Fungal α-1,3-Glucan as a New Pathogen-Associated Molecular Pattern in the Insect Model Host Galleria mellonella
Abstract
:1. Introduction
2. Results
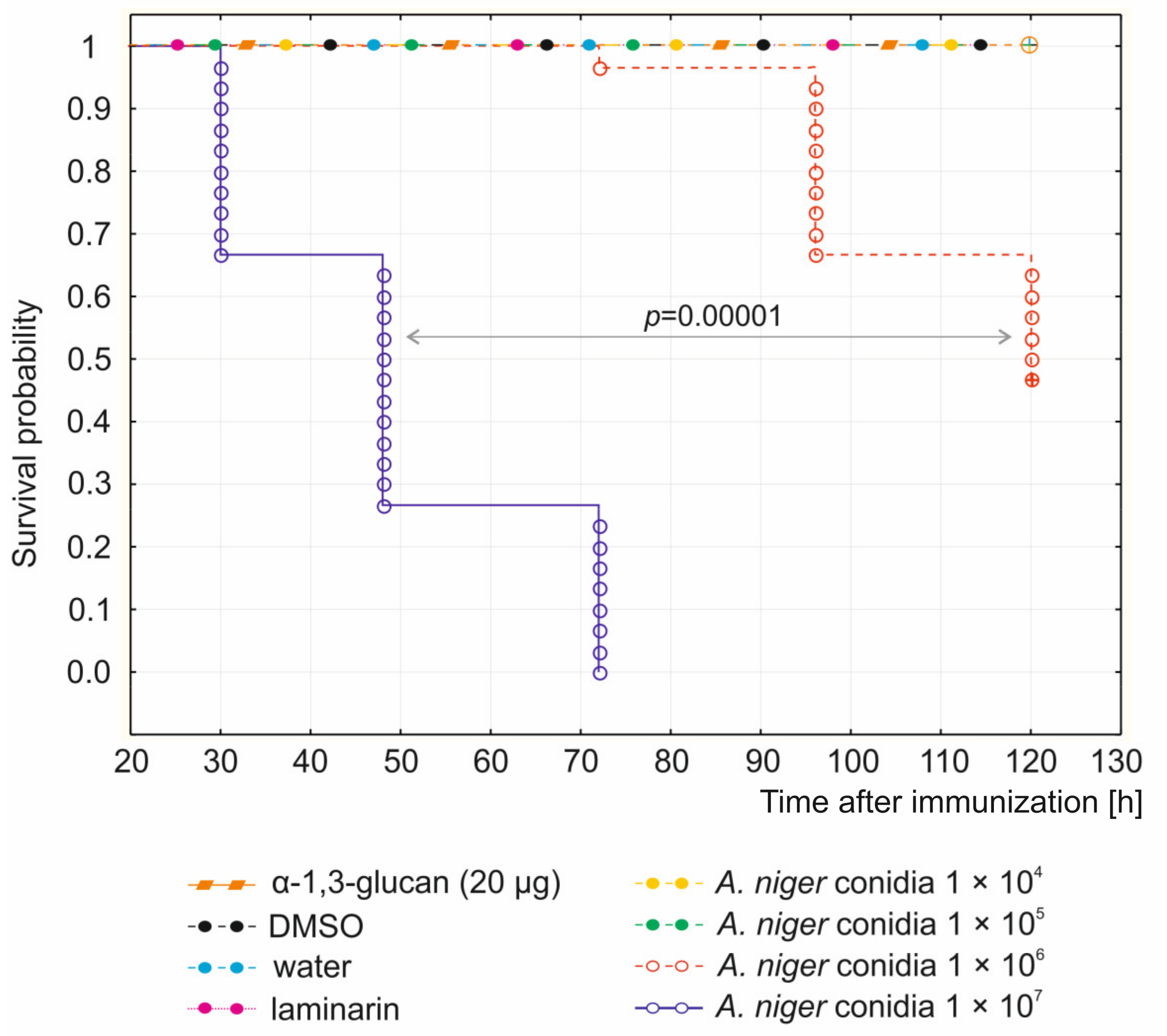
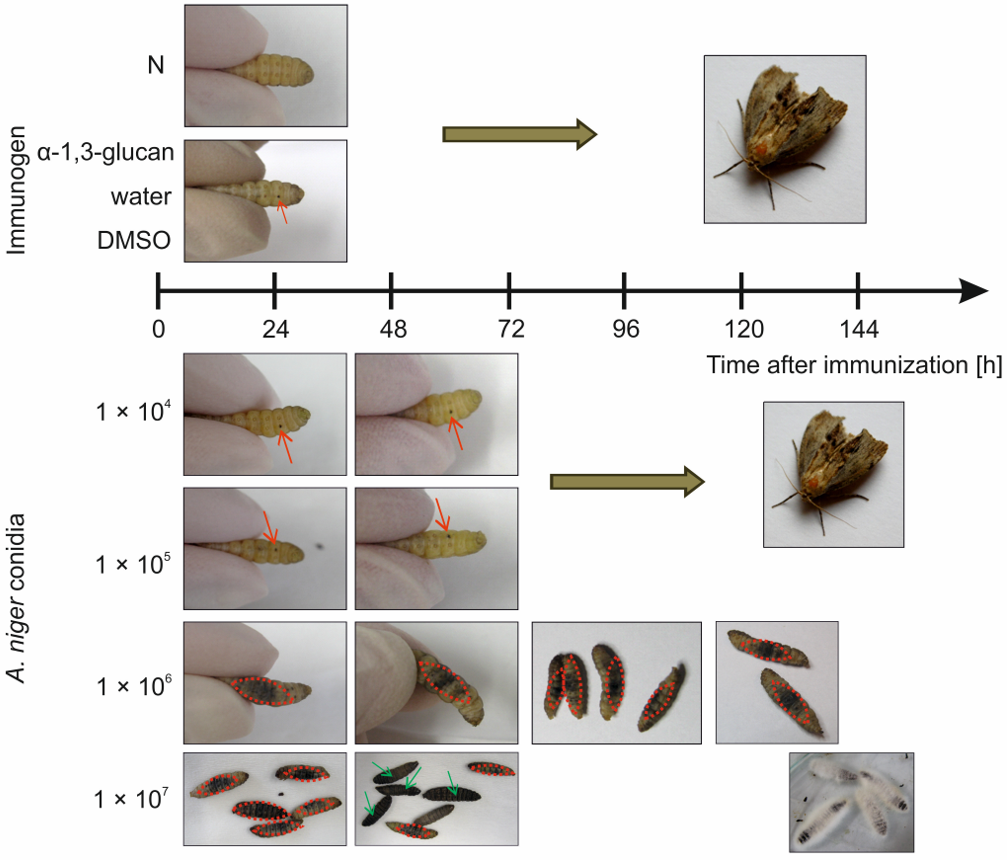
2.1. Survival Experiments
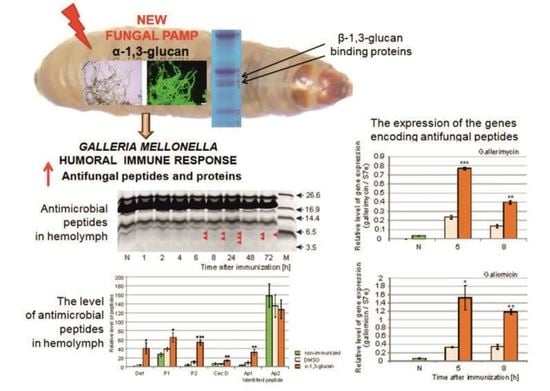
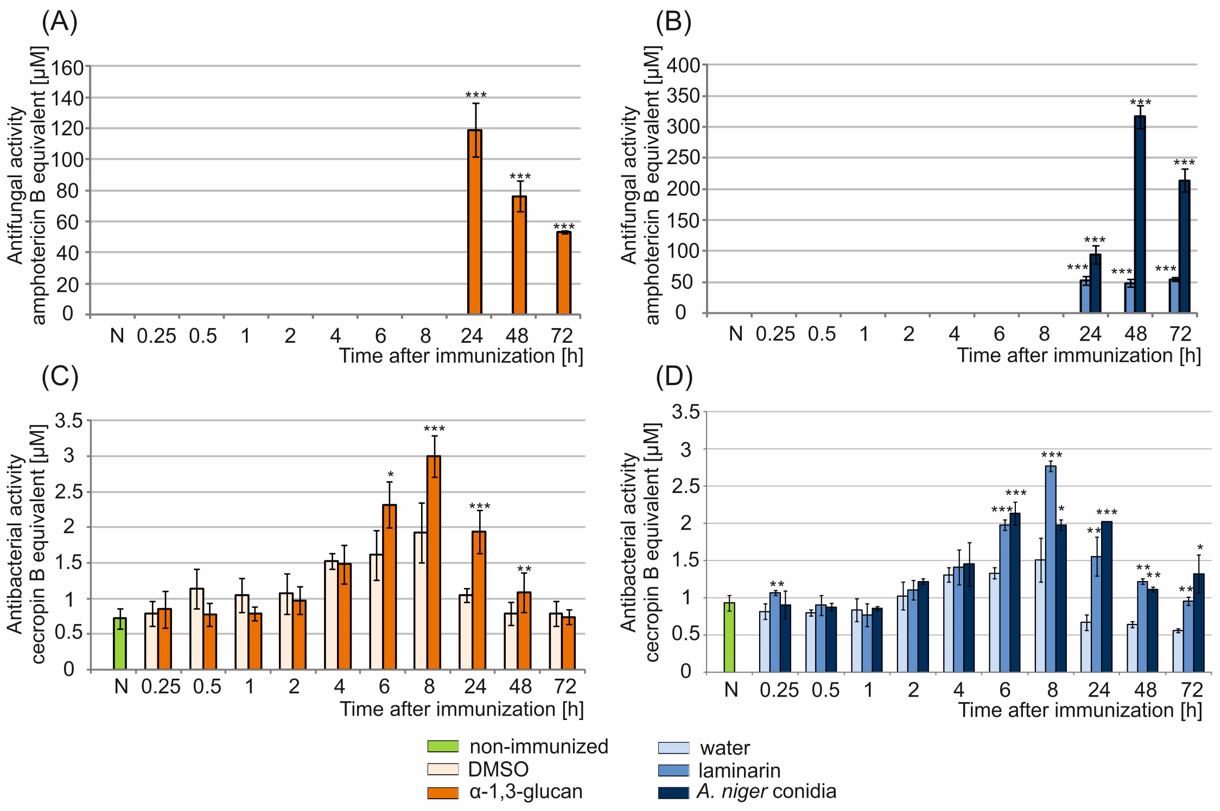
2.2. Analysis of Antimicrobial Activity in G. mellonella Hemolymph after Immunization with α-1,3-Glucan
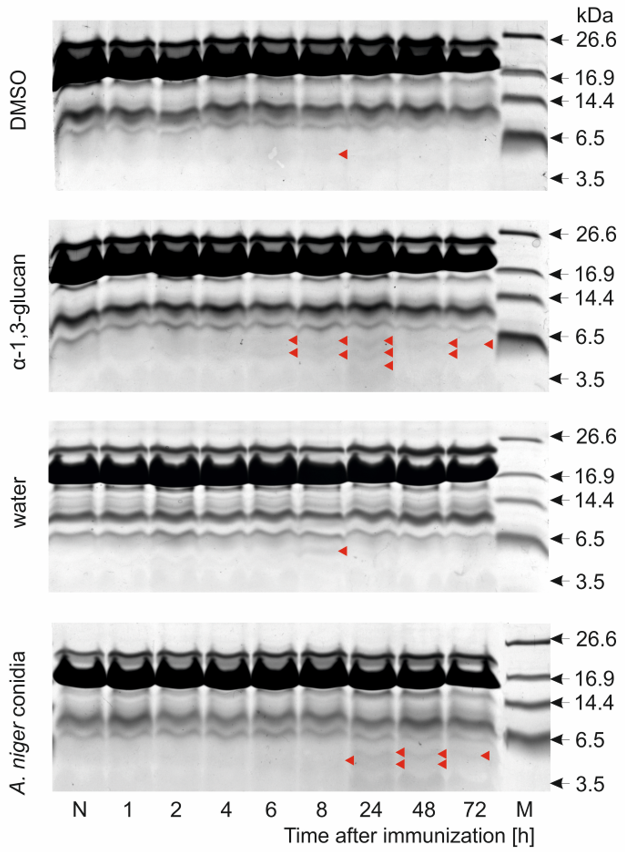
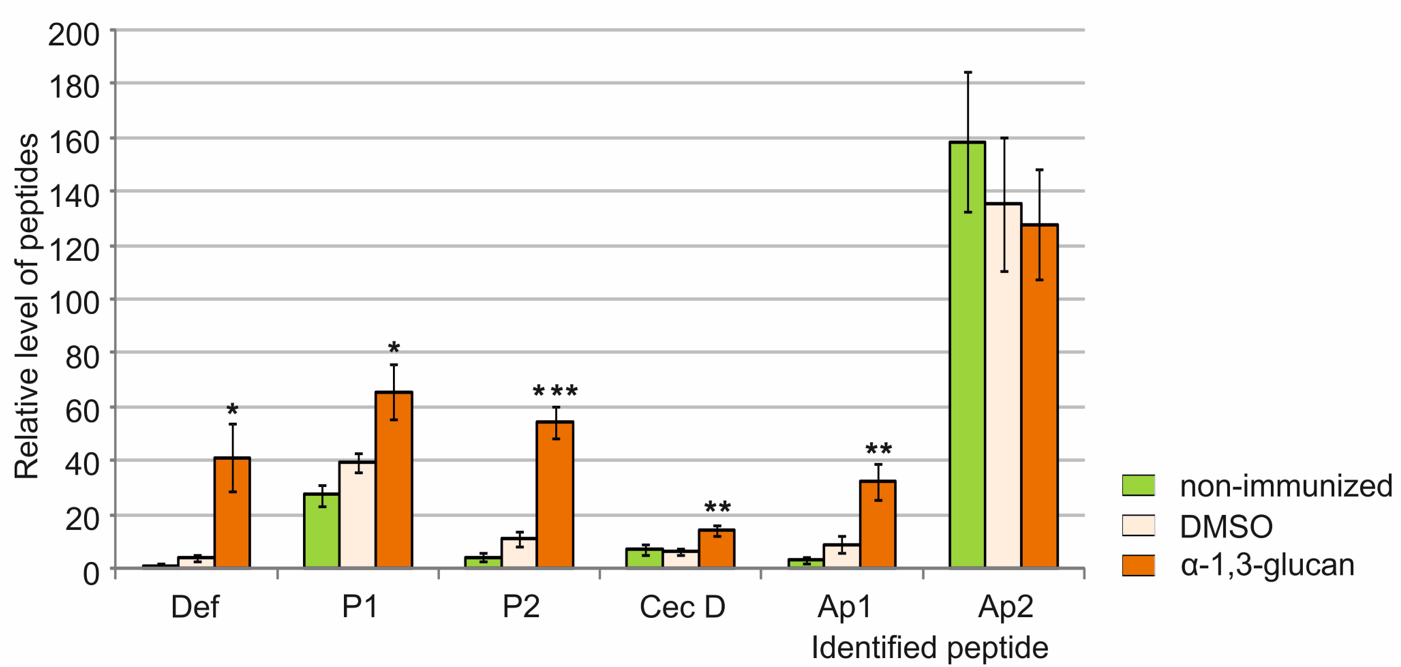
2.3. Identification of Antimicrobial Peptides in G. mellonella Hemolymph
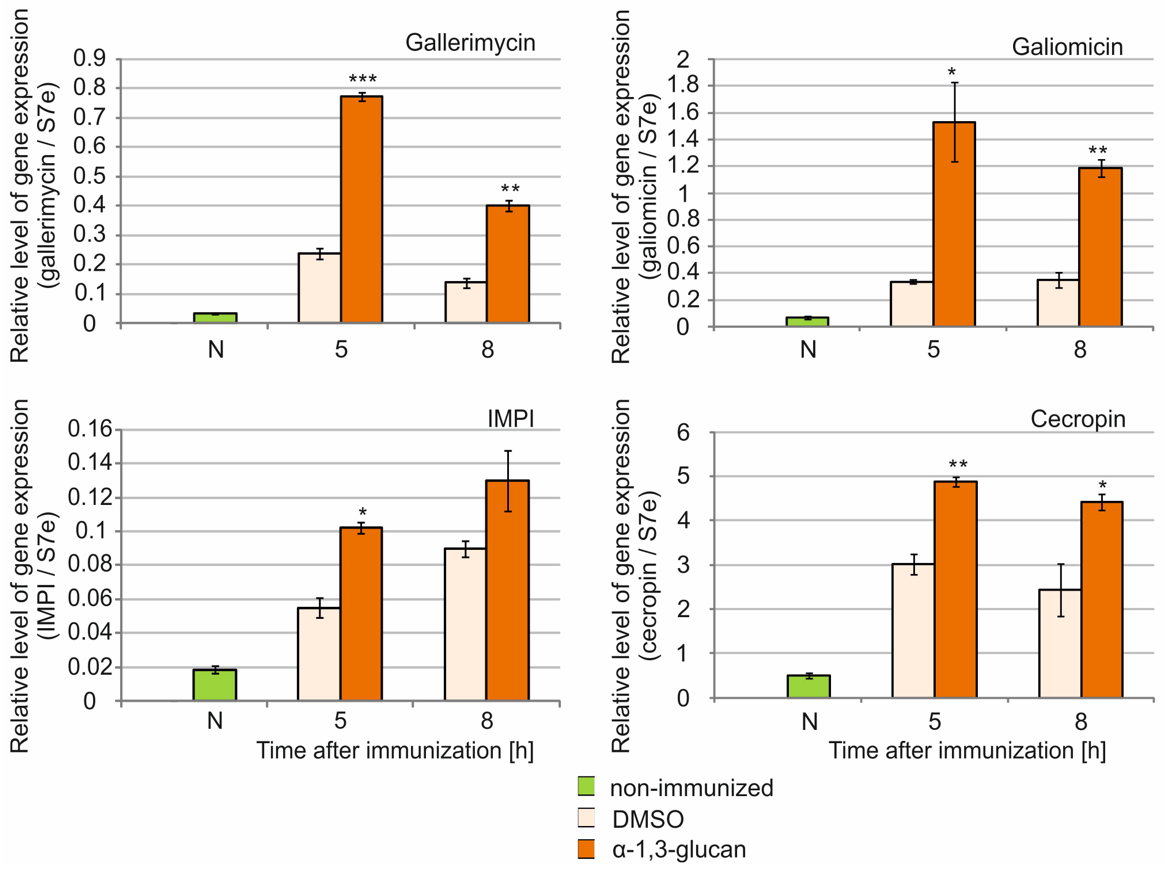
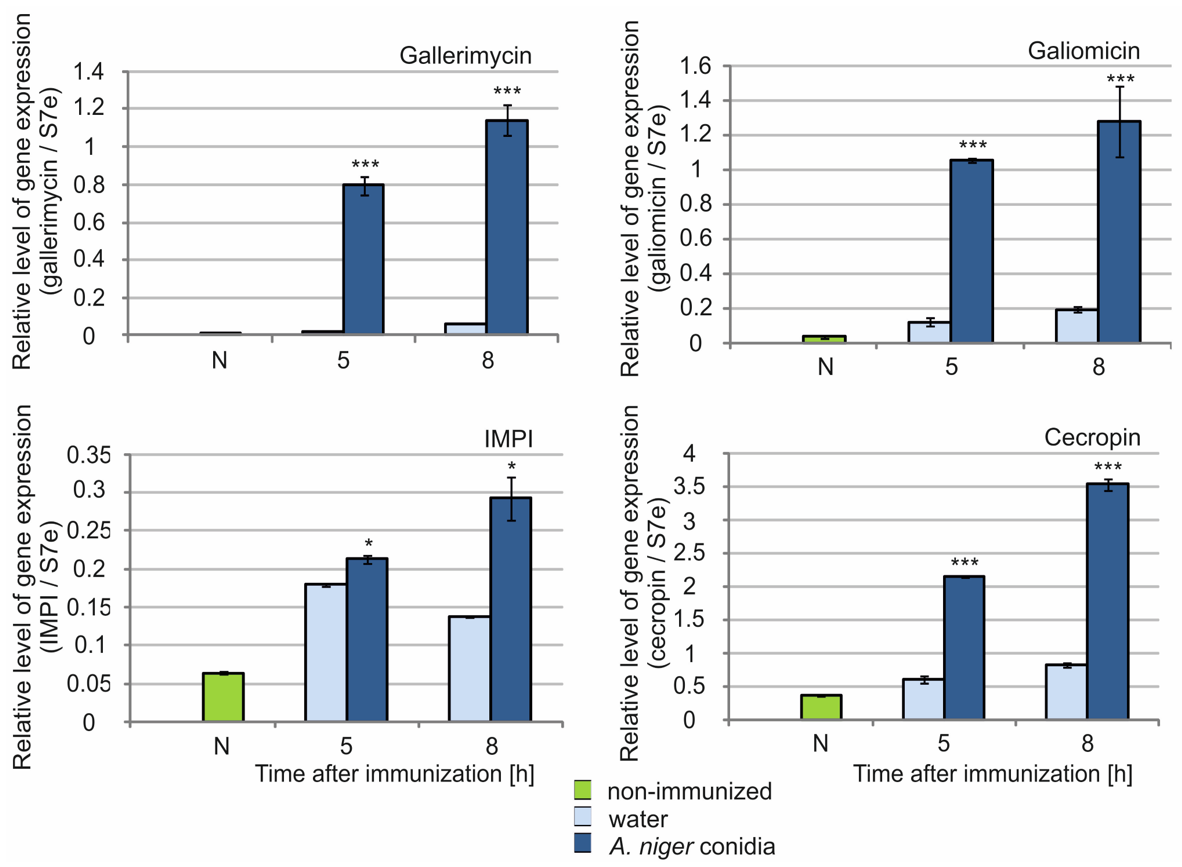
2.4. Analysis of Antimicrobial Peptide Gene Expression in G. mellonella Fat Body
2.5. Identification of G. mellonella Hemolymph Proteins Binding to A. niger α-1,3-Glucan
3. Discussion
4. Materials and Methods
4.1. Microorganisms and Growth Conditions
4.2. Immunodetection of α-1,3-Glucan in A. niger
4.3. Isolation and Characterization of α-1,3-Glucan from A. niger
4.4. Atomic Force Microscopy Imaging of α-1,3-Glucan
4.5. Insect Rearing and Immunization
4.6. Survival Experiments
4.7. Collection of Hemolymph and Preparation of Hemolymph Methanolic Extracts
4.8. Antimicrobial Activity Assays
4.9. Analysis of the Polypeptide Profile in Hemolymph
4.10. Analysis of Peptide Gene Expression in the Fat Body
4.10.1. Collection of Fat Bodies
4.10.2. Peptide Gene Expression
4.11. Identification of α-1,3-Glucan Ligands in Insect Hemolymph
4.11.1. Binding of Hemolymph Proteins to α-1,3-Glucan
4.11.2. Identification of Proteins by N-terminal Sequencing
4.11.3. Identification Using Immunodetection with Specific Antibodies
4.11.4. Mass Spectrometry
4.12. Other Methods
4.13. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Dubovskiy, I.M.; Kryukova, N.A.; Glupov, V.V.; Ratcliffe, N.A. Encapsulation and nodulation in insects. Invertebr. Surviv. J. 2016, 13, 229–246. [Google Scholar] [CrossRef]
- Sheehan, G.; Farrel, G.; Kavanagh, K. Immmune proming: The secret weapon of the insect world. Virulence 2020, 11, 238–246. [Google Scholar] [CrossRef] [Green Version]
- Beauvais, A.; Latgé, J.P. Fungal cell wall. J. Fungi 2018, 4, 91. [Google Scholar] [CrossRef] [Green Version]
- Erwig, L.P.; Gow, N.A.R. Interactions of fungal pathogens with phagocytes. Nat. Rev. Microbiol. 2016, 14, 163–176. [Google Scholar] [CrossRef] [PubMed]
- Gow, N.A.R.; Latgé, J.P.; Munro, C.A. The fungal cell wall: Structure, biosynthesis, and function. Microbiol. Spectr. 2017, 5, 1–25. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grün, C.H. Structure and Biosynthesis a Fungal Glucans. Ph.D. Thesis, Universitet Utrecht, Algmene Catalogus, Utrecht, The Netherlands, 2003; pp. 9–33. [Google Scholar]
- Wiater, A.; Paduch, R.; Choma, A.; Stączek, S.; Pleszczyńska, M.; Tomczyk, M.; Locatelli, M.; Szczodrak, J. (1→3)-α-D-glucans from Aspergillus spp.: Structural characterization and biological study on their carboxymethylated derivatives. Curr. Drug Targets 2015, 16, 1488–1494. [Google Scholar] [CrossRef]
