Exploring the Bile Stress Response of Lactobacillus mucosae LM1 through Exoproteome Analysis
Abstract
:1. Introduction
2. Results
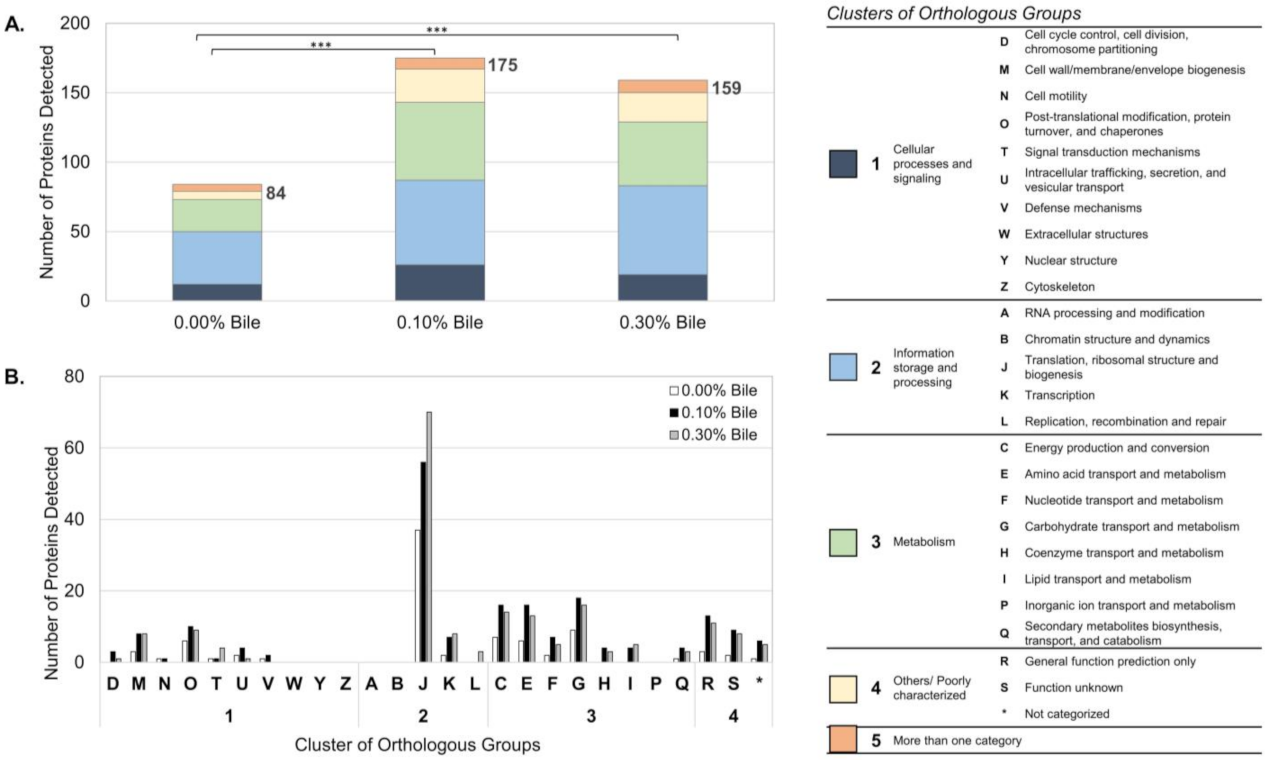
2.1. Overview of Lactobacillus mucosae LM1 Exoproteome during Bile Stress
2.2. Functional Annotation-Based Analysis of LM1 Exoproteome
2.3. Intensity-Based Abundance Analysis of the LM1 Exoproteome
3. Discussion
3.1. Bile-Induced Changes in the LM1 Exoproteome
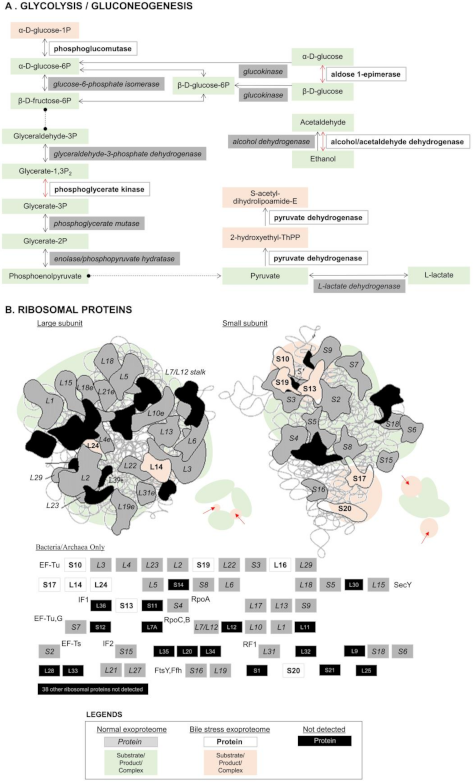
3.2. Bile Stress-Response Proteins in the LM1 Exoproteome
3.3. Consistent Abundance of Proteins in the LM1 Exoproteome under Bile Stress
3.4. Possible Secretion Mechanisms of Intracellular Proteins Moonlighting Extracellularly
3.5. Host-Specific Beneficial Proteins in the LM1 Exoproteome
4. Materials and Methods
4.1. LM1 Growth Conditions, Bile Treatment and Protein Collection
4.2. Mass Spectrometry Analysis of LM1 Extracellular Proteins
4.3. Bioinformatics and Statistical Analysis of the LM1 Exoproteome
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Holzapfel, W.H.; Schillinger, U. Introduction to pre- and probiotics. Food Res. Int. 2002, 35, 109–116. [Google Scholar] [CrossRef]
- Hill, C.; Guarner, F.; Reid, G.; Gibson, G.R.; Merenstein, D.J.; Pot, B.; Morelli, L.; Canani, R.B.; Flint, H.J.; Salminen, S.; et al. The International Scientific Association for Probiotics and Prebiotics consensus statement on the scope and appropriate use of the term probiotic. Nat. Rev. Gastroenterol. Hepatol. 2014, 11, 506–514. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Angelis, M.; Calasso, M.; Cavallo, N.; Cagno, R.D.; Gobbetti, M. Functional proteomics within the genus Lactobacillus. Proteomics 2016, 16, 946–962. [Google Scholar] [CrossRef]
- Fadda, S. Contribution of proteomics for diving into the lactic acid bacteria role and the modification of the food matrix during fermentation. Single Cell Biol. 2012, 1, 1000e113. [Google Scholar] [CrossRef] [Green Version]
- Mbye, M.; Baig, M.A.; AbuQamar, S.F.; El-Tarabily, K.A.; Obaid, R.S.; Osaili, T.M.; Al-Nabulsi, A.A.; Turner, M.S.; Shah, N.P.; Ayyash, M.M. Updates on understanding of probiotic lactic acid bacteria responses to environmental stresses and highlights on proteomic analyses. Compr. Rev. Food Sci. Food Saf. 2020, 19, 1110–1124. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pessione, A.; Lamberti, C.; Pessione, E. Proteomics as a tool for studying energy metabolism in lactic acid bacteria. Mol. BioSystems 2010, 6, 1419–1430. [Google Scholar] [CrossRef] [PubMed]
