Construction of a Tandem Repeat Peptide Sequence with Pepsin Cutting Sites to Produce Recombinant α-Melanocyte-Stimulating Hormone
Abstract
:1. Introduction
2. Results and Discussion
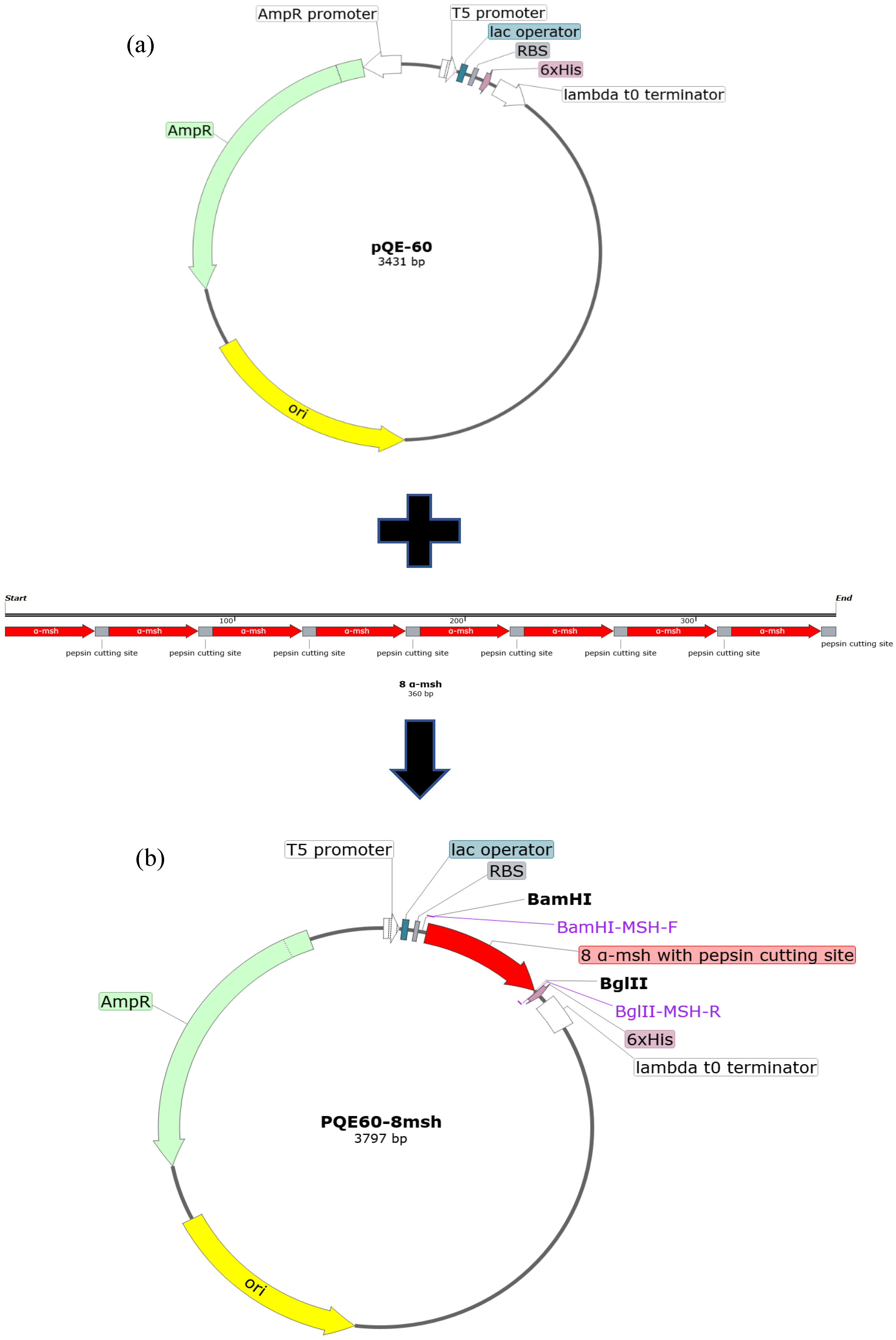
2.1. Construction, Expression, and Extraction of 8 α-MSH
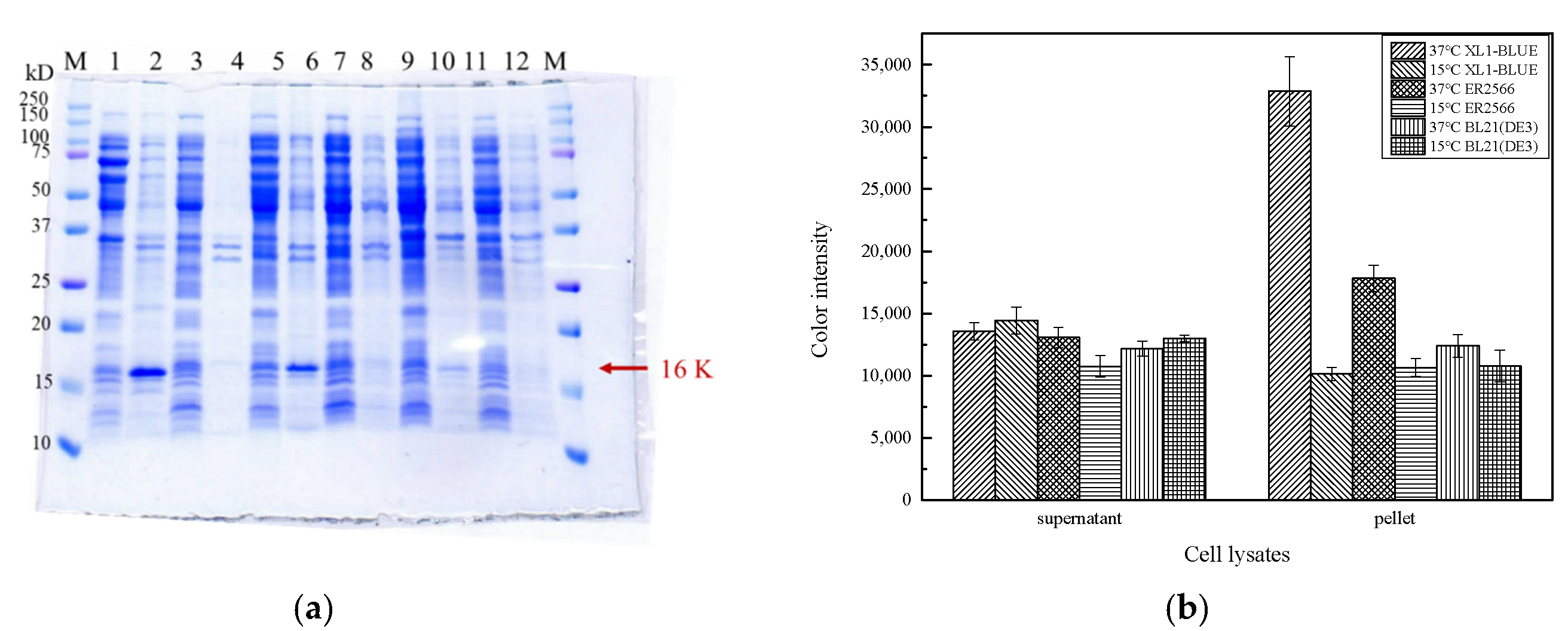
2.2. Identification of 8 α-MSH Fusion Protein
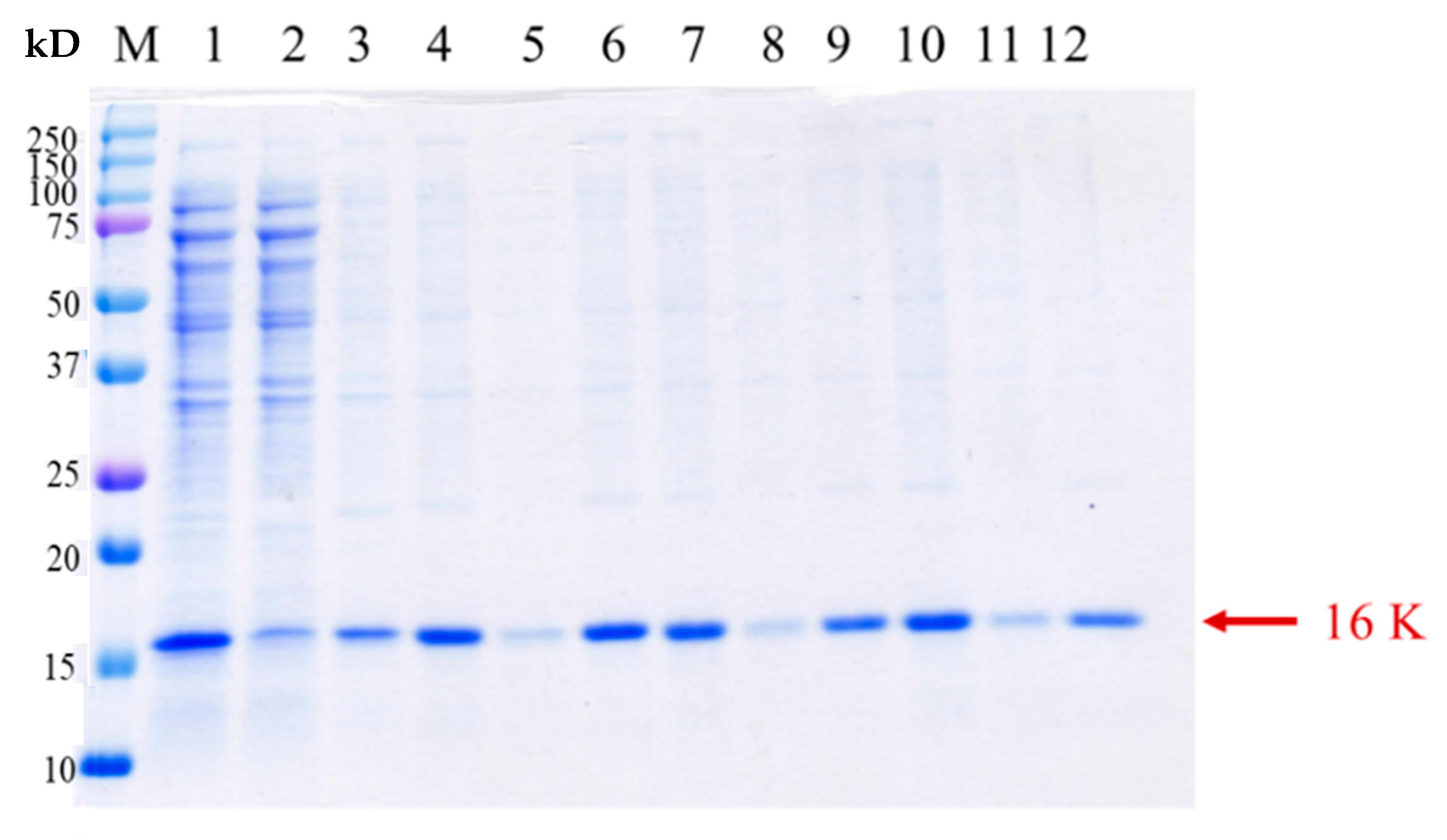
2.3. Pepsin Cleavage Reaction
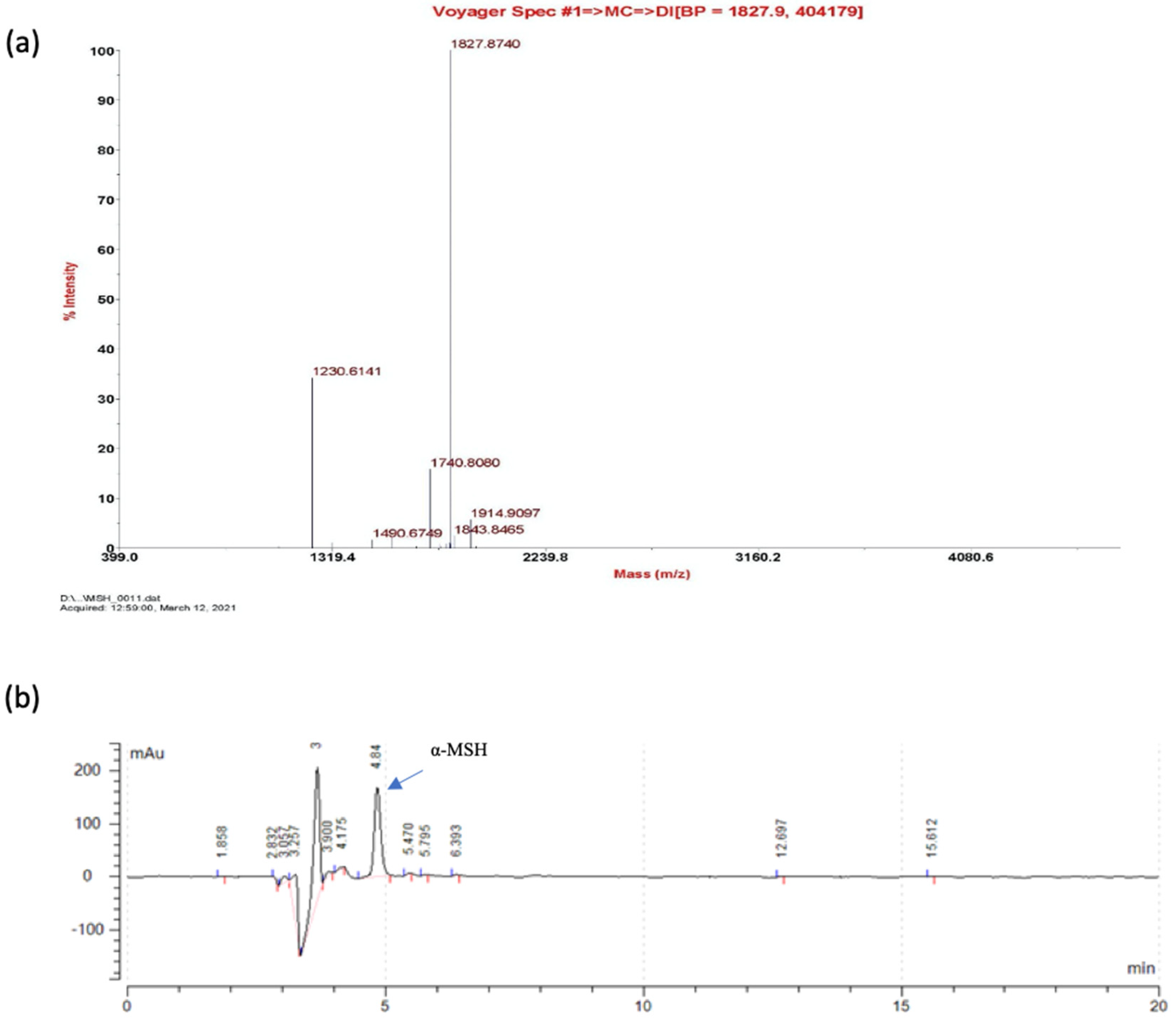
2.4. Identification of α-MSH after Pepsin Cleavage
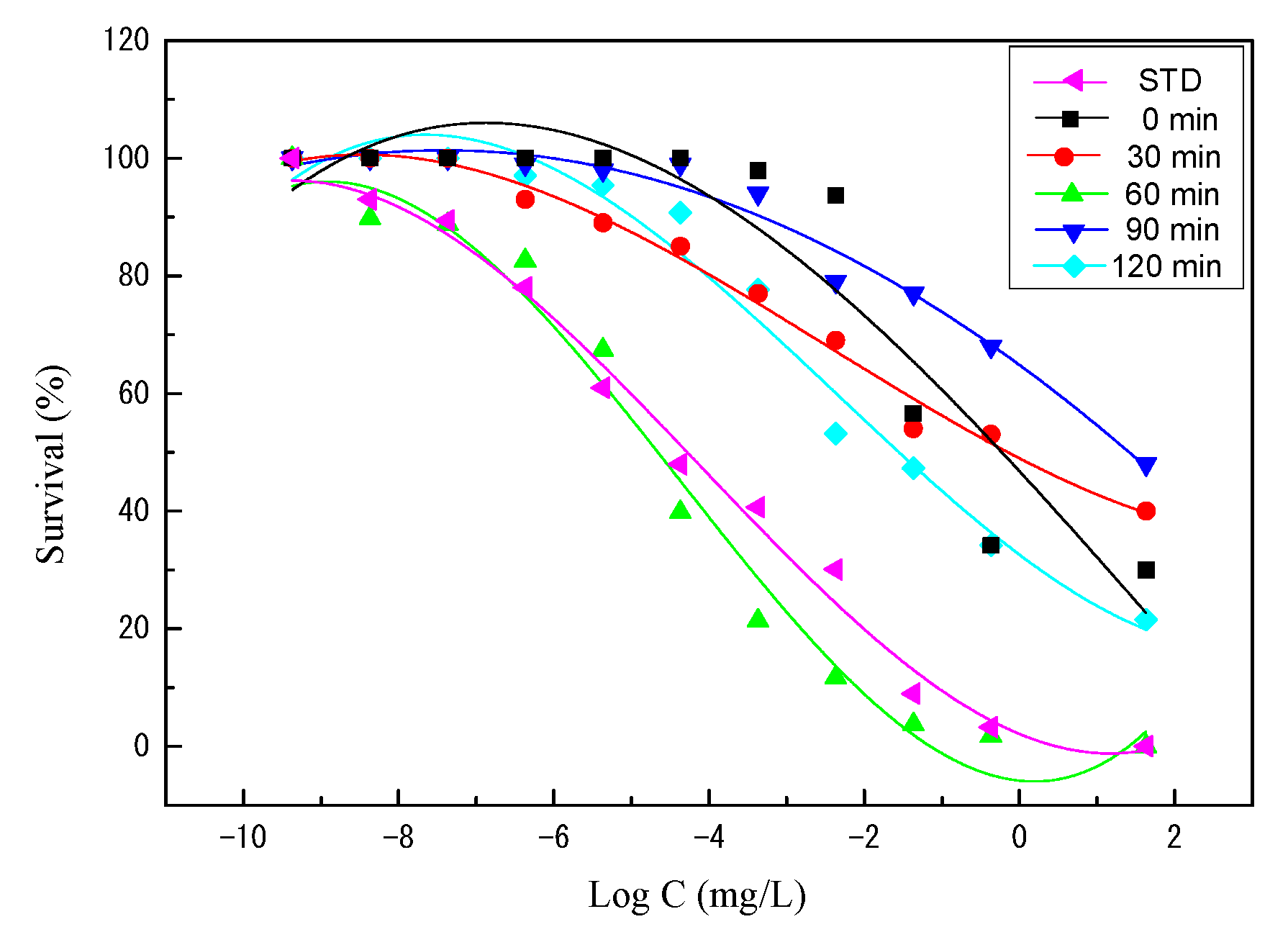
2.5. Antimicrobial Activity of α-MSH
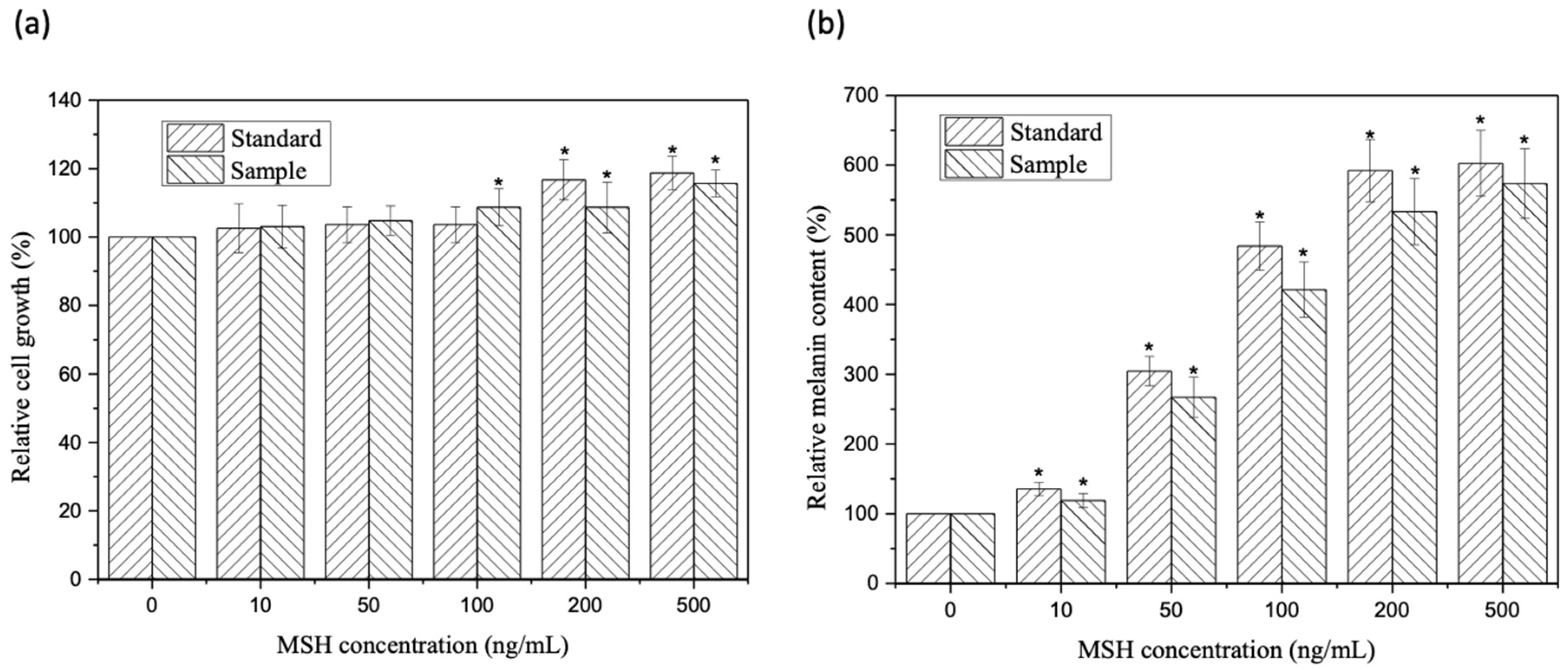
2.6. Melanin Stimulation Assay of α-MSH
2.7. Comparison of Antimicrobial Peptide Production
3. Materials and Methods
3.1. Bacterial Strains, Synthetic 8 α-MSH Plasmids, and Chemicals
3.2. E. coli Cultivation
3.3. Pepsin Cleavage
3.4. Assays
3.5. Antimicrobial Activity Assays
3.6. α-MSH Activity Assay
3.7. Statistical Analysis
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
Sample Availability
References
- Boman, H.G. Peptide antibiotics and their role in innate immunity. Annu. Rev. Immunol. 1995, 13, 61–92. [Google Scholar] [CrossRef]
- Sahl, H.G. Gene-encoded antibiotics made in bacteria. Ciba Found. Symp. 1994, 186, 27–53. [Google Scholar]
- Cammue, B.P.; De Bolle, M.F.; Schoofs, H.M.; Terras, F.R.; Thevissen, K.; Osborn, R.W.; Rees, S.B.; Broekaert, W.F. Gene-encoded antimicrobial peptides from plants. Ciba Found. Symp. 1994, 186, 91–106. [Google Scholar] [PubMed]
- Hultmark, D.; Steiner, H.; Rasmuson, T.; Boman, H.G. Insect immunity. Purification and properties of three inducible bactericidal proteins from hemolymph of immunized pupae of Hyalophora cecropia. Eur. J. Biochem. 1980, 106, 7–16. [Google Scholar] [CrossRef] [PubMed]
