Dynamic Preference for NADP/H Cofactor Binding/Release in E. coli YqhD Oxidoreductase
Abstract
1. Introduction
2. Results
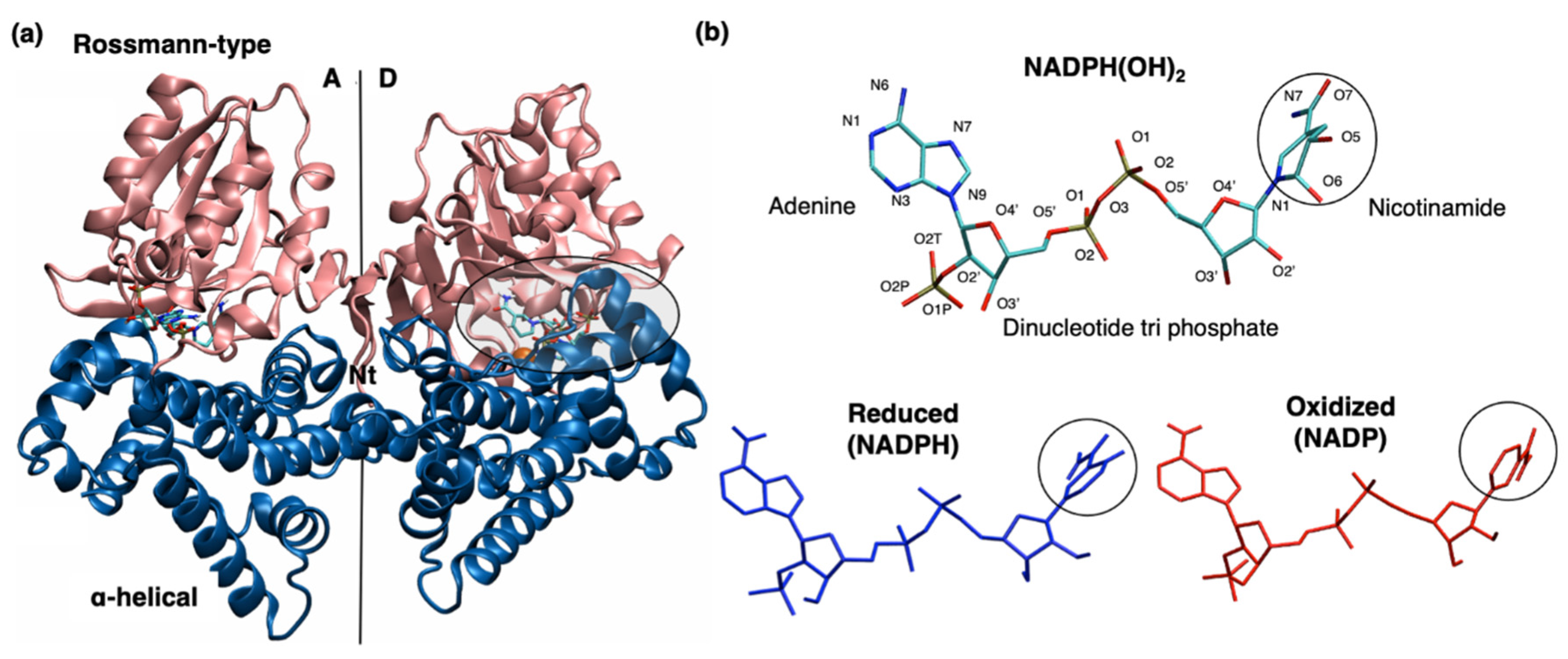
2.1. Structural and Dynamical Properties of YqhD Enzyme
2.1.1. YqhD Dimer
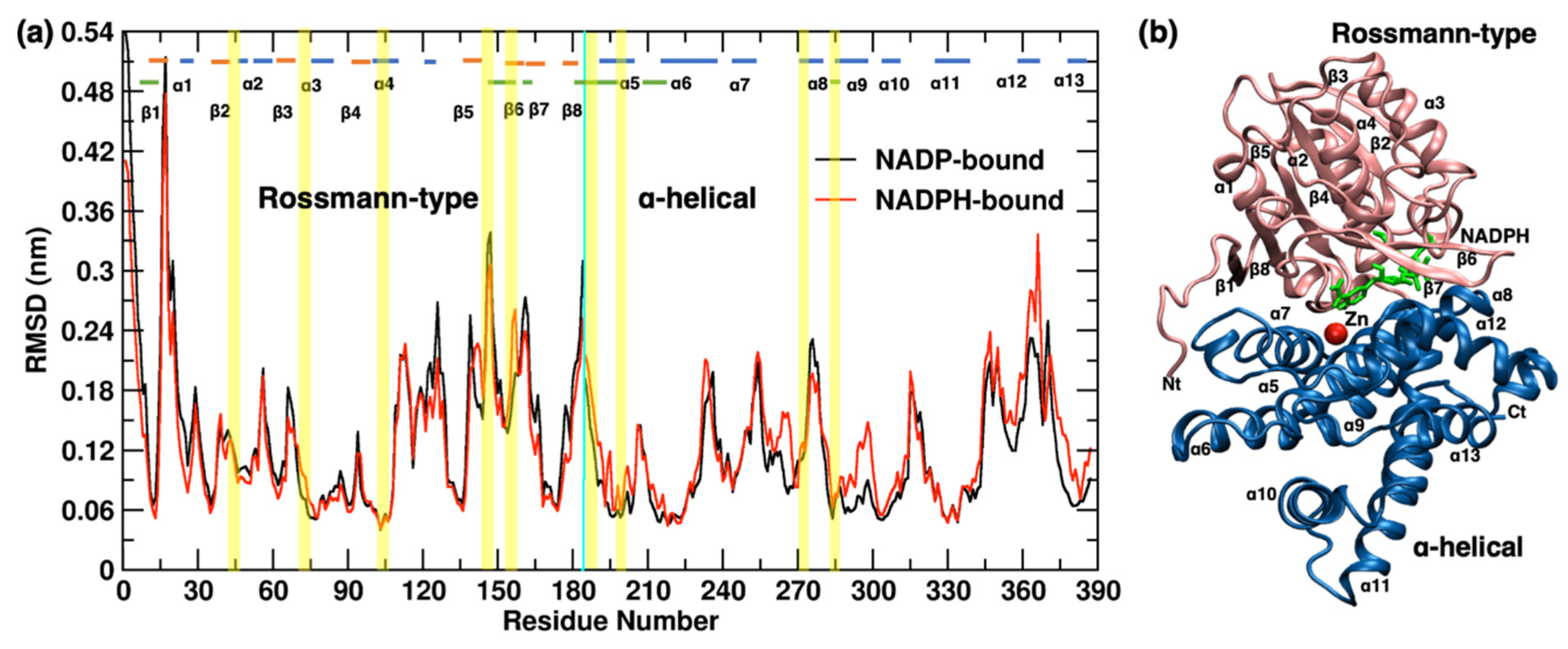
2.1.2. YqhD Domain
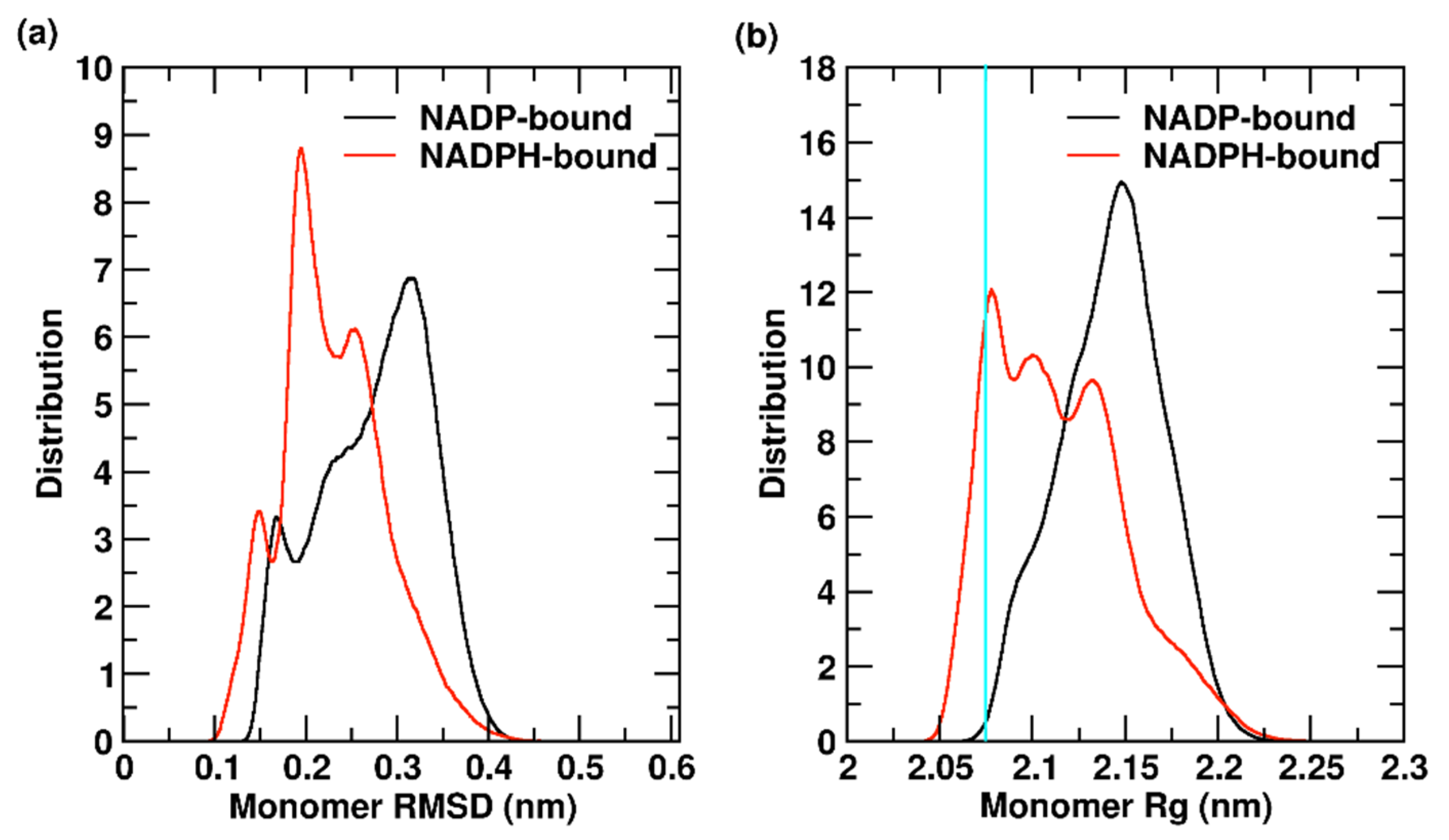
2.1.3. YqhD Monomer
2.2. Hydrogen Bonding in YqhD Enzyme
2.2.1. Inter-Monomer Hydrogen Bonding
2.2.2. Interdomain Hydrogen Bonding
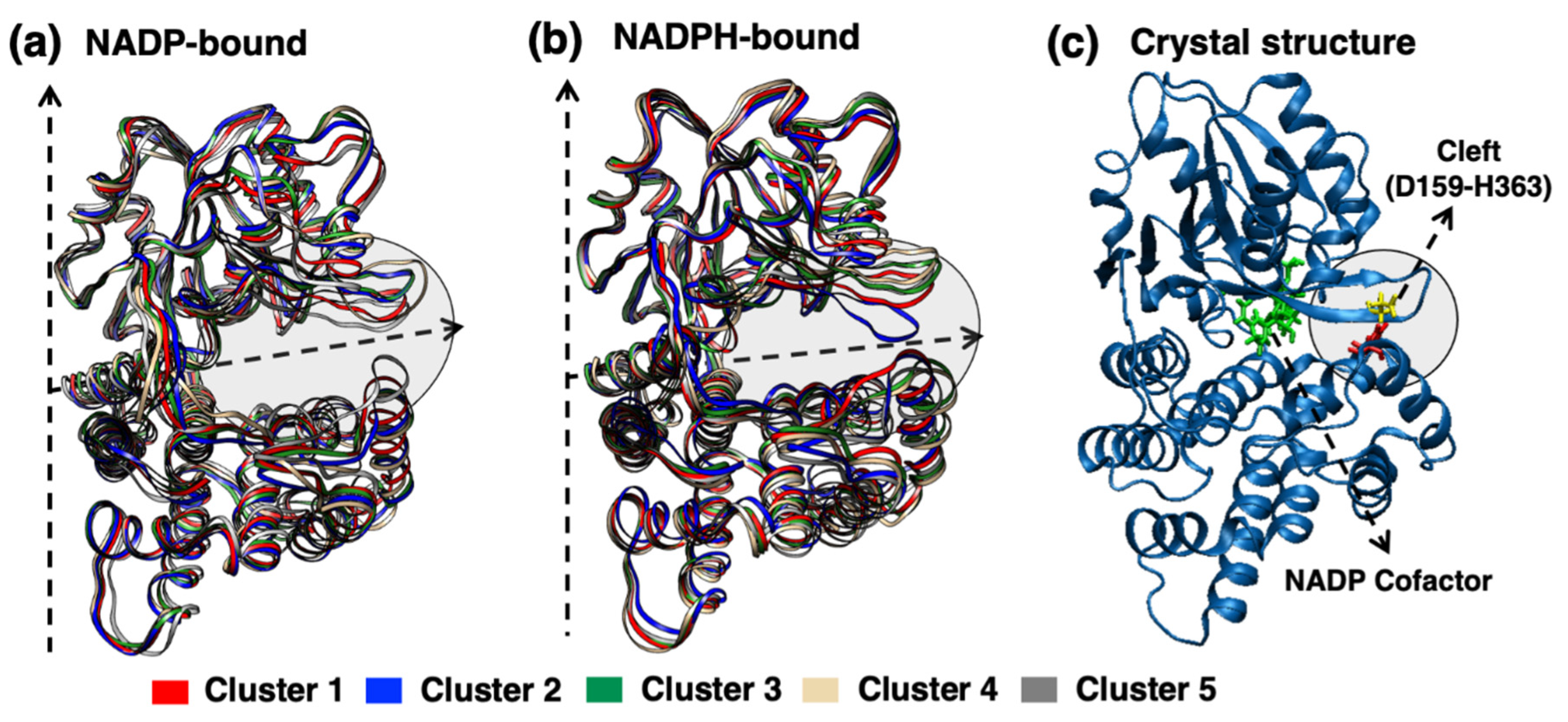
2.3. Cluster Analysis of YqhD Enzyme
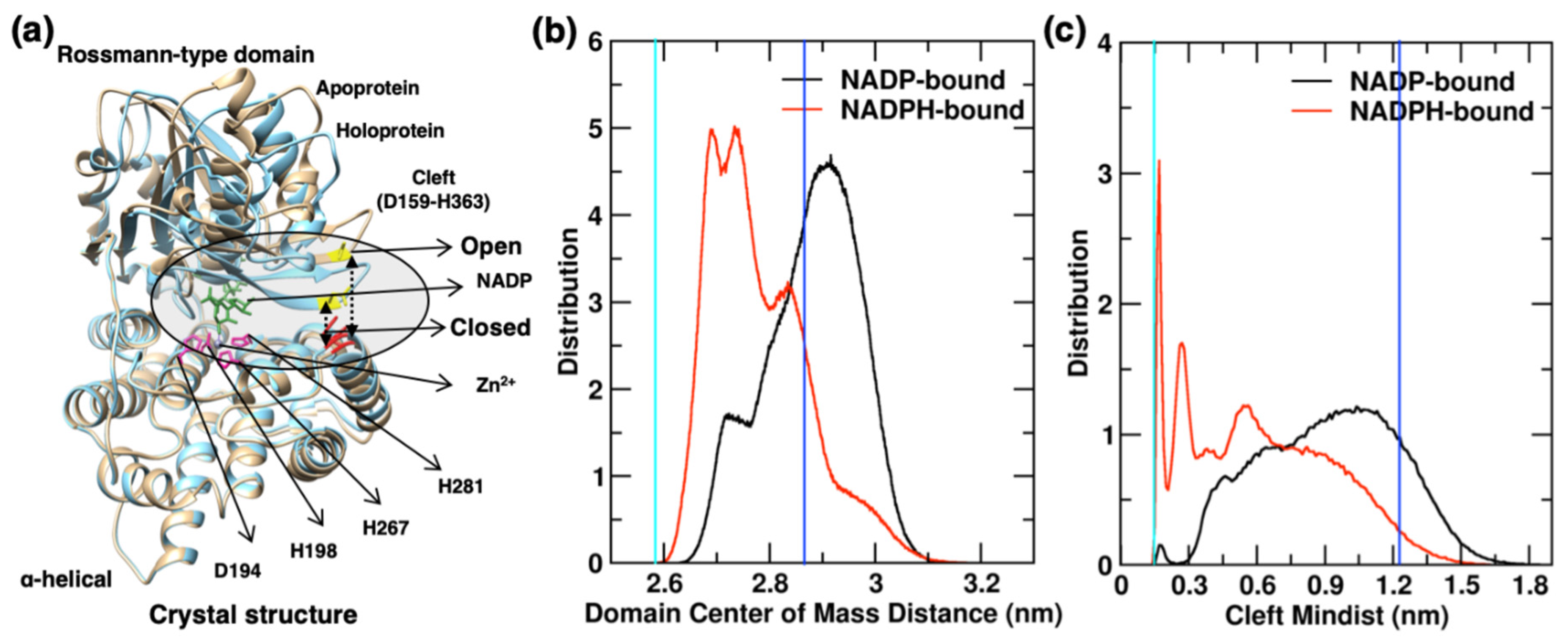
2.4. Interdomain Cleft
2.4.1. Interdomain Opening-Closing Cleft Dynamics
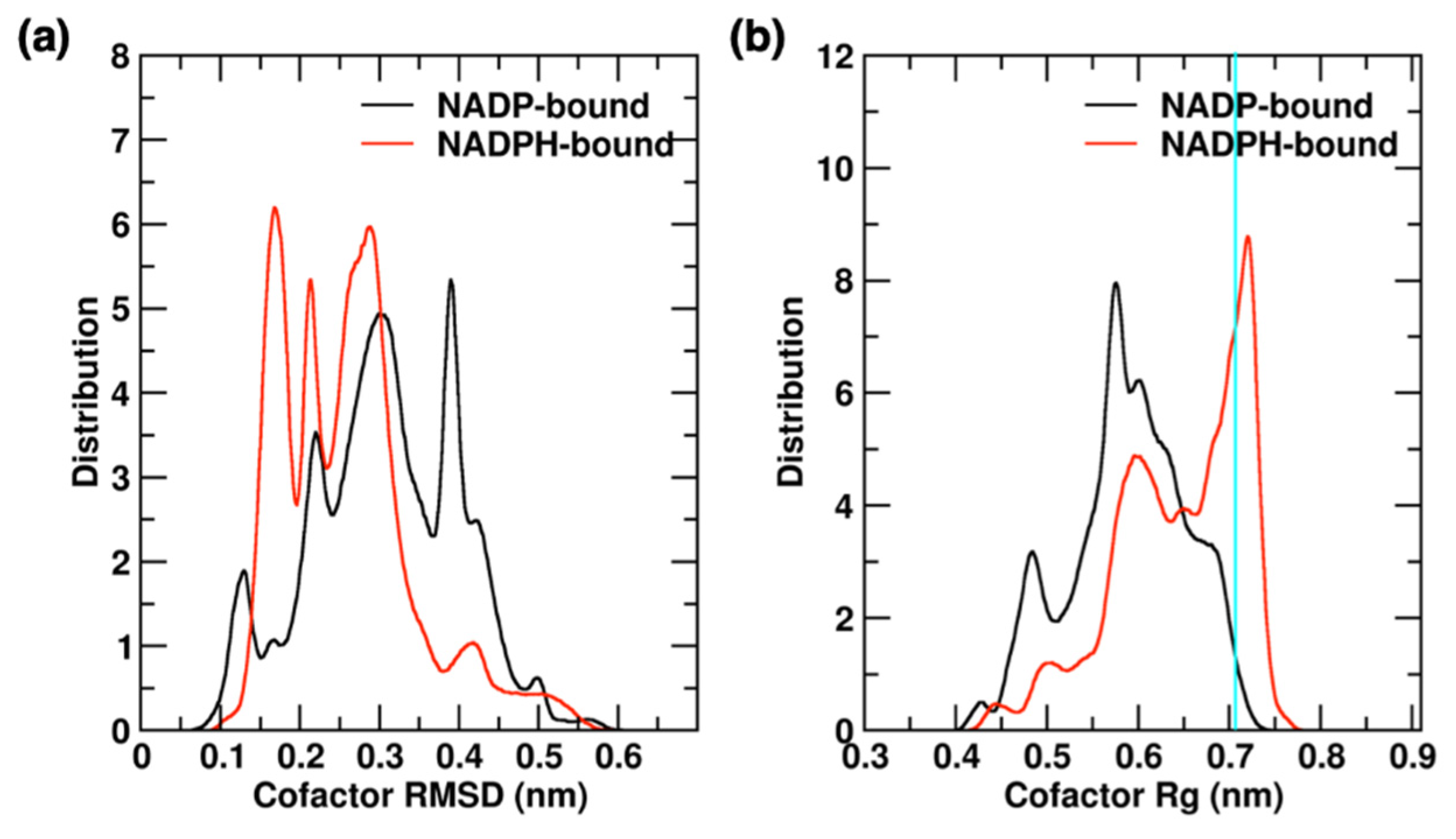
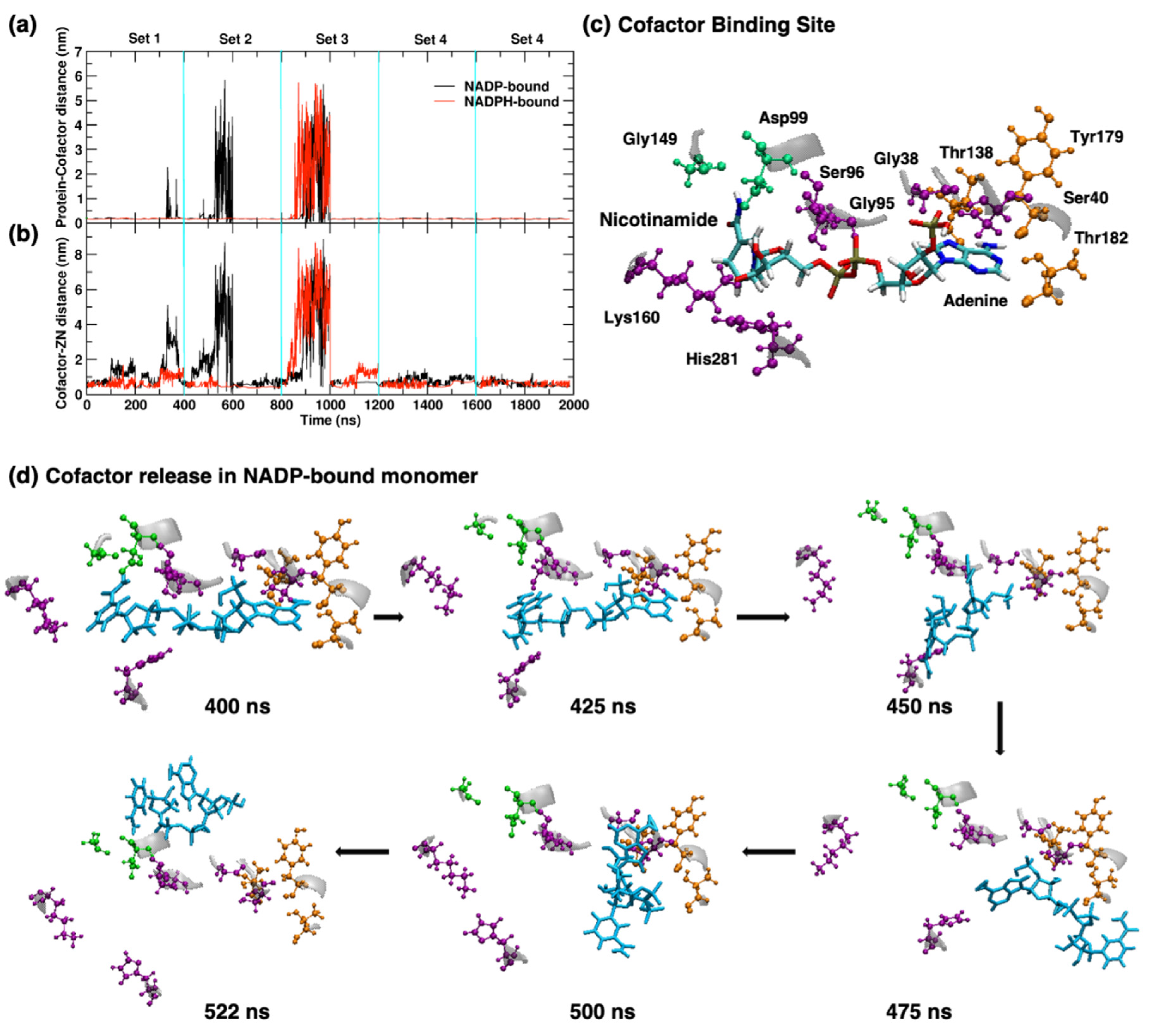
2.4.2. Cofactor Binding and Release in YqhD Enzyme
3. Discussion and Conclusions
4. Materials and Methods
4.1. Starting Coordinates
4.2. Modeling of NADP/H Cofactor
4.3. Molecular Dynamics Simulations
4.4. Cluster Analysis
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Emptage, M.; Haynie, S.L.; Laffend, L.A.; Pucci, J.P.; Whited, G. Process for the Biological Production of 1,3-Propanediol with High Titer. U.S. Patent 6,514,733B1, 4 February 2003. [Google Scholar]