- Ruiz, L.; Couté, Y.; Sánchez, B.; de los Reyes-Gavilán, C.G.; Sanchez, J.-C.; Margolles, A. The cell-envelope proteome of Bifidobacterium longum in an in vitro bile environment. Microbiology 2009, 155, 957–967. [Google Scholar] [CrossRef] [Green Version]
- Sánchez, B.; Bressollier, P.; Urdaci, M.C. Exported proteins in probiotic bacteria: Adhesion to intestinal surfaces, host immunomodulation and molecular cross-talking with the host. FEMS Immunol. Med Microbiol. 2008, 54, 1–17. [Google Scholar] [CrossRef] [Green Version]
- Sánchez, B.; Urdaci, M.C.; Margolles, A. Extracellular proteins secreted by probiotic bacteria as mediators of effects that promote mucosa-bacteria interactions. Microbiology 2010, 156 Pt 11, 3232–3242. [Google Scholar] [CrossRef] [Green Version]
- Valeriano, V.D.; Bagon, B.B.; Balolong, M.P.; Kang, D.-K. Carbohydrate-binding specificities of potential probiotic Lactobacillus strains in porcine jejunal (IPEC-J2) cells and porcine mucin. J. Microbiol. 2016, 54, 510–519. [Google Scholar] [CrossRef]
- Pajarillo, E.A.B.; Kim, S.H.; Valeriano, V.D.; Lee, J.Y.; Kang, D.-K. Proteomic view of the crosstalk between Lactobacillus mucosae and intestinal epithelial cells in co-culture revealed by Q exactive-based quantitative proteomics. Front. Microbiol. 2017, 8, 2459. [Google Scholar] [CrossRef] [Green Version]
- Bagon, B.B.; Valeriano, V.D.V.; Oh, J.K.; Pajarillo, E.A.B.; Cho, C.-S.; Kang, D.-K. Comparative exoproteome analyses of Lactobacillus spp. reveals species- and strain-specific proteins involved in their extracellular interaction and probiotic potential. LWT 2018, 93, 420–426. [Google Scholar] [CrossRef]
- Valeriano, V.D.V.; Oh, J.K.; Bagon, B.B.; Kim, H.; Kang, D.-K. Comparative genomic analysis of Lactobacillus mucosae LM1 identifies potential niche-specific genes and pathways for gastrointestinal adaptation. Genomics 2019, 111, 24–33. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Valeriano, V.D.; Parungao-Balolong, M.M.; Kang, D.-K. In vitro evaluation of the mucin-adhesion ability and probiotic potential of Lactobacillus mucosae LM1. J. Appl. Microbiol. 2014, 117, 485–497. [Google Scholar] [CrossRef]
- Celebioglu, H.U.; Svensson, B. Exo- and surface proteomes of the probiotic bacterium Lactobacillus acidophilus NCFM. Proteomics 2017, 17, 1700019. [Google Scholar] [CrossRef] [Green Version]
