Enhanced Activity by Genetic Complementarity: Heterologous Secretion of Clostridial Cellulases by Bacillus licheniformis and Bacillus velezensis
Abstract
:1. Introduction
2. Results
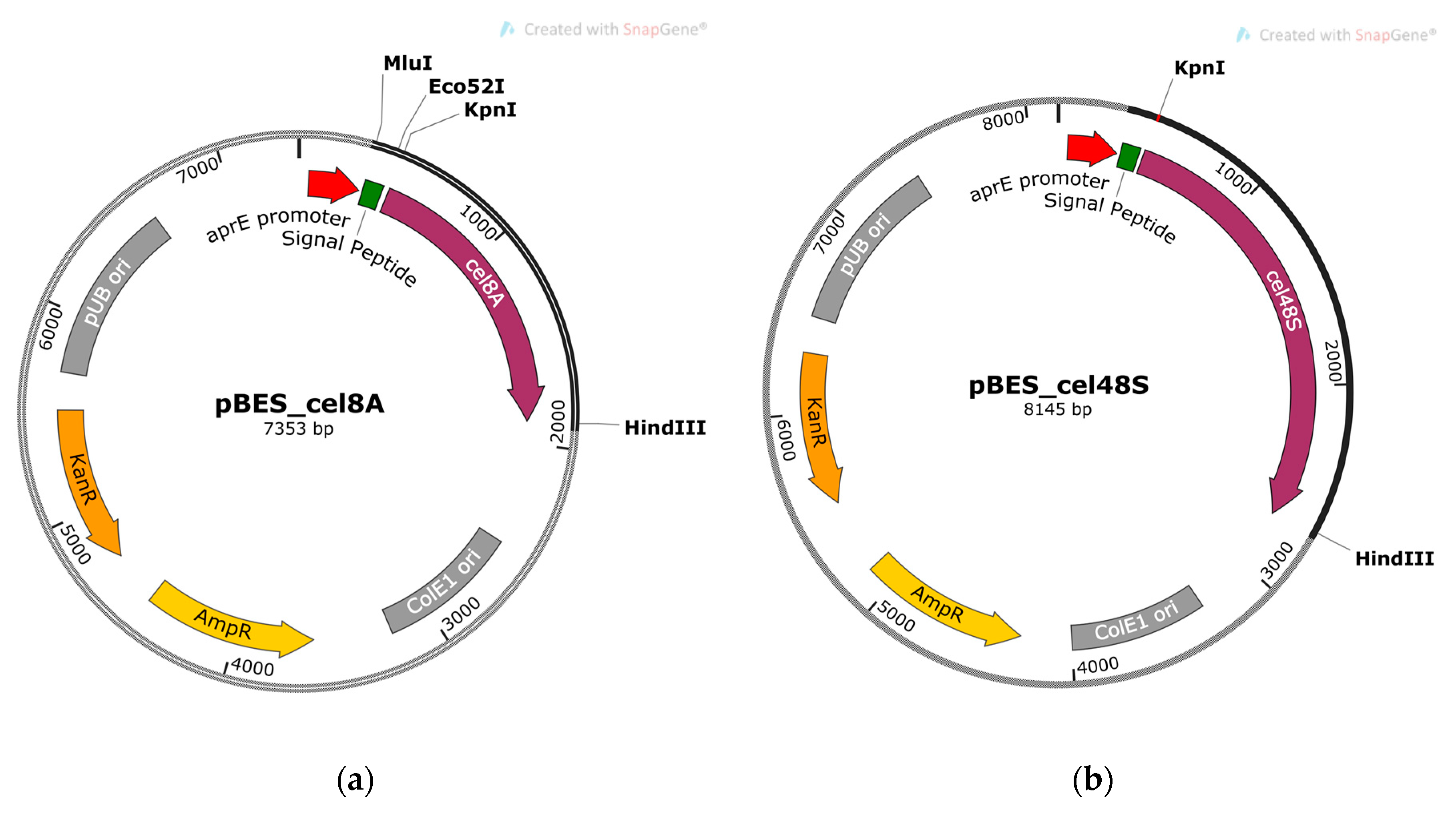
2.1. Molecular Cloning of Cel8A and Cel48S Genes of A. thermocellus
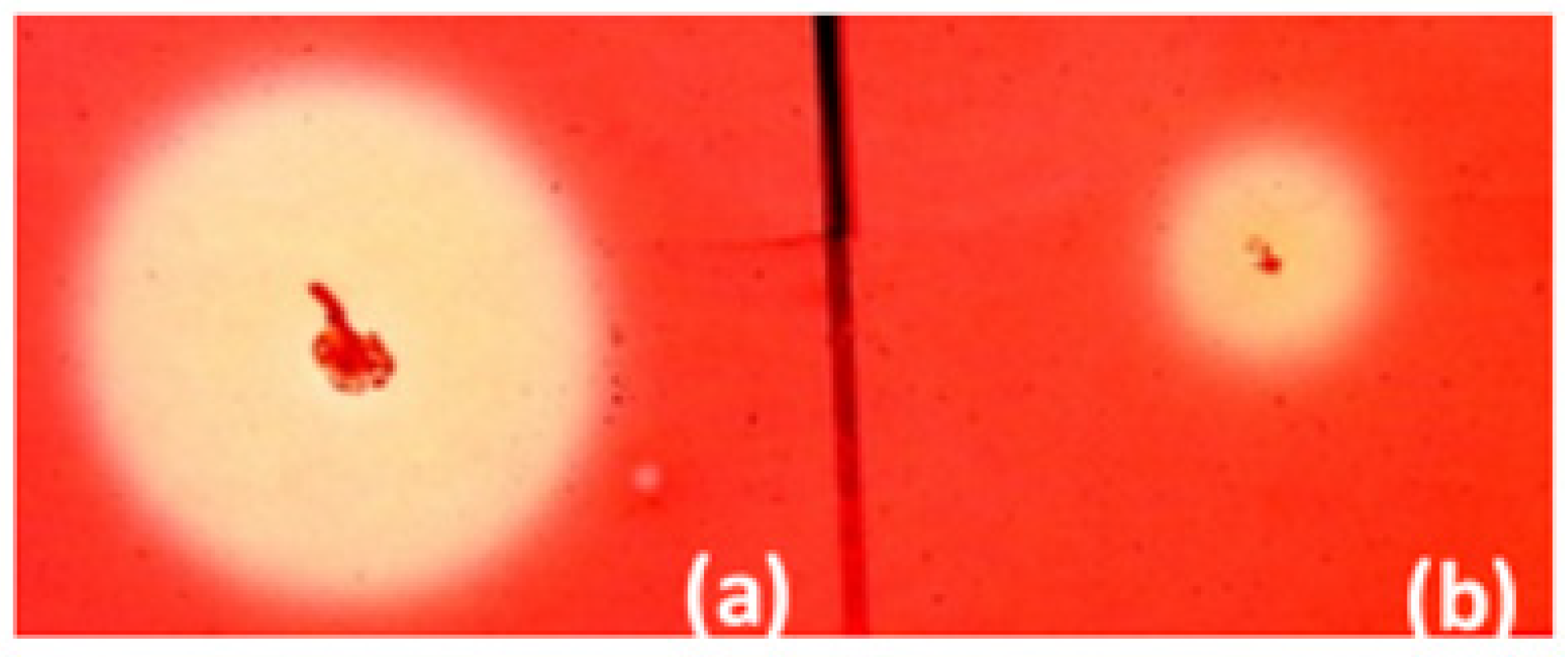
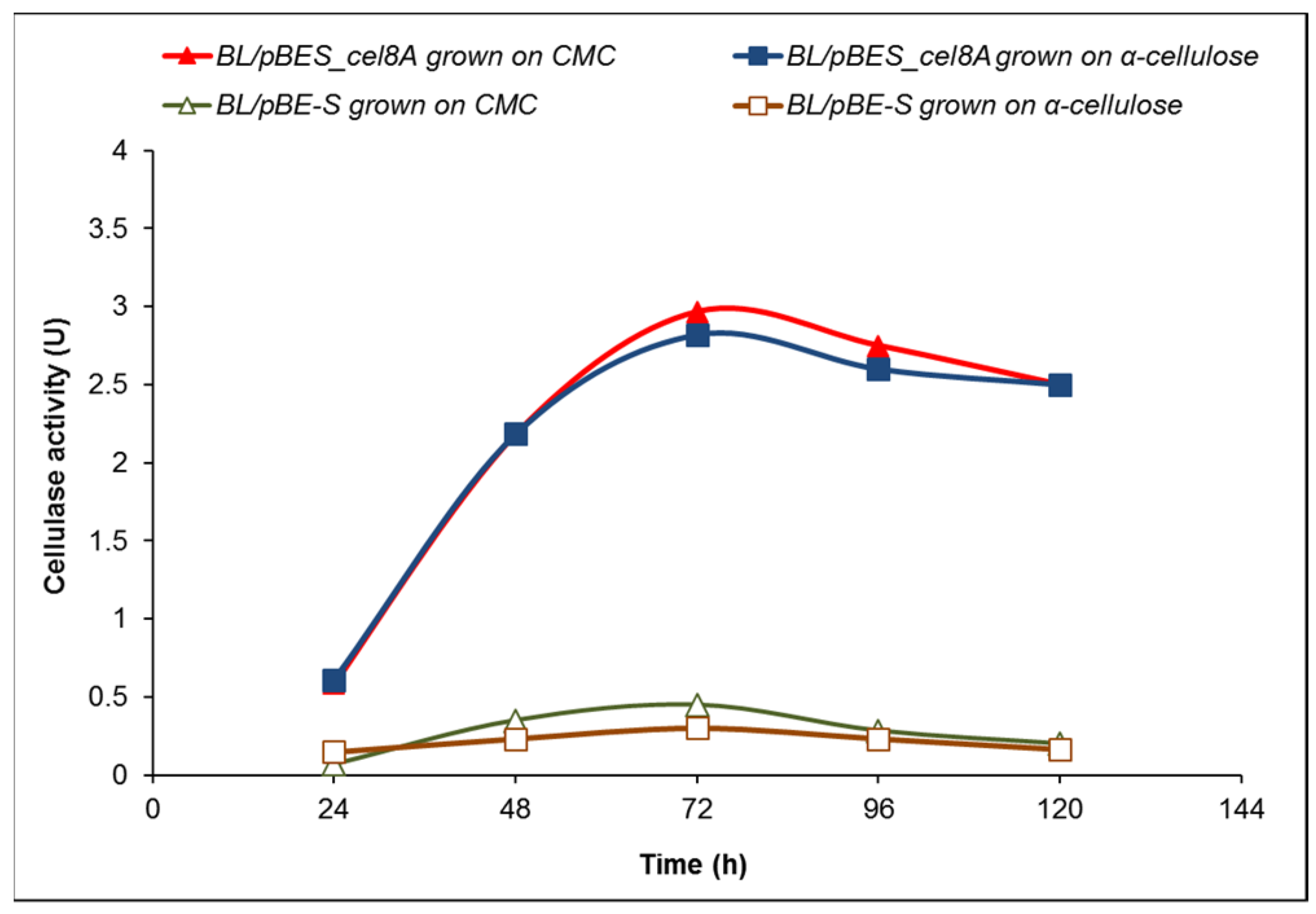
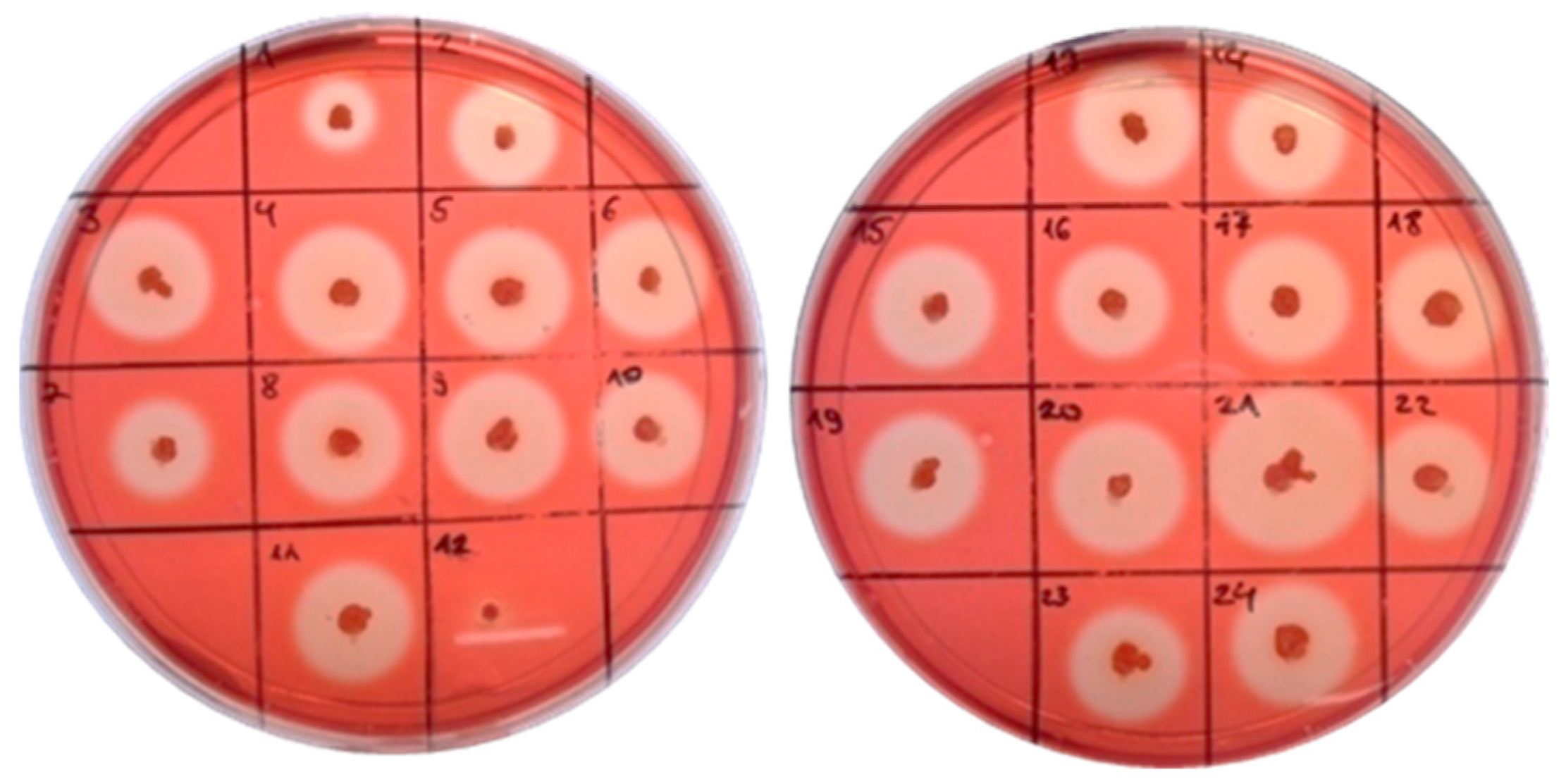
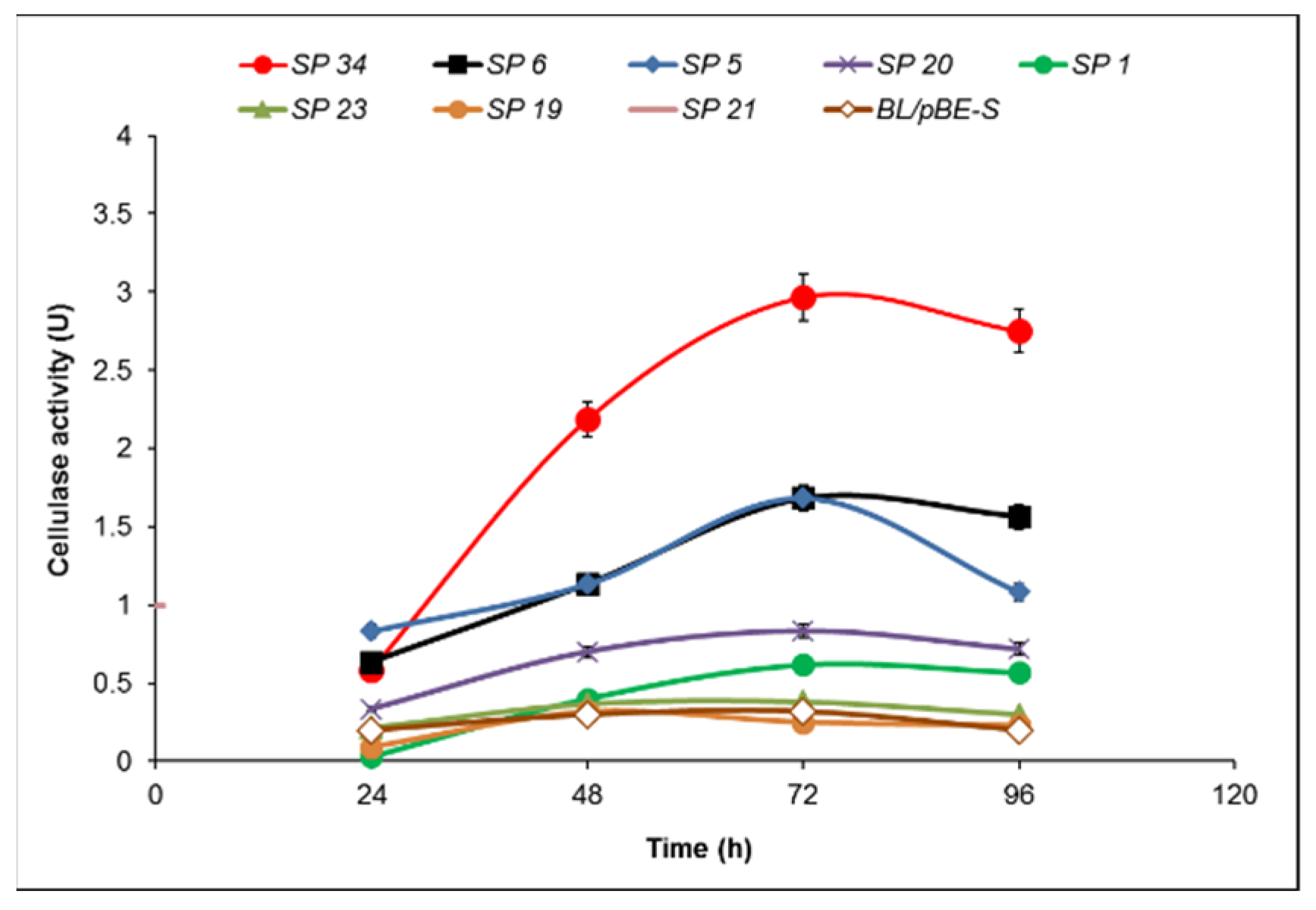
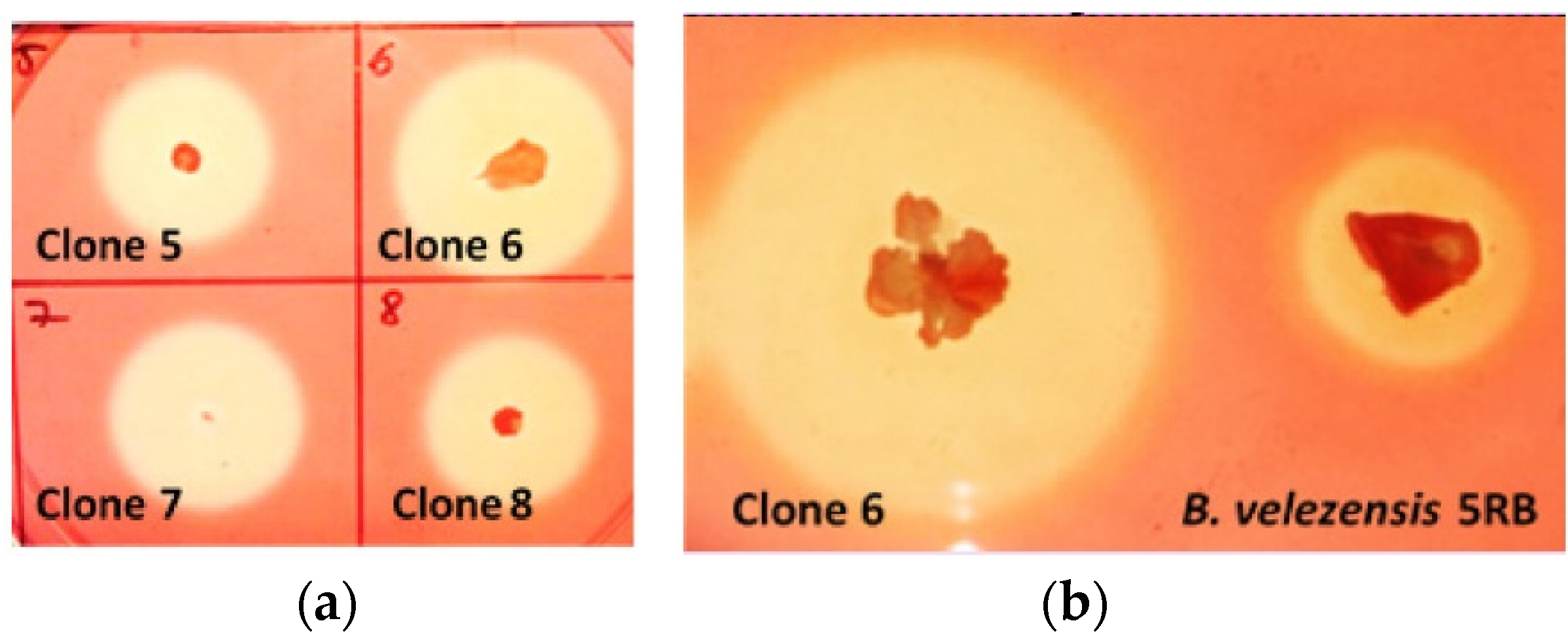
2.2. Cellulase Activity of B. licheniformis 24 and Recombinants Containing the Cel8A Gene
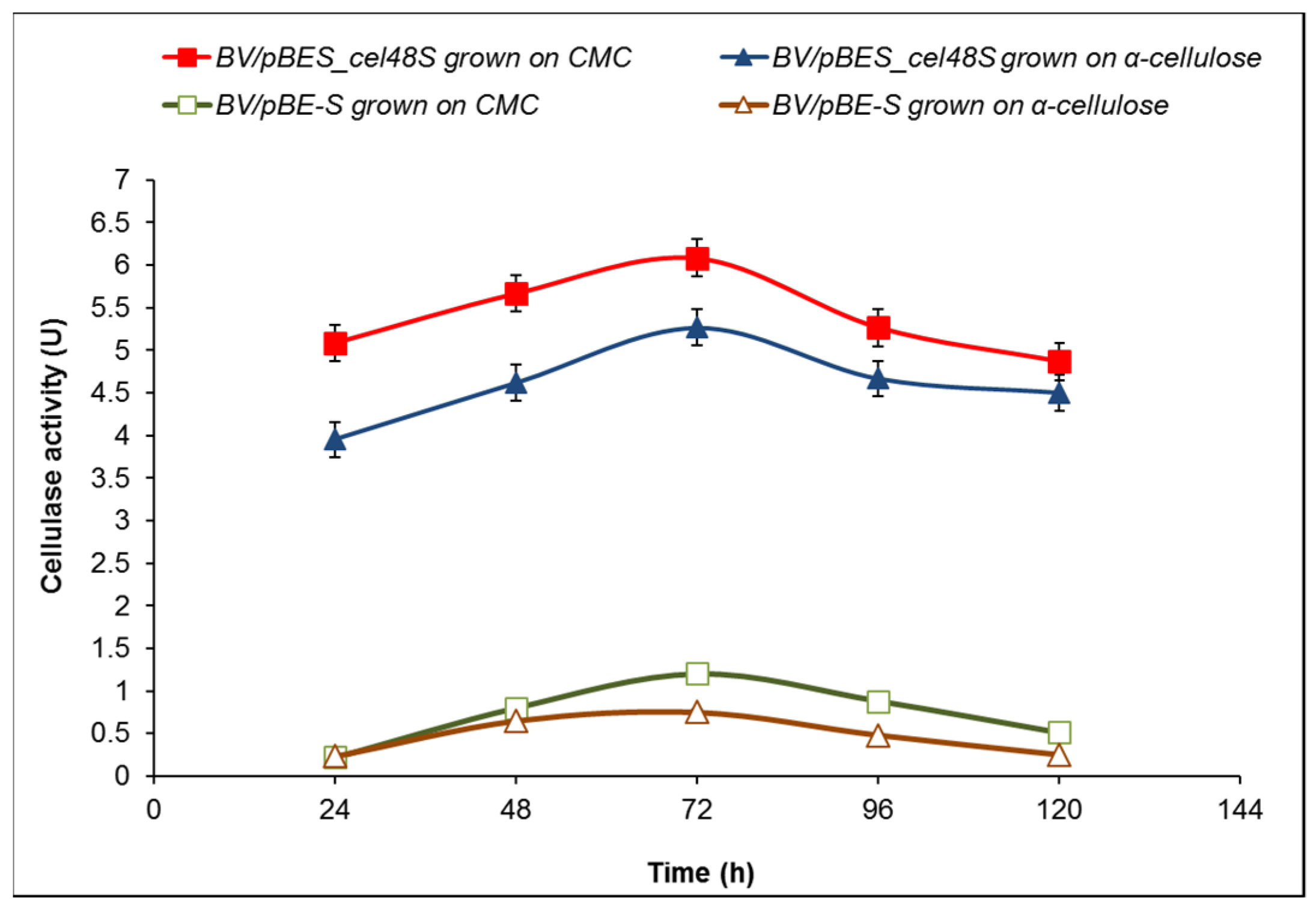
2.3. Cellulase Activity of B. velezensis 5RB Recombinants Containing the Cel48S Gene
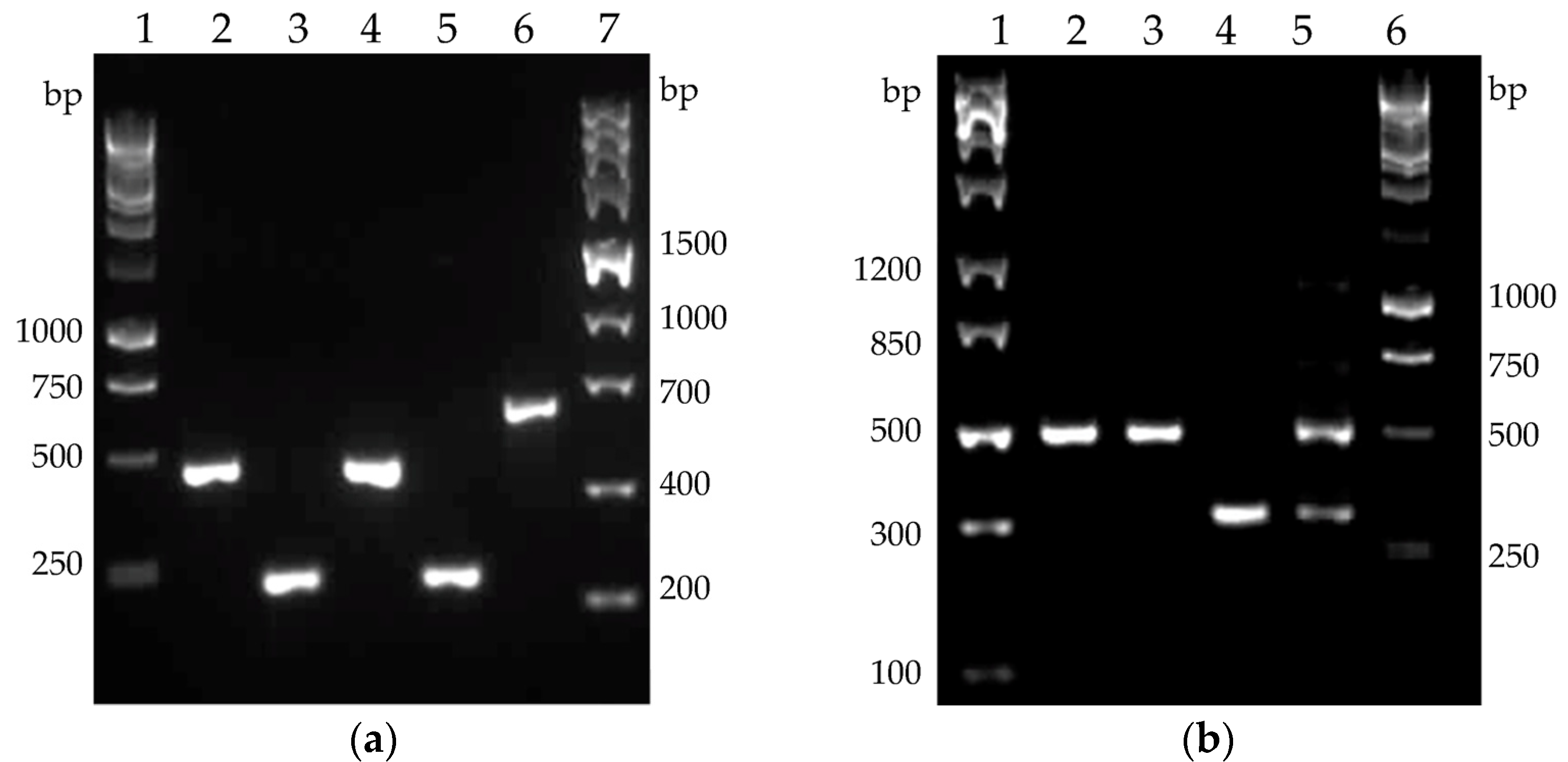
2.4. Cellulase Genes Transcription Profiles
3. Discussion
4. Materials and Methods