- Schoffelmeer, E.A.M.; Klis, F.M.; Sietsma, J.H.; Cornelissen, B.J.C. The cell wall of Fusarium oxysporum. Fungal Genet. Biol. 1999, 27, 275–282. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, J.; Jiang, H.; Du, Y.; Keyhami, N.O.; Xia, Y.; Jin, K. Members of chitin synthase family in Metarhizium acridum differentially affect fungal growth, stress tolerances, cell wall integrity and virulence. PLOS Pathog. 2019, 15, e1007964. [Google Scholar] [CrossRef]
- Zhao, T.; Tian, H.; Xia, Y.; Jin, K. MaPmt4, a protein O-mannosyltransferase, contributes to cell wall integrity, stress tolerance and virulence in Metarhizium acridum. Curr. Genet. 2019, 65, 1025–1040. [Google Scholar] [CrossRef]
- Santos, A.C.S.; Diniz, A.G.; Tiago, P.V.; de Oliveira, N.T. Entomopathogenic Fusarium species: A review of their potential for the biological control of insects, implications and prospects. Fungal Biol. Rev. 2020, 34, 41–57. [Google Scholar] [CrossRef]
- Wong, S.S.W.; Venugopalan, L.P.; Beaussart, A.; Karnam, A.; Mohammed, M.R.S.; Jayapal, J.M.; Bretagne, S.; Bayry, J.; Prajna, L.; Kuppamuthu, D.; et al. Species-specific immunological reactivities depend on the cell-wall organization of the two Aspergillus, Aspergillus fumigatus and A. flavus. Front. Cell Infect. Microbiol. 2021, 11, 643312. [Google Scholar] [CrossRef] [PubMed]
- Fontaine, T.; Simenel, C.; Dubreucq, G.; Adam, O.; Delepierre, M.; Lemoine, J.; Vorgias, C.E.; Diaquin, M.; Latgé, J.-P. Molecular organization of the alkali-insoluble fraction of Aspergillus fumigatus cell wall. J. Biol. Chem. 2000, 275, 27594–27607. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mouyna, I.; Fontaine, T. Cell wall of Aspergillus fumigatus: A dynamic structure. In Aspergillus Fumigatus and Aspergillosis; Latgé, J.-P., Steinbach, W.J., Eds.; ASM Press: Washington, DC, USA, 2009; pp. 149–158. [Google Scholar] [CrossRef]
- Latgé, J.P. Tasting the fungal cell wall. Cell. Microbiol. 2010, 12, 863–872. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.J.; Sheppard, D.C. Recent advances in the understanding of the Aspergillus fumigatus cell wall. J. Microbiol. 2016, 54, 232–242. [Google Scholar] [CrossRef]
- Yoshimi, A.; Miyazawa, K.; Abe, K. Cell wall structure and biogenesis in Aspergillus species. Biosci. Biotechnol. Biochem. 2016, 80, 1700–1711. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Beauvais, A.; Fontaine, T.; Aimanianda, V.; Latgé, J.P. Aspergillus cell wall and biofilm. Mycopathologia 2014, 178, 371–377. [Google Scholar] [CrossRef] [PubMed]
- Zonneveld, B.J.M. Inhibitory effect of 2-deoxyglucose on cell wall alpha-1,3-glucan synthesis and cleistothecium development in Aspergillus nidulans. Dev. Biol. 1973, 34, 1–8. [Google Scholar] [CrossRef]
- Yoshimi, A.; Miyazawa, K.; Abe, K. Function and biosynthesis of cell wall α-1,3-glucan in fungi. J. Fungi 2017, 3, 63. [Google Scholar] [CrossRef]
- Zhang, S.; Sato, H.; Ichinose, S.; Tanaka, M.; Miyazawa, K.; Yoshimi, A.; Abe, K.; Shintani, T.; Gomi, K. Cell wall α-1,3-glucan prevents α-amylase adsorption onto fungal cell in submerged culture of Aspergillus oryzae. J. Biosci. Bioeng. 2017, 124, 47–53. [Google Scholar] [CrossRef]