- Koskenniemi, K.; Laakso, K.; Koponen, J.; Kankainen, M.; Greco, D.; Auvinen, P.; Savijoki, K.; Nyman, T.A.; Surakka, A. Proteomics and transcriptomics characterization of bile stress response in probiotic Lactobacillus rhamnosus GG*. Mol. Cell. Proteom. 2011, 10, 18. [Google Scholar] [CrossRef] [Green Version]
- De Angelis, M.; Gobbetti, M. Environmental stress responses in Lactobacillus: A review. Proteomics 2004, 4, 106–122. [Google Scholar] [CrossRef]
- Goh, Y.J.; Klaenhammer, T.R. Functional roles of aggregation-promoting-like factor in stress tolerance and adherence of Lactobacillus acidophilus NCFM. Appl. Environ. Microbiol. 2010, 76, 5005–5012. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kelly, P.; Maguire, P.B.; Bennett, M.; Fitzgerald, D.J.; Edwards, R.J.; Thiede, B.; Treumann, A.; Collins, J.K.; O’Sullivan, G.C.; Shanahan, F.; et al. Correlation of probiotic Lactobacillus salivarius growth phase with its cell wall-associated proteome. FEMS Microbiol. Lett. 2005, 252, 153–159. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bagon, B.B.; Valeriano, V.D.V.; Oh, J.K.; Pajarillo, E.A.B.; Lee, J.Y.; Kang, D.-K. Exoproteome perspective on the bile stress response of Lactobacillus johnsonii. Proteomes 2021, 9, 10. [Google Scholar] [CrossRef]
- Zaidi, A.H.; Bakkes, P.J.; Lubelski, J.; Agustiandari, H.; Kuipers, O.P.; Driessen, A.J.M. The ABC-type multidrug resistance transporter LmrCD is responsible for an extrusion-based mechanism of bile acid resistance in Lactococcus lactis. J. Bacteriol. 2008, 190, 7357–7366. [Google Scholar] [CrossRef] [Green Version]
- Sánchez, B.; Champomier-Vergès, M.-C.; Stuer-Lauridsen, B.; Ruas-Madiedo, P.; Anglade, P.; Baraige, F.; de los Reyes-Gavilán, C.G.; Johansen, E.; Zagorec, M.; Margolles, A. Adaptation and response of Bifidobacterium animalis subsp. lactis to bile: A proteomic and physiological approach. Appl. Environ. Microbiol. 2007, 73, 6757–6767. [Google Scholar] [CrossRef] [Green Version]
- Wu, R.; Sun, Z.; Wu, J.; Meng, H.; Zhang, H. Effect of bile salts stress on protein synthesis of Lactobacillus casei Zhang revealed by 2-dimensional gel electrophoresis. J. Dairy Sci. 2010, 93, 3858–3868. [Google Scholar] [CrossRef] [PubMed]
- Bove, P.; Capozzi, V.; Garofalo, C.; Rieu, A.; Spano, G.; Fiocco, D. Inactivation of the ftsH gene of Lactobacillus plantarum WCFS1: Effects on growth, stress tolerance, cell surface properties and biofilm formation. Microbiol. Res. 2012, 167, 187–193. [Google Scholar] [CrossRef] [PubMed]