4.1. Bacterial Strains, Media and Cultivation Conditions
4.2. Molecular Cloning of Cel8A and Cel48S
4.3. Transformation and Selection of the Recombinant Clones
4.4. RT-PCR for Cel8A, Cel48S and Native Bacillus spp. Cellulases
4.5. Cellulase Activity Assay
4.6. Analytical Methods
4.7. Bioinformatics Tools
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Sample Availability
References
- Jayasekara, S.; Ratnayake, R. Microbial cellulases: An overview and applications. In Cellulose; Rodríguez Pascual, A., Eugenio Martín, M.E., Eds.; IntechOpen: London, UK, 2019. [Google Scholar] [CrossRef] [Green Version]
- Singhania, R.R.; Saini, R.; Adsul, M.; Saini, J.K.; Mathur, A.; Tuli, D. An integrative process for bio-ethanol production employing SSF produced cellulase without extraction. Biochem. Eng. J. 2015, 102, 45–48. [Google Scholar] [CrossRef]
- Md Razali, N.A.A.; Ibrahim, M.F.; Kamal Bahrin, E.; Abd-Aziz, S. Optimisation of Simultaneous Saccharification and Fermentation (SSF) for Biobutanol Production Using Pretreated Oil Palm Empty Fruit Bunch. Molecules 2018, 23, 1944. [Google Scholar] [CrossRef] [Green Version]
- Tsvetanova, F.; Petrova, P.; Petrov, K. Microbial production of 1-butanol—Recent advances and future prospects. J. Chem. Technol. Metall. 2018, 53, 683–696. Available online: https://dl.uctm.edu/journal/node/j2018-4/8_17-182_p_683-696.pdf (accessed on 3 August 2021).
- Ali, N.; Wang, F.; Xu, B.; Safdar, B.; Ullah, A.; Naveed, M.; Wang, C.; Rashid, M.T. Production and Application of Biosurfactant Produced by Bacillus licheniformis Ali5 in Enhanced Oil Recovery and Motor Oil Removal from Contaminated Sand. Molecules 2019, 24, 4448. