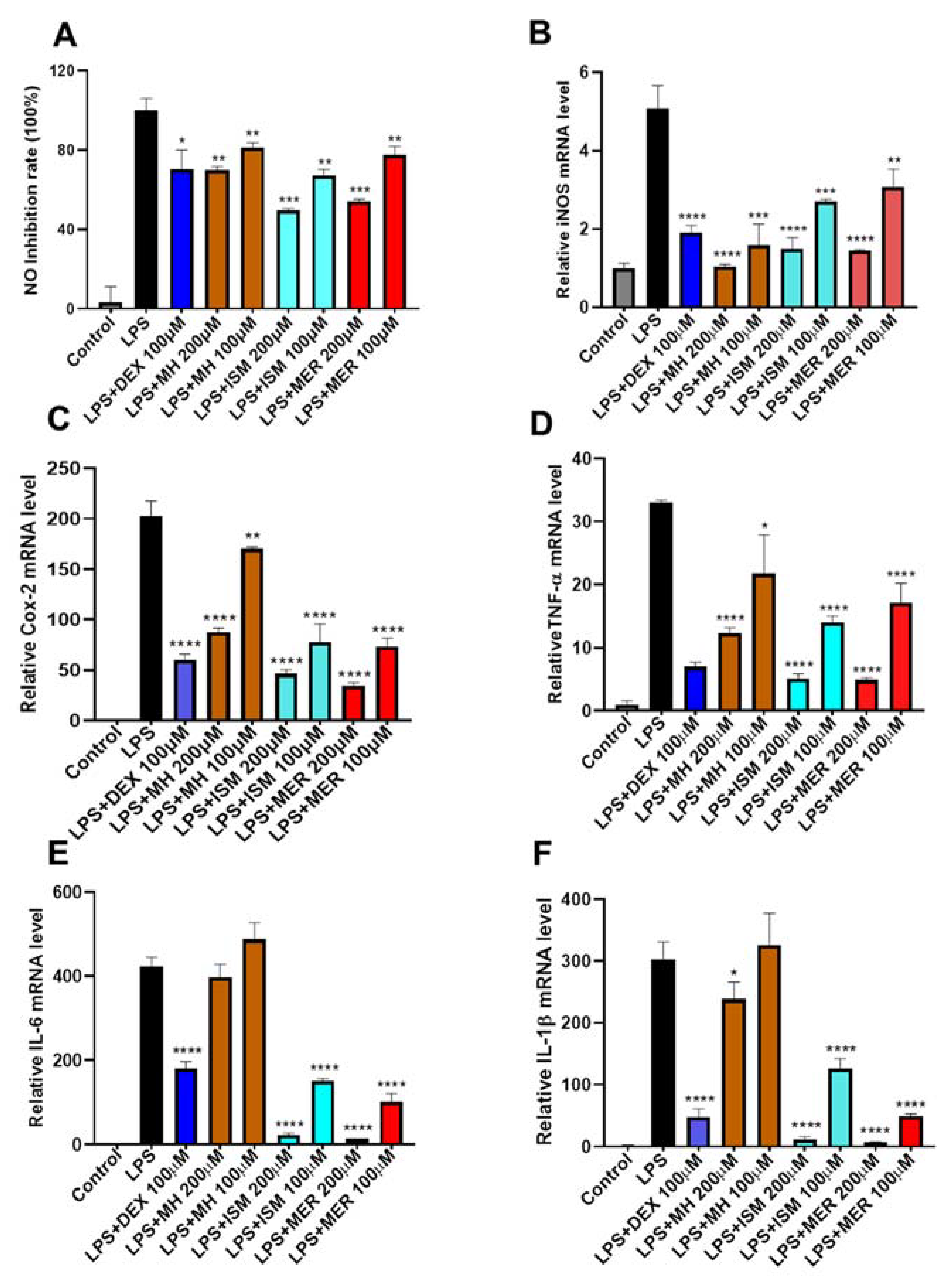
2.5. Anti-Inflammatory Activity of MH, ISM, and MER
2.5.1. Cytotoxicity of MH, ISM, and MER in RAW 264.7 Macrophages
2.5.2. Inhibition of MH, ISM, and MER on LPS-Induced NO Production in RAW 264.7 Cells
2.5.3. Inhibition of MH, ISM, and MER on LPS-Induced over Expression of Pro-Inflammatory Mediators
3. Discussions
4. Materials and Methods
4.1. Chemicals and Reagents
4.2. In Vivo Metabolism of MH, ISM, and MER
4.3. In Vitro Metabolism of Coumarins
4.4. Sample Preparation
4.5. UPLC-ESI-QTOFMS Analysis
4.6. Multivariate Data Analysis
4.7. Anti-Inflammatory Assays
4.7.1. Cell Culture
4.7.2. Cell Viability Assay
4.7.3. NO Inhibition Assay
4.7.4. Real-Time PCR
4.8. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Wang, S.; Bao, Y.R.; Li, T.J.; Yu, T.; Chang, X.; Yang, G.L.; Meng, X.S. Mechanism of Fructus aurantii Flavonoids Promoting Gastrointestinal Motility: From Organic and Inorganic Endogenous Substances Combination Point of View. Pharmacogn. Mag. 2017, 13, 372–377. [Google Scholar]
- Tan, W.X.; Li, Y.; Wang, Y.; Zhang, Z.J.; Wang, T.; Zhou, Q.; Wang, X.Y. Anti-coagulative and gastrointestinal motility regulative activities of Fructus aurantii Immaturus and its effective fractions. Biomed. Pharmacother. 2017, 90, 244–252. [Google Scholar] [CrossRef]
- Bai, Y.F.; Wang, S.W.; Wang, X.X.; Weng, Y.Y.; Fan, X.Y.; Sheng, H.; Zhu, X.T.; Lou, L.J.; Zhang, F. The flavonoid-rich Quzhou Fructus aurantii extract modulates gut microbiota and prevents obesity in high-fat diet-fed mice. Nutr. Diabetes 2019, 9, 30. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, Y.T.; Wang, G.F.; Chen, C.F.; Chen, C.C.; Hong, C.Y.; Yang, M.C. Fructus aurantii reduced portal pressure in portal hypertensive rats. Life Sci. 1995, 57, 2011–2020. [Google Scholar] [CrossRef]
- Li, X.R.; Liang, Q.; Sun, Y.J.; Diao, L.; Qin, Z.; Wang, W.; Lu, J.Z.; Fu, S.J.; Ma, B.L.; Yue, Z.J. Potential Mechanisms Responsible for the Antinephrolithic Effects of an Aqueous Extract of Fructus aurantii. Evid Based Complement Alternat Med. 2015, 491409. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Y.J.; Huang, W.; Huang, X.; Wang, Y.; Wang, Z.; Wang, C.; Zhong, B.W.; Sheng, C.X.; Wang, B.; Zhang, S.F.; et al. Fructus aurantii induced antidepressant effect via its monoaminergic mechanism and prokinetic action in rat. Phytomedicine 2012, 19, 1101–1107. [Google Scholar] [CrossRef] [PubMed]