- Jarboe, L.R. YqhD: A broad-substrate range aldehyde reductase with various applications in production of biorenewable fuels and chemicals. Appl. Environ. Microbiol. 2011, 89, 249–257. [Google Scholar] [CrossRef] [PubMed]
- Atsumi, S.; Wu, T.Y.; Eckl, E.M.; Hawkins, S.D.; Buelter, T.; Liao, J.C. Engineering the isobutanol biosynthetic pathway in Escherichia coli by comparison of three aldehyde reductase/alcohol dehydrogenase genes. Appl. Microbiol. Biotechnol. 2010, 85, 651–657. [Google Scholar] [CrossRef] [PubMed]
- Vaidyanathan, H.; Kandasamy, V.; Gopal Ramakrishnan, G.; Ramachandran, K.; Jayaraman, G.; Ramalingam, S. Glycerol conversion to 1, 3-Propanediol is enhanced by the expression of a heterologous alcohol dehydrogenase gene in Lactobacillus reuteri. AMB Express 2011, 1, 37. [Google Scholar] [CrossRef] [PubMed]
- Abergel, C.; Coutard, B.; Byrne, D.; Chenivesse, S.; Claude, J.B.; Deregnaucourt, C.; Fricaux, T.; Gianesini-Boutreux, C.; Jeudy, S.; Lebrun, R.; et al. Structural genomics of highly conserved microbial genes of unknown function in search of new antibacterial targets. J. Struct. Funct. Genomics 2003, 4, 141–157. [Google Scholar] [CrossRef] [PubMed]
- Montella, C.; Bellsolell, L.; Perez-Luque, R.; Badia, J.; Baldoma, L.; Coll, M.; Aguilar, J. Crystal structure of an iron-dependent group III dehydrogenase that interconverts l-lactaldehyde and l-1,2-propanediol in Escherichia coli. J. Bacteriol. 2005, 187, 4957–4966. [Google Scholar] [CrossRef]