- Hogan, L.H.; Klein, B.S. Altered expression of surface α-1,3-glucan in genetically related strains of Blastomyces dermatitidis that differ in virulence. Infect. Immun. 1994, 62, 3543–3546. [Google Scholar] [CrossRef] [Green Version]
- Pinto, M.R.; Barreto-Bergter, E.; Taborda, C.P. Glycoconjugates and polysaccharides of fungal cell wall and activation of immune system. Braz. J. Microbiol. 2005, 39, 195–208. [Google Scholar] [CrossRef] [Green Version]
- Rappleye, C.A.; Eissenberg, L.G.; Goldman, W.E. Histoplasma capsulatum α-(1,3)-glucan blocks innate immune recognition by the β-glucan receptor. Proc. Nati. Acad. Sci. USA 2007, 104, 1366–1370. [Google Scholar] [CrossRef] [Green Version]
- Złotko, K.; Wiater, A.; Waśko, A.; Pleszczyńska, M.; Paduch, R.; Jaroszuk-Ścieseł, J.; Bieganowski, A. A report on fungal (1→3)-α-d-glucans: Properties, functions and application. Molecules 2019, 24, 3972. [Google Scholar] [CrossRef] [Green Version]
- Edwards, J.A.; Alore, E.A.; Rappleye, C.A. The yeast-phase virulence requirement for α-glucan synthase differs among Histoplasma capsulatum chemotypes. Eucaryot. Cell 2010, 10, 87–97. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Edwards, J.A.; Rappleye, C.A. Histoplasma mechanisms of pathogenesis-one portfolio doesn’t fit all. FEMS Microbiol. Lett. 2011, 324, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Woods, J.P. Revisiting old Friends: Developments in understanding Histoplasma capsulatum pathogenesis. J. Microbiol. 2016, 54, 265–276. [Google Scholar] [CrossRef] [PubMed]
- Reese, A.J.; Doering, T.L. Cell wall α-1,3-glucan is required to anchor the Cryptococcus neoformans capsule. Mol. Microbiol. 2003, 50, 1401–1409. [Google Scholar] [CrossRef]
- Reese, A.J.; Yoneda, A.; Breger, J.A.; Beauvais, A.; Liu, H.; Griffith, C.L.; Bose, I.; Kim, M.J.; Skau, C.; Yang, S.; et al. Loss of cell wall α(1-3)glucan affects Cryptococcus neoformans from ultrastructure to virulence. Mol. Microbiol. 2007, 63, 1385–1398. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Camacho, E.; Sepulveda, V.; Goldman, W.; San-Blas, G.; Nino-Vega, G. Expression of Paracoccidioides brasiliensis AMY1 in a Histoplasma capsulatum amy 1 mutant, relates an α-(1,4)-amylase to cell wall α-1,3-glucan synthesis. PLoS ONE 2012, 7, e50201. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Beauvais, A.; Bozza, S.; Kniemeyer, O.; Formosa, C.; Balloy, V.; Henry, C.; Roberson, R.; Dague, E.; Chignard, M.; Brakhage, A.A.; et al. Deletion of the α-1,3-glucan synthase genes induces a restructuring of the conidial cell wall responsible for the avirulence of Aspergillus fumigatus. PLoS Pathog. 2013, 9, e1003716. [Google Scholar] [CrossRef]
- Polacheck, I.; Rosenberger, R.F. Aspergillus nidulans mutant lacking α-1,3-glucan, melanin and cleistothecia. J. Bacteriol. 1977, 132, 650–656. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bao, X.; Duan, J.; Fang, X.; Fang, J. Chemical modifications of the (1→3)-α-d-glucan from spores of Ganoderma lucidum and investigation of their physicochemical properties and immunological activity. Carbohydr. Res. 2001, 336, 127–140. [Google Scholar] [CrossRef]