- Huang, W.; Huang, X.; Xing, Z.; Qiu, X.; Wang, Y.; Fan, R.; Liu, W.; Ren, P.; Liu, Z.; Zhou, H. Meranzin hydrate induces similar effect to Fructus aurantii on intestinal motility through activation of H1 histamine receptors. J. Gastrointest. Surg. 2011, 15, 87–96. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Yu, A.L.; Wang, Z.L.; Chen, K.; Zheng, W.; Zhou, J.J.; Xie, Q.; Yan, H.B.; Ren, P.; Huang, X. Chaihu-Shugan-San and absorbed meranzin hydrate induce anti-atherosclerosis and behavioral improvements in high-fat diet ApoE(-/-) mice via anti-inflammatory and BDNF-TrkB pathway. Biomed. Pharmacother. 2019, 115, 108893. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.F.; Zhou, J.L.; Zhang, T.; Chen, K.; Xu, M.; Wu, L.; Liu, J.; Huang, Y.K.; Nie, B.B.; Shen, X.X.; et al. Meranzin hydrate elicits antidepressant effects and restores reward circuitry. Behav. Brain. Res. 2021, 398, 112898. [Google Scholar] [CrossRef] [PubMed]
- Xie, Y.; Huang, X.; Hu, S.Y.; Qiu, X.J.; Zhang, Y.J.; Ren, P.; Wang, Y.; Ji, H.; Zhang, C.H.; Xie, W.B.; et al. Meranzin hydrate exhibits anti-depressive and prokinetic-like effects through regulation of the shared α2-adrenoceptor in the brain-gut axis of rats in the forced swimming test. Neuropharmacology 2012, 67, 318–325. [Google Scholar] [CrossRef] [PubMed]
- Xu, G.; Feng, L.L.; Song, P.P.; Xu, F.; Li, A.; Wang, Y.B.; Shen, Y.; Wu, X.F.; Luo, Q.; Wu, X.X.; et al. Isomeranzin suppresses inflammation by inhibiting M1 macrophage polarization through the NF-κB and ERK pathway. Int. Immunopharmacol. 2016, 38, 175–185. [Google Scholar] [CrossRef] [PubMed]
- Nalin, R.; Jacqueline, A.; Khye, S.G.; Anne, D.; Emile, P.; Paul, B. Anti-mycobacterial activity of chemically defined natural substances from the Caribbean flora in Guadeloupe. FEMS Immunol. Med. Microbiol. 1998, 20, 267–273. [Google Scholar]
- Shin, E.J.; Lee, C.; Sung, S.H.; Kim, Y.C.; Hwang, B.Y.; Lee, M.K. Antifibrotic activity of coumarins from Cnidium monnieri fruits in HSC-T6 hepatic stellate cells. J. Nat. Med. 2011, 65, 370–374. [Google Scholar] [CrossRef]
- Lv, Q.Q.; Yang, X.N.; Yan, D.M.; Liang, W.Q.; Liu, H.N.; Yang, X.W.; Li, F. Metabolic profiling of dehydrodiisoeugenol using xenobiotic metabolomics. J. Pharm. Biomed. Anal. 2017, 145, 725–733. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.N.; Lv, Q.Q.; Zhao, Q.; Li, X.M.; Yan, D.M.; Yang, X.W.; Li, F. Metabolic profiling of myrislignan by UPLC-ESI-QTOFMS-based metabolomics. RSC Adv. 2017, 7, 40131–40140. [Google Scholar] [CrossRef] [Green Version]