- Sulzenbacher, G.; Alvarez, K.; Van Den Heuvel, R.H.; Versluis, C.; Spinelli, S.; Campanacci, V.; Valencia, C.; Cambillau, C.; Eklund, H.; Tegoni, M. Crystal structure of E.coli alcohol dehydrogenase YqhD: Evidence of a covalently modified NADP coenzyme. J. Mol. Biol. 2004, 342, 489–502. [Google Scholar] [CrossRef]
- Chen, Z.; Liu, H.J.; Liu, D.H. Decrease the accumulation of 3-hydroxypropionaldehyde for 1,3-propanediol production by expressing the Yqhd gene in Klebsiella pneumonia. J. Biotechnol. 2008, 136, S354. [Google Scholar] [CrossRef]
- Perez, J.M.; Arenas, F.A.; Pradenas, G.A.; Sandoval, J.M.; Vasquez, C.C. Escherichia coli YqhD exhibits aldehyde reductase activity and protects from the harmful effect of lipid peroxidation-derived aldehydes. J. Biol. Chem. 2008, 283, 7346–7353. [Google Scholar] [CrossRef]
- Turner, P.C.; Miller, E.N.; Jarboe, L.R.; Baggett, C.L.; Shanmugam, K.T.; Ingram, L.O. YqhC regulates transcription of the adjacent Escherichia coli genes yqhD and dkgA that are involved in furfural tolerance. J. Ind. Microbiol. Biotechnol. 2011, 38, 431–439. [Google Scholar] [CrossRef]
- Miller, E.N.; Jarboe, L.R.; Yomano, L.P.; York, S.W.; Shanmugam, K.T.; Ingram, L.O. Silencing of NADPH-dependent oxidoreductase genes (yqhD and dkgA) in furfural-resistant ethanologenic Escherichia coli. Appl. Environ. Microbiol. 2009, 75, 4315–4323. [Google Scholar] [CrossRef]