- Steiner, H.; Hultmark, D.; Engstrom, A.; Bennich, H.; Boman, H.G. Sequence and specificity of two antibacterial proteins involved in insect immunity. Nature 1981, 292, 246–248. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, T.; Hammami, R. Recent insights into structure–function relationships of antimicrobial peptides. J. Food Biochem. 2019, 43, e12546. [Google Scholar] [CrossRef] [Green Version]
- Bi, J.; Tian, C.; Jiang, J.; Zhang, G.L.; Hao, H.; Hou, H.M. Antibacterial Activity and Potential Application in Food Packaging of Peptides Derived from Turbot Viscera Hydrolysate. J. Agric. Food Chem. 2020, 68, 9968–9977. [Google Scholar] [CrossRef]
- Mylonakis, E.; Podsiadlowski, L.; Muhammed, M.; Vilcinskas, A. Diversity, evolution and medical applications of insect antimicrobial peptides. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2016, 371, 20150290. [Google Scholar] [CrossRef] [Green Version]
- Kang, S.-J.; Kim, D.-H.; Tsogbadrakh, M.-O.; Lee, B.-J. Antimicrobial peptides: Their physicochemical properties and therapeutic application. Arch. Pharm. Res. 2012, 35, 409–413. [Google Scholar] [CrossRef]
- Tang, S.S.; Prodhan, Z.H.; Biswas, S.K.; Le, C.F.; Sekaran, S.D. Antimicrobial peptides from different plant sources: Isolation, characterisation, and purification. Phytochem 2018, 154, 94–105. [Google Scholar] [CrossRef]
- Nawrot, R.; Barylski, J.; Nowicki, G.; Broniarczyk, J.; Buchwald, W.; Gozdzicka-Jozefiak, A. Plant antimicrobial peptides. Folia Microbiol. (Praha) 2014, 59, 181–196. [Google Scholar] [CrossRef] [Green Version]
- Li, C.; Blencke, H.M.; Paulsen, V.; Haug, T.; Stensvag, K. Powerful workhorses for antimicrobial peptide expression and characterization. Bioeng. Bugs 2010, 1, 217–220. [Google Scholar] [CrossRef] [Green Version]
- Parachin, N.S.; Mulder, K.C.; Viana, A.A.; Dias, S.C.; Franco, O.L. Expression systems for heterologous production of antimicrobial peptides. Peptides 2012, 38, 446–456. [Google Scholar] [CrossRef]