- Shetty, R.S.; Batchu, U.R.; Buddana, S.K.; Rao, K.R.S.; Penna, S. A comprehensive review on α-D-Glucans: Structural and functional diversity, derivatization and bioapplications. Carbohydr. Res. 2021, 503, 108297. [Google Scholar] [CrossRef] [PubMed]
- Huang, Q.; Zhang, L. Preparation, chain conformation and anti-tumor activities of water-soluble phosphated (1 → 3)-α-d-glucan from Poria cocos cymelia. Carbohydr. Polym. 2011, 83, 1363–1369. [Google Scholar] [CrossRef]
- Wiater, A.; Paduch, R.; Pleszczyńska, M.; Próchniak, K.; Choma, A.; Kandefer-Szerszeń, M.; Szczodrak, J. α-(1 → 3)-d-Glucans from fruiting bodies of selected macromycetes fungi and the biological activity of their carboxymethylated products. Biotechnol. Lett. 2011, 33, 787–795. [Google Scholar] [CrossRef] [Green Version]
- Bozza, S.; Clavaud, C.; Giovannini, G.; Fontaine, T.; Beauvais, A.; Sarfati, J.; D’Angelo, C.; Perruccio, K.; Bonifazi, P.; Zagarella, S.; et al. Immune sensing of Aspergillus fumigatus proteins, glycolipids, and polysaccharides and the impact on Th Immunity and vaccination. J. Immunol. 2009, 183, 2407–2414. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; Zhang, Y.; Zhang, R.; Zhang, J. The diversity of pattern recognition receptors (PRRs) involved with insect defense against pathogens. Curr. Opin. Insect Sci. 2019, 33, 105–110. [Google Scholar] [CrossRef] [PubMed]
- Rao, X.J.; Zhan, M.Y.; Pan, Y.M.; Liu, S.; Yang, P.J.; Yang, L.L.; Yu, X.Q. Immune functions of insect βGRPs and their potential application. Dev. Comp. Immunol. 2018, 83, 80–88. [Google Scholar] [CrossRef]
- Kim, Y.S.; Ryu, J.H.; Han, S.J.; Choi, K.H.; Nam, K.B.; Jang, I.H.; Lemaitre, B.; Brey, P.T.; Lee, W.J. Gram-negative bacteria-binding protein, a pattern recognition receptor for lipopolysaccharide and β-1,3-glucan that mediates the signaling for the induction of innate immune genes in Drosophila melanogaster. J. Biol. Chem. 2000, 275, 32721–32727. [Google Scholar] [CrossRef] [Green Version]
- Ma, C.; Kanost, M.R. A β-1,3-glucan recognition protein from an insect, Manduca sexta, agglutinates microorganisms and activates the phenoloxidase cascade. J. Biol. Chem. 2000, 275, 7505–7514. [Google Scholar] [CrossRef] [Green Version]
- Lee, W.J.; Lee, J.D.; Kravchenko, V.V.; Ulevitch, R.J.; Brey, P.T. Purification and molecular cloning of an inducible gram-negative bacteria-binding protein from the silkworm, Bombyx mori. Proc. Natl Acad. Sci. USA 1996, 93, 7888–7893. [Google Scholar] [CrossRef] [Green Version]
- Ochiai, M.; Ashida, M. A pattern-recognition protein for β-1,3-glucan: The binding domain and the cDNA cloning of β-1,3-glucan recognition protein from the silkworm, Bombyx mori. J. Biol. Chem. 2000, 275, 4995–5002. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Benaducci, T.; Sardi Jde, C.; Lourencetti, N.M.; Scorzoni, L.; Gullo, F.P.; Rossi, S.A.; Derissi, J.B.; de Azevedo Prata, M.C.; Fusco-Almeida, A.M.; Mendes-Giannini, M.J. Virulence of Cryptococcus sp. biofilms in vitro and in vivo using Galleria mellonella as an alternative model. Front. Microbiol. 