- Wang, X.; Yomano, L.P.; Lee, J.Y.; York, S.W.; Zheng, H.B.; Mullinnix, M.T.; Shanmugam, K.T.; Ingram, L.O. Engineering furfural tolerance in Escherichia coli improves the fermentation of lignocellulosic sugars into renewable chemicals. Proc. Natl. Acad. Sci. USA 2013, 110, 4021–4026. [Google Scholar] [CrossRef] [PubMed]
- Koma, D.; Yamanaka, H.; Moriyoshi, K.; Ohmoto, T.; Sakai, K. Production of Aromatic Compounds by Metabolically Engineered Escherichia coli with an Expanded Shikimate Pathway. Appl. Environ. Microbiol. 2012, 78, 6203–6216. [Google Scholar] [CrossRef] [PubMed]
- Iwayanagi, T.; Miyamoto, S.; Konno, T.; Mizutani, H.; Hirai, T.; Shigemoto, Y.; Gojobori, T.; Sugawara, H. TP Atlas: Integration and dissemination of advances in Targeted Proteins Research Program (TPRP)—Structural biology project phase II in Japan. J. Struct. Funct. Genomics 2012, 13, 145–154. [Google Scholar] [CrossRef] [PubMed]
- Voelker, F.; Dumon-Seignovert, L.; Soucaille, P. Mutant Yqhd Enzyme for the Production of a Biochemical by Fermentation. U.S. Patent 8,969,053B2, 3 March 2015. [Google Scholar]