- Xiao, Y.; Wang, Y.K.; Xiao, X.R.; Zhao, Q.; Huang, J.F.; Zhu, W.F.; Li, F. Metabolic profiling of coumarins by the combination of UPLC-MS-based metabolomics and multiple mass defect filter. Xenobiotica 2020, 50, 1076–1089. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Guo, Y.; Huang, W.H.; Zhang, W.; Tan., Z.R.; Peng, J.B.; Wang, Y.C.; Hu, D.L.; Ouyang, D.S.; Xiao, J. Searching the cytochrome p450 enzymes for the metabolism of meranzin hydrate: A prospective antidepressant originating from Chaihu-Shugan-San. PLoS ONE 2014, 9, e113819. [Google Scholar] [CrossRef] [PubMed]
- Hu, X.; Huang, W.; Yang, Y. Cytochrome P450 isoenzymes in rat and human liver microsomes associate with the metabolism of total coumarins in Fructus Cnidii. Eur. J. Drug Metab. Pharmacokinet. 2015, 40, 373–377. [Google Scholar] [CrossRef]
- Cao, G.Y.; Yang, X.W.; Xu, W.; Li, F. New inhibitors of nitric oxide production from the seeds of Myristica fragrans. Food. Chem. Toxicol. 2013, 62, 167–171. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.N.; Kim, J.D.; Yeo, J.H.; Son, H.J.; Park, S.B.; Eo, H.J.; Jeong, J.B. Heracleum moellendorffii roots inhibit then Access production of pro-inflammatory mediators through the inhibition of NF-κB and MAPK signaling, and activation of ROS/Nrf2/HO-1 signaling in LPS-stimulated RAW264. 7 cells. BMC Complement. Altern. Med. 2019, 19, 310. [Google Scholar]
- Han, C.; Yang, J.K.; Song, P.Y.; Wang, X.; Shi, W.Y. Effects of Salviamil tiorrhiza polysaccharides on Lipopolysaccharide-Induced Inflammatory Factor Release in RAW 264.7 Cells. J. Interferon Cytokine Res. 2018, 38, 29–37. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.; Rey, K.; Besler, K.; Wang, C.; Choy, J. Immunobiology of Nitric Oxide and Regulation of Inducible Nitric Oxide Synthase, Results. Probl. Cell. Differ. 2017, 62, 181–207. [Google Scholar]
- Manikandan, P.; Nagini, S. Cytochrome P450 Structure, Function and Clinical Significance: A Review. Curr. Drug Targets 2018, 19, 38–54. [Google Scholar] [CrossRef] [PubMed]