2016, 7, 290. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Killiny, N. Generous hosts: Why the larvae of greater wax moth, Galleria mellonella is a perfect infectious host model? Virulence 2018, 9, 860–865. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reeves, E.P.; Messina, C.G.; Doyle, S.; Kavanagh, K. Correlation between gliotoxin production and virulence of Aspergillus fumigatus in Galleria mellonella. Mycopathologia 2004, 158, 73–79. [Google Scholar] [CrossRef]
- Thomaz, L.; García-Rodas, R.; Guimarães, A.J.; Taborda, C.P.; Zaragoza, O.; Nosanchuk, J.D. Galleria mellonella as a model host to study Paracoccidioides lutzii and Histoplasma capsulatum. Virulence 2013, 4, 139–146. [Google Scholar] [CrossRef] [Green Version]
- Jacobsen, I.D. Galleria mellonella as a model host to study virulence of Candida. Virulence 2014, 5, 237–239. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brown, S.E.; Howard, A.; Kasprzak, A.B.; Gordon, K.H.; East, P.D. The discovery and analysis of a diverged family of novel antifungal moricin-like peptides in the wax moth Galleria mellonella. Insect Biochem. Mol. Biol. 2008, 38, 201–212. [Google Scholar] [CrossRef]
- Brown, S.E.; Howard, A.; Kasprzak, A.B.; Gordon, K.H.; East, P.D. A peptidomics study reveals the impressive antimicrobial peptide arsenal of the wax moth Galleria mellonella. Insect Biochem. Mol. Biol. 2009, 39, 792–800. [Google Scholar] [CrossRef] [PubMed]
- Cytryńska, M.; Mak, P.; Zdybicka-Barabas, A.; Suder, P.; Jakubowicz, T. Purification and characterization of eight peptides from Galleria mellonella immune hemolymph. Peptides 2007, 28, 533–546. [Google Scholar] [CrossRef]
- Mak, P.; Zdybicka-Barabas, A.; Cytryńska, M. A different repertoire of Galleria mellonella antimicrobial peptides in larvae challenged with bacteria and fungi. Dev. Comp. Immunol. 2010, 34, 1129–1136. [Google Scholar] [CrossRef]
- Schuhmann, B.; Seitz, V.; Vilcinskas, A.; Podsiadlowski, L. Cloning and expression of gallerimycin, an antifungal peptide expressed in immune response of greater wax moth larvae, Galleria mellonella. Arch. Insect Biochem. Physiol. 2003, 53, 125–133. [Google Scholar] [CrossRef] [PubMed]
- Vogel, H.; Altincicek, B.; Glöckner, G.; Vilcinskas, A. A comprehensive transcriptome and immune-gene repertoire of the lepidopteran model host Galleria mellonella. BMC Genom. 2011, 12, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Stączek, S.; Zdybicka-Barabas, A.; Pleszczyńska, M.; Wiater, A.; Cytryńska, M. Aspergillus niger α-1,3-glucan acts as a virulence factor by inhibiting the insect phenoloxidase system. J. Invertebr. Pathol. 2020, 171, 107341. [Google Scholar] [CrossRef]
- Stączek, S.; Zdybicka-Barabas, A.; Wiater, A.; Pleszczyńska, M.; Cytryńska, M. Activation of cellular immune response in insect model host Galleria mellonella by fungal α-1,3-glucan. Pathog. Dis. 2020, 78, ftaa062. [Google Scholar] [CrossRef] [PubMed]