- Wower, I.K.; Wower, J.; Zimmermann, R.A. Ribosomal protein L27 participates in both 50 S subunit assembly and the peptidyl transferase reaction. J. Biol. Chem. 1998, 273, 19847–19852. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ali, S.A.; Singh, P.; Tomar, S.K.; Mohanty, A.K.; Behare, P. Proteomics fingerprints of systemic mechanisms of adaptation to bile in Lactobacillus fermentum. J. Proteom. 2020, 213, 103600. [Google Scholar] [CrossRef]
- Yamamoto, H.; Kurosawa, S.; Sekiguchi, J. Localization of the vegetative cell wall hydrolases LytC, LytE, and LytF on the Bacillus subtilis cell surface and stability of these enzymes to cell wall-bound or extracellular proteases. J. Bacteriol. 2003, 185, 6666–6677. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ming, T.; Han, J.; Li, Y.; Lu, C.; Qiu, D.; Li, Y.; Zhou, J.; Su, X. A metabolomics and proteomics study of the Lactobacillus plantarum in the grass carp fermentation. BMC Microbiol. 2018, 18, 216. [Google Scholar] [CrossRef]
- Wang, G.; Zhang, M.; Zhao, J.; Xia, Y.; Lai, P.F.-H.; Ai, L. A surface protein from Lactobacillus plantarum increases the adhesion of Lactobacillus strains to human epithelial cells. Front. Microbiol. 2018, 9, 2858. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hu, P.-L.; Yuan, Y.-H.; Yue, T.-L.; Guo, C.-F. Bile acid patterns in commercially available oxgall powders used for the evaluation of the bile tolerance ability of potential probiotics. PLoS ONE 2018, 13, e0192964. [Google Scholar] [CrossRef] [Green Version]
- Wiśniewski, J.R.; Zougman, A.; Nagaraj, N.; Mann, M. Universal sample preparation method for proteome analysis. Nature Methods 2009, 6, 359–362. [Google Scholar] [CrossRef] [PubMed]
- Cox, J.; and Mann, M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 2008, 26, 1367–1372. [Google Scholar] [CrossRef] [PubMed]
- Zhou, M.; Boekhorst, J.; Francke, C.; Siezen, R.J. LocateP: Genome-scale subcellular-location predictor for bacterial proteins. BMC Bioinform. 2008, 9, 173. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Petersen, T.N.; Brunak, S.; von Heijne, G.; Nielsen, H. SignalP 4.0: Discriminating signal peptides from transmembrane regions. Nature Methods 2011, 8, 785–786. [Google Scholar] [CrossRef]
- Kanehisa, M.; Sato, Y.; Morishima, K. BlastKOALA and GhostKOALA: KEGG tools for functional characterization of genome and metagenome sequences. J. Mol. Biol. 2016, 428, 726–731. [Google Scholar] [CrossRef] [Green Version]