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, J.; Li, J.; Gao, H.; Zhou, D.; Xu, H.; Cong, Y.; Zhang, W.; Xin, F.; Jiang, M. Recent progress on bio-succinic acid production from lignocellulosic biomass. World J. Microbiol. Biotechnol. 2021, 37, 16. [Google Scholar] [CrossRef]
- Shen, X.; Xia, L. Lactic acid production from cellulosic material by synergetic hydrolysis and fermentation. Appl. Biochem. Biotechnol. 2006, 33, 251–262. [Google Scholar] [CrossRef]
- Cho, E.J.; Trinh, L.T.P.; Song, Y.; Lee, Y.G.; Bae, H.J. Bioconversion of biomass waste into high value chemicals. Bioresour. Technol. 2020, 298, 122386. [Google Scholar] [CrossRef] [PubMed]
- Song, C.W.; Rathnasingh, C.; Park, J.M.; Lee, J.; Song, H. Isolation and evaluation of Bacillus strains for industrial production of 2,3-butanediol. J. Microbiol. Biotechnol. 2018, 28, 409–417. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vivek, N.; Hazeena, S.H.; Alphy, M.P.; Kumar, V.; Magdouli, S.; Sindhu, R.; Pandey, A.; Binod, P. Recent advances in microbial biosynthesis of C3–C5 diols: Genetics and process engineering approaches. Bioresour. Technol. 2021, 322, 124527. [Google Scholar] [CrossRef]
- Koutinas, A.A.; Yepez, B.; Kopsahelis, N.; Freire, D.M.G.; de Castro, A.M.; Papanikolaou, S.; Kookos, I.K. Techno-economic evaluation of a complete bioprocess for 2,3-butanediol production from renewable resources. Bioresour. Technol. 2016, 204, 55–64. [Google Scholar] [CrossRef]
- NREL—Biomass Compositional Analysis Laboratory Procedures. Available online: https://www.nrel.gov/bioenergy/biomass-compositional-analysis.html (accessed on 23 August 2021).