- Stączek, S.; Zdybicka-Barabas, A.; Mak, P.; Sowa-Jasiłek, A.; Kędracka-Krok, S.; Jankowska, U.; Suder, P.; Wydrych, J.; Grygorczuk, K.; Jakubowicz, T.; et al. Studies on localization and protein ligands of Galleria mellonella apolipophorin III during immune response against different pathogens. J. Insect Physiol. 2018, 105, 18–27. [Google Scholar] [CrossRef] [PubMed]
- Brown, G.D.; Gordon, S. Fungal beta-glucans and mammalian immunity. Immunity 2003, 19, 311–315. [Google Scholar] [CrossRef] [Green Version]
- Ferrandon, D.; Imler, J.L.; Hoffmann, J.A. Sensing infection in Drosophila: Toll and beyond. Semin. Immunol. 2004, 16, 43–53. [Google Scholar] [CrossRef]
- Kavanagh, K.; Reeves, E.P. Exploiting the potential of insects for in vivo pathogenicity testing of microbial pathogens. FEMS Microbiol. Rev. 2004, 28, 101–112. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fabrick, J.A.; Baker, J.E.; Kanost, M.R. cDNA cloning, purification, properties, and function of a β-1,3-glucan recognition protein from a pyralid moth, Plodia interpunctella. Insect Biochem. Mol. Biol. 2003, 33, 579–594. [Google Scholar] [CrossRef]
- Sun, Z.; Wu, W.; Zhang, G. Characterization of novel β-1,3-glucan recognition proteins from a Tibetan Plateau ghost moth Thitarodes pui (Lepidoptera, Hepialidae). Physiol. Entomol. 2019, 44, 20–32. [Google Scholar] [CrossRef]
- Zhang, R.; Cho, H.Y.; Kim, H.S.; Ma, Y.G.; Osaki, T.; Kwabata, S.; Söderhäll, K.; Lee, B.L. Characterization and properties of a 1,3-β-D-glucan pattern recognition protein of Tenebrio molitor larvae that is specifically degraded by serine protease during prophenoloxidase activation. J. Biol. Chem. 2003, 278, 42072–42079. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fallon, J.P.; Troy, N.; Kavanagh, K. Pre-exposure of Galleria mellonella larvae to different doses of Aspergillus fumigatus conidia causes differential activation of cellular and humoral immune responses. Virulence 2011, 2, 413–421. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Halwani, A.E.; Niven, D.F.; Dunphy, G.B. Apolipophorin-III and the interactions of lipoteichoic acids with the immediate immune responses of Galleria mellonella. J. Invertebr. Pathol. 2000, 76, 233–241. [Google Scholar] [CrossRef] [PubMed]
- Kato, Y.; Motoi, Y.; Taniai, K.; Kadono-Okuda, K.; Yamamoto, M.; Higashino, Y.; Shimabukuro, M.; Chowdhury, S.; Xu, J.; Sugiyama, M. Lipopolysaccharide-lipophorin complex formation in insect hemolymph: A common pathway of lipopolysaccharide detoxification both in insects and in mammals. Insect Biochem. Mol. Biol. 1994, 24, 547–555. [Google Scholar] [CrossRef]
- Whitten, M.M.A.; Tew, I.F.; Lee, B.L.; Ratcliffe, N.A. A novel role for an insect apolipoprotein (apolipophorin III) in β-1,3-glucan pattern recognition and cellular encapsulation reactions. J. Immunol. 2004, 172, 2177–2185. [Google Scholar] [CrossRef] [Green Version]
- Yu, H.Z.; Wang, J.; Zhang, S.Z.; Toufeeq, S.; Li, B.; Li, Z.; Yang, L.A.; Hu, P.; Xu, J.P. Molecular characterisation of apolipophorin-III gene in Samia cynthia ricini and its roles in response to bacterial infection. J. Invertebr. Pathol. 2018, 159, 61–70. [Google Scholar] [CrossRef]