- Souza, P.F.N.; Marques, L.S.M.; Oliveira, J.T.A.; Lima, P.G.; Dias, L.P.; Neto, N.A.S.; Lopes, F.E.S.; Sousa, J.S.; Silva, A.F.B.; Caneiro, R.F.; et al. Synthetic antimicrobial peptides: From choice of the best sequences to action mechanisms. Biochimie 2020, 175, 132–145. [Google Scholar] [CrossRef]
- Li, C.; Haug, T.; Styrvold, O.B.; Jorgensen, T.O.; Stensvag, K. Strongylocins, novel antimicrobial peptides from the green sea urchin, Strongylocentrotus droebachiensis. Dev. Comp. Immunol. 2008, 32, 1430–1440. [Google Scholar] [CrossRef]
- Gantz, I.; Fong, T.M. The melanocortin system. Am. J. Physiol. Endocrinol. Metab. 2003, 284, E468–E474. [Google Scholar] [CrossRef] [PubMed]
- Raffin-Sanson, M.L.; de Keyzer, Y.; Bertagna, X. Proopiomelanocortin, a polypeptide precursor with multiple functions: From physiology to pathological conditions. Eur. J. Endocrinol. 2003, 149, 79–90. [Google Scholar] [CrossRef] [Green Version]
- Bertolini, A.; Tacchi, R.; Vergoni, A.V. Brain effects of melanocortins. Pharmacol. Res. 2009, 59, 13–47. [Google Scholar] [CrossRef] [PubMed]
- Singh, J.; Mumtaz, S.; Joshi, S.; Mukhopadhyay, K. In Vitro and Ex Vivo Efficacy of Novel Trp-Arg Rich Analogue of alpha-MSH against Staphylococcus aureus. ACS Omega 2020, 5, 3258–3270. [Google Scholar] [CrossRef]
- Thody, A.J. alpha-MSH and the regulation of melanocyte function. Ann. N. Y. Acad. Sci. 1999, 885, 217–229. [Google Scholar] [CrossRef] [PubMed]
- Catani, A.; Lonati, C.; Sordi, A.; Carlin, A.; Leonardi, P.; Gatti, S. The Melanocortin System in Control of Inflammation. Sci. World J. 2010, 10, 1840–1853. [Google Scholar] [CrossRef] [PubMed]
- Madhuri; Shireen, T.; Venugopal, S.K.; Ghosh, D.; Gadepalli, R.; Dhawan, B.; Mukhopadhyay, K. In vitro antimicrobial activity of alpha-melanocyte stimulating hormone against major human pathogen Staphylococcus aureus. Peptides 2009, 30, 1627–1635. [Google Scholar] [CrossRef] [PubMed]