- Meyer, U.A. Overview of enzymes of drug metabolism. J. Pharmacokinet. Biopharm. 1996, 24, 449–459. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.H.; Lu, A.Y. Role of pharmacokinetics and metabolism in drug discovery and development. Pharmacol. Rev. 1997, 49, 403–449. [Google Scholar]
- Alavijeh, M.S.; Palmer, A.M. The pivotal role of drug metabolism and pharmacokinetics in the discovery and development of new medicines. IDrugs 2004, 7, 755–763. [Google Scholar] [PubMed]
- Zhang, Z.P.; Tang, W. Drug metabolism in drug discovery and development. Acta Pharm. Sin. B 2018, 8, 721–732. [Google Scholar] [CrossRef]
- Fang, Z.Z.; Krausz, K.W.; Li, F.; Cheng, J.; Tanaka, N.; Gonzalez, F.J. Metabolic map and bioactivation of the anti-tumour drug noscapine. Br. J. Pharmacol. 2012, 167, 1271–1286. [Google Scholar] [CrossRef]
- ZH, Z.M.; Wang, Y.K.; Yan, D.M.; Fang, J.H.; Xiao, X.R.; Zhang, T.; Cheng, Y.; Xu, K.P.; Li, F. Metabolic profiling of tyrosine kinase inhibitor nintedanib using metabolomics. J. Pharm. Biomed. Anal. 2020, 180, 113045. [Google Scholar] [CrossRef] [PubMed]
- Korhonen, R.; Lahti, A.; Hamalainen, M.; Kankaanranta, H.; Moilanen, E. Dexamethasome inhibits Inducible Nitric-Oxide Synthase Expression and Nitric Oxide Production by Destabilizing mRNA in Lipopolysaccharide-Treated Macrophages. Mol. Pharmacol. 2002, 62, 698–704. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Q.Q.; Shan, H.; Li, X.X.; Gao, H.W.; Yang, S.L. Anti-inflammatory effect and mechanism of verbascoside on LPS-induced RAW 264.7 cells. Chin. Tradit. Herb. Drug. 2020, 51, 4217–4222. [Google Scholar]
- Tian, Y.; Zhou, S.Q.; Takeda, R.; Okazaki, K.; Sekita, M.; Sakamoto, K. Anti-inflammatory activities of amber extract in lipopolysaccharide-induced RAW 264.7 macrophages. Biomed. Pharm. 2021, 141, 111854. [Google Scholar] [CrossRef] [PubMed]




| Metabolite | Molecular Formula | RT (min) | m/z [M + H]+ | Mass Error (ppm) | MS/MS Fragments | Identification | Source |
|---|---|---|---|---|---|---|---|
| Me0 | C15H16O4 | 8.15 | 261.1121 | 0.27 | 243, 231, 217, 189 | Meranzin | P, F, U, H, M |
| Me1 * | C14H14O4 | 6.79 | 247.0965 | −1.13 | 229, 221, 201, 187 | Mer-CH2 | U, F, H, M |
| Me2 * | C15H14O4 | 6.47 | 259.0965 | 3.94 | 243, 231, 217, 201, 189 | Mer-2H | H, M |
| Me2a * | C15H14O4 | 6.85 | 259.0965 | −8.03 | 243, 231, 217, 189 | Mer-2H | H, M |
| Me2b * | C15H14O4 | 7.68 | 259.0965 | −6.49 | 243, 229, 217, 203, 189 | Mer-2H | F, U, H, M |