- McHugh, M.L. Multiple comparison analysis testing in ANOVA. Biochem. Med. 2011, 21, 203–209. [Google Scholar] [CrossRef] [PubMed]

| Predicted Localization | Number of Proteins | Cell Wall and Membrane-Associated Proteins | |||||
|---|---|---|---|---|---|---|---|
| 0.00% Bile | 0.10% Bile | 0.30% Bile | 0.00% Bile | 0.10% Bile | 0.30% Bile | ||
| Cellular Destination | Cytoplasmic | 79 | 91 | 44 | - | - | - |
| Membrane | 0 | 2 | 4 | - | LBLM1_16160 MurD, LBLM1_00750 PepD | LBLM1_19820 HflB, LBLM1_21140 HisM, LBLM1_08510 MelB, LBLM1_16160 MurD * | |
| Cell Wall | 1 | 0 | 0 | LBLM1_10850 hypothetical protein EmrA | - | - | |
| Extracellular | 4 | 0 | 3 | - | - | - | |
| Subcellular Localization | Intracellular | 79 | 91 | 44 | - | - | - |
| Multi-transmembrane | 0 | 0 | 3 | - | - | LBLM1_19820 HflB, LBLM1_21140 HisM, LBLM1_08510 MelB | |
| LPXTG Cell wall anchored | 1 | 0 | 0 | LBLM1_10850 hypothetical protein EmrA | - | - | |
| N-terminally anchored | 0 | 2 | 1 | - | LBLM1_16160 MurD, LBLM1_00750 PepD | LBLM1_16160 MurD * | |
| Lipid-anchored | 0 | 0 | 1 | - | - | LBLM1_04250 hypothetical protein | |
| Secretory | 4 | 0 | 2 | - | - | - | |
| Localization Class | Cytoplasm | 76 | 84 | 41 | - | - | - |
| Inner membrane | 0 | 4 | 3 | - | LBLM1_15150 AbiF, LBLM1_04880 GlnQ, LBLM1_10270 GlnQ, LBLM1_21130 GlnQ | LBLM1_19820 HflB, LBLM1_21140 HisM, LBLM1_08510 MelB | |
| Periplasm | 2 | 3 | 4 | - | - | - | |
| Secreted | 6 | 2 | 3 | - | - | - | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bagon, B.B.; Oh, J.K.; Valeriano, V.D.V.; Pajarillo, E.A.B.; Kang, D.-K. Exploring the Bile Stress Response of Lactobacillus mucosae LM1 through Exoproteome Analysis. Molecules 2021, 26, 5695. https://doi.org/10.3390/molecules26185695
Bagon BB, Oh JK, Valeriano VDV, Pajarillo EAB, Kang D-K. Exploring the Bile Stress Response of Lactobacillus mucosae LM1 through Exoproteome Analysis. Molecules. 2021; 26(18):5695. https://doi.org/10.3390/molecules26185695
Chicago/Turabian StyleBagon, Bernadette B., Ju Kyoung Oh, Valerie Diane V. Valeriano, Edward Alain B. Pajarillo, and Dae-Kyung Kang. 2021. "Exploring the Bile Stress Response of Lactobacillus mucosae LM1 through Exoproteome Analysis" Molecules 26, no. 18: 5695. https://doi.org/10.3390/molecules26185695
APA StyleBagon, B. B., Oh, J. K., Valeriano, V. D. V., Pajarillo, E. A. B., & Kang, D.-K. (2021). Exploring the Bile Stress Response of Lactobacillus mucosae LM1 through Exoproteome Analysis. Molecules, 26(18), 5695. https://doi.org/10.3390/molecules26185695