- Zdybicka-Barabas, A.; Sowa-Jasiłek, A.; Stączek, S.; Jakubowicz, T.; Cytryńska, M. Different forms of apolipophorin III in Galleria mellonella larvae challenged with bacteria and fungi. Peptides 2015, 68, 105–112. [Google Scholar] [CrossRef]
- Zdybicka-Barabas, A.; Cytryńska, M. Involvement of apolipophorin III in antimicrobial defense of Galleria mellonella larvae. Comp. Biochem. Physiol. B Bioch. Mol. Biol. 2011, 158, 90–98. [Google Scholar] [CrossRef]
- Bolouri Moghaddam, M.R.; Tonk, M.; Schreiber, C.; Salzig, D.; Czermak, P.; Vilcinskas, A.; Rahnamaeian, M. The potential of the Galleria mellonella innate immune system is maximized by the co-presentation of diverse antimicrobial peptides. Biol. Chem. 2016, 397, 939–945. [Google Scholar] [CrossRef]
- Lee, Y.S.; Yun, E.K.; Jang, W.S.; Kim, I.; Lee, J.H.; Park, S.Y.; Ryu, K.S.; Seo, S.J.; Kim, C.H.; Lee, I.H. Purification, cDNA cloning and expression of an insect defensin from the great wax moth, Galleria mellonella. Insect Mol. Biol. 2004, 13, 65–72. [Google Scholar] [CrossRef] [PubMed]
- Yi, H.Y.; Chowdhury, M.; Huang, Y.D.; Yu, X.Q. Insect antimicrobial peptides and their applications. Appl. Microbiol. Biotechnol. 2014, 98, 5807–5822. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vertyporokh, L.; Wojda, I. Expression of the insect metalloproteinase inhibitor IMPI in the fat body of Galleria mellonella exposed to infection with Beauveria bassiana. Acta Biochim. Pol. 2017, 64, 273–278. [Google Scholar] [CrossRef] [PubMed]
- Wojda, I.; Jakubowicz, T. Humoral immune response upon mild heat-shock conditions in Galleria mellonella. J. Insect Physiol. 2007, 53, 1134–1144. [Google Scholar] [CrossRef]
- Dekkerová-Chupáčová, J.; Borghi, E.; Morace, G.; Bujdáková, H. Up-regulation of antimicrobial peptides gallerimycin and galiomicin in Galleria mellonella infected with Candida yeasts displaying different virulence traits. Mycopathologia 2018, 183, 935–940. [Google Scholar] [CrossRef]
- Rossoni, R.D.; Fuchs, B.B.; de Barros, P.P.; Velloso, M.D.S.; Jorge, A.O.C.; Junquiera, J.C. Lactobacillus paracasei modulates the immune system of Galleria mellonella and protects against Candida albicans infection. PLoS ONE 2017, 12, e0173332. [Google Scholar] [CrossRef]
- Ekengren, S.; Hultmark, D. Drosophila cecropin as an antifungal agent. Insect Biochem. Mol. Biol. 1999, 29, 965–972. [Google Scholar] [CrossRef]
- Lu, D.; Geng, T.; Hou, C.; Huang, Y.; Qin, G.; Guo, X. Bombyx mori cecropin A has a high antifungal activity to entomopathogenic fungus Beauveria bassiana. Gene 2016, 583, 29–35. [Google Scholar] [CrossRef]
- Yun, J.E.; Lee, D.G. Cecropin A-induced apoptosis is regulated by ion balance and glutathione antioxidant system in Candida albicans. IUBMB Life 2016, 68, 652–662. [Google Scholar] [CrossRef] [Green Version]
- Mukherjee, K.; Vilcinskas, A. The entomopathogenic fungus Metarhizium robertsii communicates with the insect host Galleria mellonella during infection. Virulence 2018, 9, 402–413. [Google Scholar] [CrossRef] [Green Version]
- Yu, B.; Sang, Q.; Pan, G.; Li, C.; Zhou, Z. A Toll-Spätzle pathway in the in the immune response of Bombyx mori. Insects 2020, 11, 586. [Google Scholar] [CrossRef]