- Singh, M.; Mukhopadhyay, K. C-terminal amino acids of alpha-melanocyte-stimulating hormone are requisite for its antibacterial activity against Staphylococcus aureus. Antimicrob. Agents Chemother. 2011, 55, 1920–1929. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carotenuto, A.; Saviello, M.R.; Auriemma, L.; Campiglia, P.; Catania, A.; Novellino, E.; Grieco, P. Structure-function relationships and conformational properties of alpha-MSH(6-13) analogues with candidacidal activity. Chem. Biol. Drug Des. 2007, 69, 68–74. [Google Scholar] [CrossRef]
- Cutuli, M.; Cristiani, S.; Lipton, J.M.; Catania, A. Antimicrobial effects of alpha-MSH peptides. J. Leukoc. Biol. 2000, 67, 233–239. [Google Scholar] [CrossRef]
- Yang, Y.C.S.; Hruby, V.J.; Heward, C.B.; Hadley, M.E. Synthesis of α-and β-melanocyte stimulating hormones. Int. J. Peptide Protein Res. 1980, 15, 130–138. [Google Scholar] [CrossRef]
- Hruby, V.J.; Sawyer, T.K.; Yang, Y.C.; Bregman, M.D.; Hadley, M.E.; Heward, C.B. Synthesis and structure-function studies of melanocyte stimulating hormone analogues modified in the 2 and 4(7) positions: Comparison of activities on frog skin melanophores and melanoma adenylate cyclase. J. Med. Chem. 1980, 23, 1432–1437. [Google Scholar] [CrossRef]
- Florkin, M. Discovery of pepsin by Theodor Schwann. Rev. Med. Liege 1957, 12, 139–144. [Google Scholar] [PubMed]
- Burrell, M.M. Enzymes of Molecular Biology; Humana Press: Totowa, NJ, USA, 1993. [Google Scholar]
- Rosiello, A.P.; Essigmann, J.M.; Wogan, G.N. Rapid and accurate determination of the median lethal dose (LD50) and its error with a small computer. J. Toxicol. Environ. Health 1977, 3, 797–809. [Google Scholar] [CrossRef] [PubMed]
- Wu, L.C.; Chang, L.H.; Chen, S.H.; Fan, N.C.; Ho, J.A. Antioxidant activity and melanogenesis inhibitory effect of the acetonic extract of Osmanthus fragrans: A potential natural and functional food flavor additive. LWT-Food Sci. Technol. 2009, 42, 1513–1519. [Google Scholar] [CrossRef]