- Low Solids Enzymatic Saccharification of Lignocellulosic Biomass. Laboratory Analytical Procedure (LAP). Available online: https://www.nrel.gov/docs/fy15osti/63351.pdf (accessed on 23 August 2021).
- Chen, X.; Shekiro, J.; Pschorn, T.; Sabourin, M.; Tao, L.; Elander, R.; Park, S.; Jennings, E.; Nelson, R.; Trass, O.; et al. A highly efficient dilute alkali deacetylation and mechanical (disc) refining process for the conversion of renewable biomass to lower cost sugars. Biotechnol. Biofuels 2014, 7, 98. [Google Scholar] [CrossRef] [Green Version]
- Resch, M.G.; Donohoe, B.S.; Baker, J.O.; Decker, S.R.; Bayer, E.A.; Beckham, G.T.; Himmel, M.E. Fungal cellulases and complexed cellulosomal enzymes exhibit synergistic mechanisms in cellulose deconstruction. Energy Environ. Sci. 2013, 6, 1858–1867. [Google Scholar] [CrossRef]
- Jurchescu, I.M.; Hamann, J.; Zhou, X.; Ortmann, T.; Kuenz, A.; Prusse, U.; Lang, S. Enhanced 2,3-butanediol production in fed batch cultures of free and immobilized Bacillus licheniformis DSM 8785. Appl. Microbiol. Biotechnol. 2013, 97, 6715–6723. [Google Scholar] [CrossRef] [PubMed]
- Seo, J.K.; Park, T.S.; Kwon, I.H.; Piao, M.Y.; Lee, C.H.; Ha, J.K. Characterization of Cellulolytic and Xylanolytic Enzymes of Bacillus licheniformis JK7 Isolated from the Rumen of a Native Korean Goat. Asian-Australas. J. Anim. Sci. 2013, 26, 50–58. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Gu, W.; Xu, H.-Y.; Yang, G.-L.; Shan, X.-F.; Chen, G.; Kang, Y.-H.; Wang, C.-F.; Qian, A.-D. Comparative genome analysis of Bacillus velezensis reveals a potential for degrading lignocellulosic biomass. 3 Biotech 2018, 8, 253. [Google Scholar] [CrossRef]
- Petrova, P.; Petlichka, S.; Petrov, K. New Bacillus spp. with potential for 2,3-butanediol production from biomass. J. Biosci. Bioeng. 2020, 130, 20–28. [Google Scholar] [CrossRef] [PubMed]
- Gong, G.; Kim, S.; Lee, S.M.; Woo, H.M.; Park, T.H.; Um, Y. Complete genome sequence of Bacillus sp. 275, producing extracellular cellulolytic, xylanolytic and ligninolytic enzymes. J. Biotechnol. 2017, 254, 59–62. [Google Scholar] [CrossRef]
- Robson, L.M.; Chambliss, G.H. Cellulases of bacterial origin. Enzym. Microb. Technol. 1989, 11, 626–644. [Google Scholar] [CrossRef]
- Rey, M.W.; Ramaiya, P.; Nelson, B.A.; Brody-Karpin, S.D.; Zaretsky, E.J.; Tang, M.; Lopez de Leon, A.; Xiang, H.; Gusti, V.; Clausen, I.G.; et al. Complete genome sequence of the industrial bacterium Bacillus licheniformis and comparisons with closely related Bacillus species. Genome Biol. 2004, 5, R77. [Google Scholar] [CrossRef] [Green Version]
- Choi, J.; Nam, J.; Seo, M.H. Complete genome sequence of Bacillus velezensis NST6 and comparison with the species belonging to operational group B. amyloliquefaciens. Genomics 2021, 113, 380–386. [Google Scholar] [CrossRef]
- Petrova, P.; Velikova, P.; Petrov, K. Genome sequence of Bacillus velezensis 5RB, an overproducer of 2,3-butanediol. Microbiol. Res. Announc. 2019, 8, e01475-18. [Google Scholar] [CrossRef] [Green Version]
- Ejaz, U.; Sohail, M.; Ghanemi, A. Cellulases: From Bioactivity to a Variety of Industrial Applications. Biomimetics 2021, 6, 44. [Google Scholar] [CrossRef] [PubMed]
- Leis, B.; Held, C.; Bergkemper, F.; Dennemarck, K.; Steinbauer, R.; Reiter, A.; Mechelke, M.; Moerch, M.; Graubner, S.; Liebl, W.; et al. Comparative characterization of all cellulosomal cellulases from Clostridium thermocellum reveals high diversity in endoglucanase product formation essential for complex activity. Biotechnol. Biofuels 2017, 10, 240. [Google Scholar] [CrossRef] [Green Version]
- Hirano, K.; Kurosaki, M.; Nihei, S.; Hasegawa, H.; Shinoda, S.; Haruki, M.; Hirano, N. Enzymatic diversity of the Clostridium thermocellum cellulosome is crucial for the degradation of crystalline cellulose and plant biomass. Sci. Rep. 2016, 6, 35709. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Olson, D.G.; Tripathi, S.A.; Giannone, R.J.; Lo, J.; Caiazza, N.C.; Hogsett, D.A.; Hettich, R.L.; Guss, A.M.; Dubrovsky, G.; Lynd, L.R. Deletion of the Cel48S cellulase from Clostridium thermocellum. Proc. Natl. Acad. Sci. USA 2010, 107, 17727–17732. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lombard, V.; Golaconda Ramulu, H.; Drula, E.; Coutinho, P.M.; Henrissat, B. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 2014, 42, D490–D495. [Google Scholar] [CrossRef] [Green Version]
- Sidar, A.; Albuquerque, E.D.; Voshol, G.P.; Ram, A.F.J.; Vijgenboom, E.; Punt, P.J. Carbohydrate Binding Modules: Diversity of Domain Architecture in Amylases and Cellulases from Filamentous Microorganisms. Front. Bioeng. Biotechnol. 2020, 8, 871. [Google Scholar] [CrossRef]
- Guimaraes, B.G.; Souchon, H.; Lytle, B.L.; David Wu, J.H.; Alzari, P.M. The crystal structure and catalytic mechanism of cellobiohydrolase CelS, the major enzymatic component of the Clostridium thermocellum cellulosome. J. Mol. Biol. 2002, 320, 587–596. [Google Scholar] [CrossRef]
- Saharay, M.; Guo, H.; Smith, J.C. Catalytic mechanism of cellulose degradation by a cellobiohydrolase, CelS. PLoS ONE 2010, 5, e1294. [Google Scholar] [CrossRef]
- Glycoside Hydrolase Family Classification. Available online: http://www.cazy.org/Glycoside-Hydrolases.html (accessed on 11 September 2021).