- Zhu, H.L.; Yi, X.Y.; Liu, Y.; Hu, H.B.; Wood, T.K.; Zhang, X.H. Production of acetol from glycerol using engineered Escherichia coli. Bioresour. Technol. 2013, 149, 238–243. [Google Scholar] [CrossRef] [PubMed]
- Clomburg, J.M.; Gonzalez, R. Metabolic Engineering of Escherichia coli for the Production of 1,2-Propanediol From Glycerol. Biotechnol. Bioeng. 2011, 108, 867–879. [Google Scholar] [CrossRef] [PubMed]
- Lan, E.I.; Liao, J.C. ATP drives direct photosynthetic production of 1-butanol in cyanobacteria. Proc. Natl. Acad. Sci. USA 2012, 109, 6018–6023. [Google Scholar] [CrossRef]
- Li, H.; Chen, J.; Li, Y. Enhanced activity of yqhD oxidoreductase in synthesis of 1,3-propanediol by error-prone PCR. Pro. Nat. Sci. 2008, 18, 1519–1524. [Google Scholar] [CrossRef]
- Rao, Z.; Ma, Z.; Shen, W.; Fang, H.; Zhuge, J.; Wang, X. Engineered Saccharomyces cerevisiae that produces 1,3-propanediol from d-glucose. J. Appl. Microbiol. 2008, 105, 1768–1776. [Google Scholar] [CrossRef]
- Tang, X.M.; Tan, Y.S.; Zhu, H.; Zhao, K.; Shen, W. Microbial Conversion of Glycerol to 1,3-Propanediol by an Engineered Strain of Escherichia coli. Appl. Environ. Microbiol. 2009, 75, 1628–1634. [Google Scholar] [CrossRef]
- Wang, F.H.; Qu, H.J.; Zhang, D.W.; Tian, P.F.; Tan, T.W. Production of 1,3-propanediol from glycerol by recombinant E. coli using incompatible plasmids system. Mol. Biotechnol. 2007, 37, 112–119. [Google Scholar] [CrossRef]