- Strober, W. Trypan blue exclusion test of cell viability. Curr. Protoc. Immunol. 2015, 111, A3.B.1–A3.B.3. [Google Scholar] [CrossRef] [PubMed]
- Fang, Y.T.; Lai, W.S.; Liu, J.H.; Liu, Y.C. Enhanced cecropin B2 production via chitin-binding domain and intein self-cleavage system. Biotechnol. Appl. Biochem. 2019, 66, 209–215. [Google Scholar] [CrossRef]
- Fang, Y.T.; Li, S.Y.; Hu, N.J.; Yang, J.; Liu, J.H.; Liu, Y.C. Study on Cecropin B2 Production via Construct Bearing Intein Oligopeptide Cleavage Variants. Molecules 2020, 25, 1005. [Google Scholar] [CrossRef] [Green Version]
- Wei, X.; Wu, R.; Zhang, L.; Ahmad, B.; Si, D.; Zhang, R. Expression, purification, and characterization of a novel hybrid peptide with potent antibacterial activity. Molecules 2018, 23, 1491. [Google Scholar] [CrossRef] [Green Version]
- Feng, X.; Liu, C.; Guo, J.; Song, X.; Li, J.; Xu, W.; Li, Z. Recombinant expression, purification, and antimicrobial activity of a novel hybrid antimicrobial peptide LFT33. Appl. Microbiol. Biotechnol. 2012, 95, 1191–1198. [Google Scholar] [CrossRef]
- Montfort-Gardeazabal, J.M.; Balderas-Renteria, I.; Casillas-Vega, N.G.; Zarate, X. Expression and purification of the antimicrobial peptide Bin1b in Escherichia coli tagged with the fusion proteins CusF3H+ and SmbP. Protein Expr. Purif. 2021, 178, 105784. [Google Scholar] [CrossRef]
- Xu, X.; Jin, F.; Yu, X.; Ji, S.; Wang, J.; Cheng, H.; Wang, C.; Zhang, W. Expression and purification of a recombinant antibacterial peptide, cecropin, from Escherichia coli. Protein Expr. Purif. 2007, 53, 293–301. [Google Scholar] [CrossRef] [PubMed]
- Jonsson, M. Isoelectric spectra of native and base denatured crystallized swine pepsin. Acta Chem. Scand. 1972, 26, 3435–3440. [Google Scholar] [CrossRef] [Green Version]
- Knowles, J.R.; Sharp, H.; Greenwell, P. The pH-dependence of the binding of competitive inhibitors to pepsin. Biochem. J. 1969, 113, 343–351. [Google Scholar] [CrossRef] [Green Version]