- Alzari, P.M.; Souchon, H.; Dominguez, R. The crystal structure of endoglucanase CelA, a family 8 glycosyl hydrolase from Clostridium thermocellum. Structure 1996, 4, 265–275. [Google Scholar] [CrossRef] [Green Version]
- Kerényiová, L.; Janeček, Š. Extension of the taxonomic coverage of the family GH126 outside Firmicutes and in silico characterization of its non-catalytic terminal domains. 3 Biotech 2020, 10, 420. [Google Scholar] [CrossRef]
- Imran, M.; Anwar, Z.; Irshad, M.; Asad, M.; Ashfaq, H. Cellulase Production from Species of Fungi and Bacteria from Agricultural Wastes and Its Utilization in Industry: A Review. Adv. Enzym. Res. 2016, 4, 44–55. [Google Scholar] [CrossRef] [Green Version]
- Karim, A.; Asif Nawaz, M.; Aman, A.; Qader, S.A.U. Hyper production of cellulose degrading endo (1,4) β-d-glucanase from Bacillus licheniformis KIBGE-IB2. J. Radiat. Res. Appl. Sci. 2015, 8, 160–165. [Google Scholar] [CrossRef] [Green Version]
- Fan, B.; Blom, J.; Klenk, H.-P.; Borriss, R. Bacillus amyloliquefaciens, Bacillus velezensis, and Bacillus siamensis form an “operational group B. amyloliquefaciens” within the B. subtilis species complex. Front. Microbiol. 2017, 8, 22. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Borriss, R.; Chen, X.H.; Rueckert, C.; Blom, J.; Becker, A.; Baumgarth, B.; Fan, B.; Pukall, R.; Schumann, P.; Spröer, C. Relationship of Bacillus amyloliquefaciens clades associated with strains DSM 7T and FZB42T: A proposal for Bacillus amyloliquefaciens subsp. amyloliquefaciens subsp. nov. and Bacillus amyloliquefaciens subsp. plantarum subsp. nov. based on complete genome sequence comparisons. Int. J. Syst. Evol. Microbiol. 2011, 61, 1786–1801. [Google Scholar] [CrossRef] [PubMed]
- Rabbee, M.F.; Ali, M.S.; Choi, J.; Hwang, B.S.; Jeong, S.C.; Baek, K.H. Bacillus velezensis: A Valuable Member of Bioactive Molecules within Plant Microbiomes. Molecules 2019, 24, 1046. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tsigoriyna, L.; Ganchev, D.; Petrova, P.; Petrov, K. Highly Efficient 2,3-Butanediol Production by Bacillus licheniformis via Complex Optimization of Nutritional and Technological Parameters. Fermentation 2021, 7, 118. [Google Scholar] [CrossRef]
- Zhang, X.-Z.; Zhang, Y.-H.P. Cellulases: Characteristics, sources, production, and applications. In Bioprocessing Technologies in Biorefinery for Sustainable Production of Fuels, Chemicals, and Polymers, 1st ed.; Yang, S.-T., El-Enshasy, H.A., Thongchul, N., Eds.; John Wiley & Sons Inc.: Hoboken, NJ, USA, 2013; pp. 131–146. [Google Scholar]
- Shajahan, S.; Ganesh Moorthy, I.; Sivakumar, N.; Selvakumar, G. Statistical modeling and optimization of cellulase production by Bacillus licheniformis NCIM 5556 isolated from the hot spring, Maharashtra, India. J. King Saud Univ.-Sci. 2017, 29, 302–310. [Google Scholar] [CrossRef] [Green Version]
- O’Hair, J.A.; Hui, L.; Mahesh, R.; Thapa, S.; Yang, Y.; Fish, T.; Bhatti, S.; Thannhauser, T.W.; Zhou, S. Proteomic Effects of Magnesium Stress on Biofilm Associated Proteins Isolated from Cellulolytic Bacillus licheniformis YNP5-TSU. J. Proteom. Bioinform. 2019, 12, 113–121. [Google Scholar] [CrossRef] [Green Version]
- Da Silva, R.N.; Melo, L.F.A.; Finkler, C.L.L. Optimization of the cultivation conditions of Bacillus licheniformis BCLLNF-01 for cellulase production. Biotechnol. Rep. 2021, 29, e00599. [Google Scholar] [CrossRef]
- De Araújo, E.A.; de Oliveira Neto, M.; Polikarpov, I. Biochemical characterization and low-resolution SAXS structure of two-domain endoglucanase BlCel9 from Bacillus licheniformis. Appl. Microbiol. Biotechnol. 2019, 103, 1275–1287. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Ku, S. Bacillus Cellulase Molecular Cloning, Expression, and Surface Display on the Outer Membrane of Escherichia coli. Molecules 2018, 23, 503. [Google Scholar] [CrossRef] [Green Version]
- Kataeva, I.; Li, X.-L.; Chen, H.; Choi, S.-K.; Ljungdahl, L.G. Cloning and sequence analysis of a new cellulase gene encoding CelK, a major cellulosome component of Clostridium thermocellum: Evidence for gene duplication and recombination. J. Bacteriol. 1999, 181, 5288–5295. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Z.Y.; Raza, M.F.; Zheng, Z.; Zhang, X.; Dong, X.; Zhang, H. Complete genome sequence of Bacillus velezensis ZY-1-1 reveals the genetic basis for its hemicellulosic/cellulosic substrate-inducible xylanase and cellulase activities. 3 Biotech 2018, 8, 465. [Google Scholar] [CrossRef]
- Nair, A.S.; Al-Battashi, H.; Al-Akzawi, A.; Annamalai, N.; Gujarathi, A.; Al-Bahry, S.; Dhillon, G.S.; Sivakumar, N. Waste office paper: A potential feedstock for cellulase production by a novel strain Bacillus velezensis ASN1. Waste Manag. 2018, 79, 491–500. [Google Scholar] [CrossRef]
- Abd Elhameed, E.; Sayed, A.R.M.; Radwan, T.E.E.; Hassan, G. Biochemical and Molecular Characterization of Five Bacillus Isolates Displaying Remarkable Carboxymethyl Cellulase Activities. Curr. Microbiol. 2020, 77, 3076–3084. [Google Scholar] [CrossRef]