| Me3 * | C14H14O5 | 6.67 | 263.0914 | 4.20 | 245, 207, 191, 175 | Mer + O-CH2 | U |
| Me4 * | C15H14O5 | 5.43 | 275.0914 | 5.83 | 259, 247, 231, 219, 205 | Mer + CH2 | U |
| Me5 * | C15H16O5 | 6.47 | 277.1070 | −3.05 | 259, 247, 219, 205 | Mer + O | F, H, M |
| Me5a * | C15H16O5 | 6.83 | 277.1070 | 7.78 | 259, 243, 227, 217, 205 | Mer + O | M |
| Me5b * | C15H16O5 | 7.67 | 277.1070 | 4.90 | 259, 233, 217, 205 | Mer + O | F, U, H, M |
| Me5c * | C15H16O5 | 7.84 | 277.1070 | −7.02 | 261, 243, 215, 205, 191 | Mer + O | U |
| Me6 * | C15H18O5 | 6.29 | 279.1227 | 9.70 | 261, 243, 217, 207, 189 | Mer + H2O | H |
| Me6a * | C15H18O5 | 7.70 | 279.1227 | −7.14 | 261, 235, 221, 207, 189 | Mer + H2O | H, M |
| Me7 * | C15H14O6 | 6.68 | 291.0863 | 0.65 | 275, 263, 245, 217, 203 | Mer + O + CH2 | U |
| Me8 * | C15H18O6 | 5.62 | 295.1176 | 7.42 | 259, 231, 221, 203, 189 | Mer + 2O + 2H | H |
| Me9 * | C18H21NO5S | 5.36 | 380.1162 | 1.24 | 334, 305 | Mer + Cysteine | U |
| Me9a * | C18H21NO5S | 5.84 | 380.1162 | 3.08 | 364, 334, 291 | Mer + Cysteine | U |
| Me10 * | C20H23NO7S | 7.11 | 422.1268 | 0.02 | 404, 380, 376, 362, 334 | Mer + N-acetylcysteine | F, U |
| Me10a * | C20H23NO7S | 7.35 | 422.1268 | 7.02 | 404, 380, 358, 376, 334 | Mer + N-acetylcysteine | F |
| Me11 * | C21H24O10 | 6.12 | 437.1442 | 4.08 | 359, 261, 243, 189 | Mer + Gluc | U |
| Me12 * | C21H24O11 | 4.95 | 453.1391 | 2.36 | 437, 419, 395, 291, 237 | Mer + O + Gluc | U |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cheng, Y.; Ma, X.; Zhao, Q.; Wang, C.; Yan, D.; Li, F. Metabolic Profile of C-Prenyl Coumarins Using Mass Spectrometry-Based Metabolomics. Molecules 2021, 26, 6558. https://doi.org/10.3390/molecules26216558
Cheng Y, Ma X, Zhao Q, Wang C, Yan D, Li F. Metabolic Profile of C-Prenyl Coumarins Using Mass Spectrometry-Based Metabolomics. Molecules. 2021; 26(21):6558. https://doi.org/10.3390/molecules26216558
Chicago/Turabian StyleCheng, Yan, Xiaofang Ma, Qi Zhao, Chunyan Wang, Dongmei Yan, and Fei Li. 2021. "Metabolic Profile of C-Prenyl Coumarins Using Mass Spectrometry-Based Metabolomics" Molecules 26, no. 21: 6558. https://doi.org/10.3390/molecules26216558
APA StyleCheng, Y., Ma, X., Zhao, Q., Wang, C., Yan, D., & Li, F. (2021). Metabolic Profile of C-Prenyl Coumarins Using Mass Spectrometry-Based Metabolomics. Molecules, 26(21), 6558. https://doi.org/10.3390/molecules26216558