- Laemmli, U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 1970, 227, 680–685. [Google Scholar] [CrossRef] [PubMed]
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. NIH image to ImageJ: 25 years of image analysis. Nat. Methods 2012, 9, 671–675. [Google Scholar] [CrossRef] [PubMed]
- Barry, A.L.; Coyle, M.B.; Thornsberry, C.; Gerlach, E.H.; Hawkinson, R.W. Methods of measuring zones of inhibition with the Bauer-Kirby disk susceptibility test. J. Clin. Microbiol. 1979, 10, 885–889. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- DePass, L.R. Alternative approaches in median lethality (LD50) and acute toxicity testing. Toxicol. Lett. 1989, 49, 159–170. [Google Scholar] [CrossRef]
- Lee, Y.S.; Kim, H.K.; Lee, K.J.; Jeon, H.W.; Cui, S.; Lee, Y.M.; Moon, B.J.; Kim, Y.H.; Lee, Y.S. Inhibitory effect of glyceollin isolated from soybean against melanogenesis in B16 melanoma cells. BMB Rep. 2010, 43, 461–467. [Google Scholar] [CrossRef]

| Name | Sequence (5′-3′) | Restriction Enzyme |
|---|---|---|
| BamHI-msh-F a | CGC GGA TCC TGG CGC GCC GAG CTC TCT TAT AGC | BamHI |
| BglII-msh-R | GGA AGA TCT AAA ACC CAC TG | BglII |
| Host | PCR | Restriction Site |
|---|---|---|
| DH5 | − a | − |
| JM109 (DE3) | + | − |
| BW25113 | + | − |
| EPI300 | + | − |
| W3110 | + | − |
| HB101 | + | − |
| ER2566 | + | + |
| XL1-Blue | + | + |
| BL21 (DE3) | + | + |
| Reaction Time (min) | Group I Buffer a | Group II Pepsin b | Group III 8α-MSH with Pepsin c |
|---|---|---|---|
| 0 | − d | − | + |
| 30 | − | − | + |
| 60 | − | − | ++ |
| 90 | − | − | + |
| 120 | − | − | − |
| Reaction pH | Group I Buffer a | Group II Pepsin b | Group III 8α-MSH with Pepsin c |
|---|---|---|---|
| 2 | − d | − | ++ |
| 4 | − | − | + |
| 7 | − | − | − |
| Cleavage Time | LD0 | LD20 | LD50 | LD100 |
|---|---|---|---|---|