- Elleuche, S.; Fodor, K.; von der Heyde, A.; Klippel, B.; Wilmanns, M.; Antranikian, G. Group III alcohol dehydrogenase from Pectobacterium atrosepticum: Insights into enzymatic activity and organization of the metal ion-containing region. Appl. Environ. Microbiol. 2014, 98, 4041–4051. [Google Scholar] [CrossRef] [PubMed]
- Elleuche, S.; Klippel, B.; von der Heyde, A.; Antranikian, G. Comparative analysis of two members of the metal ion-containing group III-alcohol dehydrogenases from Dickeya zeae. Biotechnol. Lett. 2013, 35, 725–733. [Google Scholar] [CrossRef] [PubMed]
- Verma, R.; Schwaneberg, U.; Roccatano, D. Conformational Dynamics of the FMN-Binding Reductase Domain of Monooxygenase P450BM-3. J. Chem. Theory Comput. 2013, 9, 96–105. [Google Scholar] [CrossRef] [PubMed]
- Sellés Vidal, L.; Kelly, C.L.; Mordaka, P.M.; Heap, J.T. Review of NAD(P)H-dependent oxidoreductases: Properties, engineering and application. Biochimica Biophys. Acta (BBA) 2018, 1866, 327–347. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.-G.; Li, S.; Ji, X.-J.; Huang, H.; Hu, N. Enhanced 1,3-propanediol production in recombinant Klebsiella pneumoniae carrying the gene yqhD encoding 1,3-propanediol oxidoreductase isoenzyme. World J. Microbiol. Biotechnol. 2009, 25, 1217. [Google Scholar] [CrossRef]