- Stephenson, K.; Jensen, C.L.; Jørgensen, S.T.; Lakey, J.H.; Harwood, C.R. The influence of secretory-protein charge on late stages of secretion from the Gram-positive bacterium Bacillus subtilis. Biochem. J. 2000, 350, 31–39. [Google Scholar] [CrossRef]
- Kovács, K.; Willson, B.J.; Schwarz, K.; Heap, J.T.; Jackson, A.; Bolam, D.N.; Winzer, K.; Minton, N.P. Secretion and assembly of functional mini-cellulosomes from synthetic chromosomal operons in Clostridium acetobutylicum ATCC 824. Biotechnol. Biofuels 2013, 6, 117. [Google Scholar] [CrossRef] [Green Version]
- Okonkwo, C.C.; Ujor, V.; Ezeji, T.C. Investigation of relationship between 2,3-butanediol toxicity and production during growth of Paenibacillus polymyxa. New Biotechnol. 2017, 34, 23–31. [Google Scholar] [CrossRef] [PubMed]
- Xue, G.-P.; Johnson, J.S.; Dalrymple, B.P. High osmolarity improves the electro-transformation efficiency of the gram-positive bacteria Bacillus subtilis and Bacillus licheniformis. J. Microbiol. Methods 1999, 34, 183–191. [Google Scholar] [CrossRef]
- Miller, G.L. Use of dinitrosalicylic acid reagent for determination of reducing sugar. Anal. Chem. 1959, 31, 426–428. [Google Scholar] [CrossRef]
- Yan, S.; Wu, G. Signal peptide of cellulase. Appl. Microbiol. Biotechnol. 2014, 98, 5329–5362. [Google Scholar] [CrossRef] [PubMed]
| Clone | Signal Peptide Sequence * | Sec/SPI Likelihood | AA | Cleavage Site |
|---|---|---|---|---|
| 34 | VRSKKLWISLLFALTLIFTMAFSNMSAQA | 0.916 | 29 | 29–30 |
| 6 | MFKKLLLATSALTFSLSLVLPLDGHAKA | 0.932 | 28 | 28–29 |
| 5 | MKKTIMSFVAVAALSTTAFGAHA | 0.960 | 23 | 23–24 |
| 21 | MKKKKTWKRFLHFSSAALAAGLIFTSAAPAEA | 0.933 | 32 | 32–33 |
| 23 | MKRLFMKASLVLFAVVFVFAVKGAPAKA | 0.906 | 28 | 30–31 |
| 20 | MKKRLIGFLVLVPALIMSGITLIEA | 0.863 | 25 | 27–28 |
| 1 | MKVCQKSIVRFLVSLIIGTFVISVPFMANA | 0.858 | 30 | 30–31 |
| 19 | MKRLLSTLLIGIMLLTCAPSAFA | 0.850 | 23 | 23–24 |
| Target | Gene | Primer Pair 1 | Product Size (bp) |
|---|---|---|---|
| B. licheniformis 24 | celB (GH5) | BLcelB_F, BLcelB_R | 451 |
| B. licheniformis 24 | celA (GH9) | BLcelA_F, BLcelA_R | 232 |
| B. licheniformis 24 | celC (GH5) | BLcelC_F, BLcelC_R | 407 |
| B. velezensis 5RB | eglS (GH5) | eglS_F, eglS_R | 502 |
| Clone 34 | cel8A (GH8) | RTCel8AF, RTCel8AR | 614 |
| Clone 6 | cel48S (GH48) | RTCel48SF, RTCel48SR | 327 |
| Primer | Sequence (5′–3′) 1 | Target Sequence | Positions in Gene | Tm (°C) | Purpose |
|---|---|---|---|---|---|
| Cel8A_F | CAGTCGGTACCGTGAAGAACGTAAAAAAAAGAGTAGGTGTGG | CP000568 | 1–31 | 77.9 | Cloning |
| Cel8A_R | CTAGTAAGCTTCTAATAAGGTAGGTGGGGTATGCTCTTTATC | CP000568 | 1403–1434 | 76 | Cloning |
| Cel48S_F | CAGTCGGTACCATGGTAAAAAGCAGAAAGATTTCTATTCTG | CP000568 | 1–30 | 74.9 | Cloning |
| Cel48S_R | CTAGTAAGCTTTTAGTTCTTGTACGGCAATGTATCTATTTC | CP000568 | 2196–2226 | 72.9 | Cloning |
| SP_F | CTTAAGCAAAAGGAGAGGGACGCGT | pBES_cel8A | 312–337 | 67.4 | Sequencing |
| SP_R | GTAGGTGTGGTTTTGCTGATTCTTGC | pBES_cel8A | 475–502 | 64 | Sequencing |
| RTCel8AF | TCTTGCAGTGTTGGGGGTTT | CP000568 | 42–61 | 58.4 | RT-PCR |
| RTCel8AR | CAGGCGCAAAATATGACGGG | CP000568 | 655–636 | 60.5 | RT-PCR |
| RTCel48SF | CTGCATTCGCAGGTCCTACA | CP000568 | 71–90 | 60.5 | RT-PCR |
| RTCel48SR | AAGAAGACACATCCGGCTGC | CP000568 | 397–378 | 60.5 | RT-PCR |
| eglS_F | AACAAAGATTCAACGAAGGACGCC | NZ_CP011937 | 994–1017 | 63.5 | RT-PCR |
| eglS_R | TGGGTTCTGTTCCCCAAATCA | NZ_CP011937 | 1495–1475 | 59.4 | RT-PCR |
| BLcelB_F | GCCAATGAAAAGCATTCAAAATGG | WP_011197981 | 901–924 | 60.1 | RT-PCR |
| BLcelB_R | CCATAACCTGGCTGGCTCC | WP_011197981 | 1351–1333 | 61.7 | RT-PCR |
| BLcelC_F | GGTTCGTCAGGAGACTCTGG | CP034569 | 1000–1019 | 62.5 | RT-PCR |
| BLcelC_R | ATCTGGATTTCGCCAGTATCAG | CP034569 | 1406–1385 | 60.3 | RT-PCR |
| BLcelA_F | CGATTTAACGGGAGGGTGGT | LR134392 | 252–271 | 60.5 | RT-PCR |
| BLcelA_R | GTCTAGAGCGCCATTTCCGA | LR134392 | 483–464 | 60.5 | RT-PCR |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Arsov, A.; Petrov, K.; Petrova, P. Enhanced Activity by Genetic Complementarity: Heterologous Secretion of Clostridial Cellulases by Bacillus licheniformis and Bacillus velezensis. Molecules 2021, 26, 5625. https://doi.org/10.3390/molecules26185625
Arsov A, Petrov K, Petrova P. Enhanced Activity by Genetic Complementarity: Heterologous Secretion of Clostridial Cellulases by Bacillus licheniformis and Bacillus velezensis. Molecules. 2021; 26(18):5625. https://doi.org/10.3390/molecules26185625
Chicago/Turabian StyleArsov, Alexander, Kaloyan Petrov, and Penka Petrova. 2021. "Enhanced Activity by Genetic Complementarity: Heterologous Secretion of Clostridial Cellulases by Bacillus licheniformis and Bacillus velezensis" Molecules 26, no. 18: 5625. https://doi.org/10.3390/molecules26185625
APA StyleArsov, A., Petrov, K., & Petrova, P. (2021). Enhanced Activity by Genetic Complementarity: Heterologous Secretion of Clostridial Cellulases by Bacillus licheniformis and Bacillus velezensis. Molecules, 26(18), 5625. https://doi.org/10.3390/molecules26185625