| Standard | a 6.35 × 10−5 | 2.00 × 10−3 | 1.28 × 10−2 | 1.22 × 101 |
| 0 | 8.93 × 10−4 | 1.54 × 10−1 | 2.46 × 100 | 6.82 × 104 |
| 30 | 1.23 × 10−4 | 3.60 × 10−2 | 7.66 × 10−1 | 6.07 × 104 |
| 60 | 3.85 × 10−5 | 1.15 × 10−3 | 7.19 × 10−3 | 6.16 × 100 |
| 90 | 4.89 × 10−4 | 3.59 × 10−1 | 1.25 × 101 | 6.21 × 106 |
| 120 | 2.62 × 10−4 | 2.66 × 10−2 | 3.20 × 10−1 | 3.10 × 103 |
| Antimicrobial Peptide | Molecular Weight | Cultivation Conditions | Post-Treatments | Weight Yield (mg/L) | Molar Yield (μmol/L) | Ref. |
|---|---|---|---|---|---|---|
| Cecropin B2 | 4 kDa | OD600 = 0.8 0.1 mM IPTG 25 °C, 16 h | 1. Intein cleavage 2. Centrifugal filtration 3. Column separation | 58.7 | 14.68 | [33,34] |
| Cecropin A (C–L) | 2.9 kDa | OD600 = 0.6~0.8 1.5 mM IPTG 37 °C, 5 h | 1. Ni-IMAC 2. SUMO cleavage | 17.54 | 6.05 | [35] |
| LFT33 (LfcinB-thanatin) | 4.2 kDa | OD600 = 0.6 0.2 mM IPTG 30 °C, 4 h | 1. Ni-IMAC 2. HPLC | 0.5 | 0.12 | [36] |
| Bin1b | 5.2 kDa | OD600 = 0.5 1 mM IPTG 25 °C, 16 h | Ni-IMAC | 4.4 | 0.85 | [37] |
| Mdmcec | 4 kDa | OD600 = 0.8 0.4 mM IPTG 25 °C, 9 h | 1. Ni-IMAC 2. Centrifugal filtration 3. HPLC | 11.2 | 2.80 | [38] |
| α-MSH | 1.8 kDa | OD600 = 0.4 0.9 mM IPTG 37 °C, 16 h | Pepsin cleavage | 42.9 | 23.83 | This study |
| Strain or Plasmid | Genotype and Relevant Characteristics | Source |
|---|---|---|
| BL21 (DE3) | F− ompT gal dcm lon hsdSB (rB−mB−) λ− (DE3 [lacI lacUV5-T7 gene ind1 sam7 nin5]) | Novagen, WI, USA |
| BW25113 | F− DE(araD-araB)567 lacZ4787(del)::rrnB-3 LAM− rph-1 DE(rhaD-rhaB)568 hsdR514 | Coli Genetics Stock Center, New Haven, CT, USA |
| DH5α | F− endA1 glnV44 thi-1 recA1 relA1 gyrA96 deoR nupG Φ80dlacZΔM15 Δ(lacZYA-argF)U169 hsdR17(rK−mK+) λ− | Novagen, WI, USA |
| ER2566 | F− λ− fhuA2 [lon] ompT lacZ::T7 gene1 gal sulA11 Δ(mcrC-mrr)114::IS10R(mcr-73::miniTn10-TetS)2 R(zgb-210::Tn10)(TetS) endA1[dcm] | New England Biolabs, Ipswich, MA, USA |