- Verma, R.; Mitchell-Koch, K. In Silico Studies of Small Molecule Interactions with Enzymes Reveal Aspects of Catalytic Function. Catalysts 2017, 7, 212. [Google Scholar] [CrossRef]
- Cummins, P.L.; Ramnarayan, K.; Singh, U.C.; Gready, J.E. Molecular dynamics/free energy perturbation study on the relative affinities of the binding of reduced and oxidized NADP to dihydrofolate reductase. J. Am. Chem. Soc. 1991, 113, 8247–8256. [Google Scholar] [CrossRef]
- Blikstad, C.; Dahlstrom, K.M.; Salminen, T.A.; Widersten, M. Substrate scope and selectivity in offspring to an enzyme subjected to directed evolution. FEBS J. 2014, 281, 2387–2398. [Google Scholar] [CrossRef]
- Luo, J.; Bruice, T.C. Dynamic Structures of Horse Liver Alcohol Dehydrogenase (HLADH): Results of Molecular Dynamics Simulations of HLADH-NAD+-PhCH2OH, HLADH-NAD+-PhCH2O−, and HLADH-NADH-PhCHO. J. Am. Chem. Soc. 2001, 123, 11952–11959. [Google Scholar] [CrossRef]
- Oyen, D.; Fenwick, R.B.; Stanfield, R.L.; Dyson, H.J.; Wright, P.E. Cofactor-Mediated Conformational Dynamics Promote Product Release From Escherichia coli Dihydrofolate Reductase via an Allosteric Pathway. J. Am. Chem. Soc. 2015, 137, 9459–9468. [Google Scholar] [CrossRef]
- The PyMOL Molecular Graphics System, Version 1.8; Schrodinger LLC.: New York, NY, USA, 2015.
- Best, R.B.; Zhu, X.; Shim, J.; Lopes, P.E.; Mittal, J.; Feig, M.; Mackerell, A.D., Jr. Optimization of the additive CHARMM all-atom protein force field targeting improved sampling of the backbone phi, psi and side-chain chi(1) and chi(2) dihedral angles. J. Chem. Theory Comput. 2012, 8, 3257–3273. [Google Scholar] [CrossRef] [PubMed]
- Hess, B.; Kutzner, C.; van der Spoel, D.; Lindahl, E. GROMACS 4: Algorithms for highly efficient, load-balanced, and scalable molecular simulation. J. Chem. Theory. Comput. 2008, 4, 435–447. [Google Scholar] [CrossRef] [PubMed]
- Jorgensen, W.L.; Chandrasekhar, J.; Madura, J.D.; Impey, R.W.; Klein, M.L. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983, 79, 926–935. [Google Scholar] [CrossRef]
- Neria, E.; Fischer, S.; Karplus, M. Simulation of activation free energies in molecular systems. J. Chem. Phys. 1996, 105, 1902–1921. [Google Scholar] [CrossRef]
- Hess, B.; Bekker, H.; Berendsen, H.J.C.; Fraaije, J.G.E.M. LINCS: A linear constraint solver for molecular simulations. J. Comput. Chem. 1997, 18, 1463–1472. [Google Scholar] [CrossRef]
- Miyamoto, S.; Kollman, P.A. Settle - an Analytical Version of the Shake and Rattle Algorithm for Rigid Water Models. J. Comput. Chem. 1992, 13, 952–962. [Google Scholar] [CrossRef]
- Darden, T.; York, D.; Pedersen, L. Particle Mesh Ewald - an N.Log(N) Method for Ewald Sums in Large Systems. J. Chem. Phys. 1993, 98, 10089–10092. [Google Scholar] [CrossRef]
- Bussi, G.; Donadio, D.; Parrinello, M. Canonical sampling through velocity rescaling. J. Chem. Phys. 2007, 126, 014101. [Google Scholar] [CrossRef]
- Berendsen, H.J.C.; Postma, J.P.M.; Vangunsteren, W.F.; Dinola, A.; Haak, J.R. Molecular-Dynamics with Coupling to an External Bath. J. Chem. Phys. 1984, 81, 3684–3690. [Google Scholar] [CrossRef]
- Daura, X.; Gademann, K.; Jaun, B.; Seebach, D.; van Gunsteren, W.F.; Mark, A.E. Peptide folding: When simulation meets experiment. Angew. Chem. Int. Ed. 1999, 38, 236–240. [Google Scholar] [CrossRef]

| Enzyme | Cofactor | Atoms | Water Molecules | Counter Ions | No. of Simulations | Time (ns) |
|---|---|---|---|---|---|---|
| YqhD Dimer | NADP | 128,728 | 40,363 | 14 Na+ | 5 | 200 |
| YqhD Dimer | NADPH | 128,723 | 40,360 | 16 Na+ | 5 | 200 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Verma, R.; Ellis, J.M.; Mitchell-Koch, K.R. Dynamic Preference for NADP/H Cofactor Binding/Release in E. coli YqhD Oxidoreductase. Molecules 2021, 26, 270. https://doi.org/10.3390/molecules26020270
Verma R, Ellis JM, Mitchell-Koch KR. Dynamic Preference for NADP/H Cofactor Binding/Release in E. coli YqhD Oxidoreductase. Molecules. 2021; 26(2):270. https://doi.org/10.3390/molecules26020270
Chicago/Turabian StyleVerma, Rajni, Jonathan M. Ellis, and Katie R. Mitchell-Koch. 2021. "Dynamic Preference for NADP/H Cofactor Binding/Release in E. coli YqhD Oxidoreductase" Molecules 26, no. 2: 270. https://doi.org/10.3390/molecules26020270
APA StyleVerma, R., Ellis, J. M., & Mitchell-Koch, K. R. (2021). Dynamic Preference for NADP/H Cofactor Binding/Release in E. coli YqhD Oxidoreductase. Molecules, 26(2), 270. https://doi.org/10.3390/molecules26020270