| EPI300 | F´[proAB+ lacIq lacZΔM15 zzf::Tn10(TetR)] fhuA2 glnV Δ(lac-proAB) thi-1 Δ(hsdS-mcrB)5 | Lucigen, WI, USA |
| HB101 | F−, thi-1, hsdS20 (rB–, mB–), supE44, recA13, ara-14, leuB6, proA2, lacY1, galK2, rpsL20 (strr), xyl-5, mtl-1 | Promega, WI, USA |
| JM109 (DE3) | endA1recA1 gyrA96 hsdR17 (rk−, rk+) relA1 supE44 λ− Δ(lac-proAB) [F´ traD36 proAB lacIq ZΔM15], lDE3 | Promega, WI, USA |
| W3110 | F− λ− rph-1 INV(rrnD, rrnE) | New England Biolabs, Ipswich, MA, USA |
| XL1-Blue | recA1 endA1 gyrA96 thi-1 hsdR17 supE44 relA1 lac [F´ proAB lacIq Z∆M15 Tn10 (Tetr)] | Agilent, Santa Clara, CA, USA |
| pQE60 | bacterial vectors for inducible expression of N-terminally T5-tagged protein | Addgene, Teddington, UK |
| pUC19 | bacterial vectors with lac promoter and ampicillin resistant marker | Addgene, Teddington, UK |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jiang, D.-L.; Yao, C.-L.; Hu, N.-J.; Liu, Y.-C. Construction of a Tandem Repeat Peptide Sequence with Pepsin Cutting Sites to Produce Recombinant α-Melanocyte-Stimulating Hormone. Molecules 2021, 26, 6207. https://doi.org/10.3390/molecules26206207
Jiang D-L, Yao C-L, Hu N-J, Liu Y-C. Construction of a Tandem Repeat Peptide Sequence with Pepsin Cutting Sites to Produce Recombinant α-Melanocyte-Stimulating Hormone. Molecules. 2021; 26(20):6207. https://doi.org/10.3390/molecules26206207
Chicago/Turabian StyleJiang, Dai-Lin, Chao-Ling Yao, Nien-Jen Hu, and Yung-Chuan Liu. 2021. "Construction of a Tandem Repeat Peptide Sequence with Pepsin Cutting Sites to Produce Recombinant α-Melanocyte-Stimulating Hormone" Molecules 26, no. 20: 6207. https://doi.org/10.3390/molecules26206207
APA StyleJiang, D.-L., Yao, C.-L., Hu, N.-J., & Liu, Y.-C. (2021). Construction of a Tandem Repeat Peptide Sequence with Pepsin Cutting Sites to Produce Recombinant α-Melanocyte-Stimulating Hormone. Molecules, 26(20), 6207. https://doi.org/10.3390/molecules26206207