- Valsecchi, I.; Lai, J.I.; Stephen-Victor, E.; Pillé, A.; Beaussart, A.; Lo, V.; Pham, C.L.L.; Aimanianda, V.; Kwan, A.H.; Duchateau, M.; et al. Assembly and disassembly of Aspergillus fumigatus conidial rodlets. Cell Surf. 2019, 5, 100023. [Google Scholar] [CrossRef] [PubMed]
- Wojda, I.; Kowalski, P.; Jakubowicz, T. Humoral immune response of Galleria mellonella larvae after infection by Beauveria bassiana under optimal and heat-shock conditions. J. Insect Physiol. 2009, 55, 525–531. [Google Scholar] [CrossRef] [PubMed]
- Andrejko, M.; Mizerska-Dudka, M.; Jakubowicz, T. Antibacterial activity in vivo and in vitro in the hemolymph of Galleria mellonella infected with Pseudomonas aeruginosa. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 2009, 152, 118–123. [Google Scholar] [CrossRef]
- Andrejko, M.; Zdybicka-Barabas, A.; Cytryńska, M. Diverse effects of Galleria mellonella infection with entomopathogenic and clinical strains of Pseudomonas aeruginosa. J. Invertebr. Pathol. 2014, 115, 14–25. [Google Scholar] [CrossRef]
- Kim, C.H.; Lee, J.H.; Kim, I.; Seo, S.J.; Son, S.M.; Lee, K.Y.; Lee, I.H. Purification and cDNA cloning of a cecropin-like peptide from the great wax moth, Galleria mellonella. Mol. Cells 2004, 17, 262–266. [Google Scholar]
- Stephen-Victor, E.; Karnam, A.; Fontaine, T.; Beauvais, A.; Das, M.; Hegde, P.; Prakhar, P.; Holla, S.; Balaji, K.N.; Kaveri, S.V.; et al. Aspergillus fumigatus cell wall α-(1,3)-glucan stimulates regulatory T-cell polarization by inducing PD-L1 expression on human dendritic cells. J. Infect. Dis. 2017, 216, 1281–1294. [Google Scholar] [CrossRef]
- Valsecchi, I.; Dupres, V.; Michel, J.P.; Duchateau, M.; Matondo, M.; Chamilos, G.; Saveanu, C.; Guijarro, J.I.; Aimanianda, V.; Lafont, F.; et al. The puzzling construction of the conidial outer layer of Aspergillus fumigatus. Cell Microbiol. 2019, 21, e12994. [Google Scholar] [CrossRef]
- Hasegawa, S.; Nordin, J.H. Enzymes that hydrolyze fungal cell wall polysaccharides. J. Biol. Chem. 1969, 244, 5460–5470. [Google Scholar] [CrossRef]
- Boman, H.G.; Nilsson-Faye, I.; Paul, K.; Rasmuson, T., Jr. Insect immunity I. Characteristics of an inducible cell-free antibacterial reaction in hemolymph of Samia cythia pupae. Infect. Immun. 1974, 10, 136–145. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fujikawa, T.; Kuga, Y.; Yano, S.; Yoshimi, A.; Tachiki, T.; Abe, K.; Nishimura, M. Dynamics of cell wall components of Magnaporthe grisea during infectious structure development. Mol. Microbiol. 2009, 73, 553–570. [Google Scholar] [CrossRef]
- Kiho, T.; Yoshida, I.; Katsuragawa, M.; Sakushima, M.; Usui, S.; Ukai, S. Polysaccharides in Fungi XXXIV: A polysaccharide from fruiting bodies of Amanita muscaria and the antitumor activities of its carboxymethylated product. Biol. Pharm. Bull. 1994, 17, 1460–1462. [Google Scholar] [CrossRef] [Green Version]
- Horisberger, M.; Lewis, B.A.; Smith, F. Structure of a (1→3)-α-d-glucan (pseudonigeran) of Aspergillus niger NNRL 326 cell wall. Carbohydr. Res. 1972, 23, 183–188. [Google Scholar] [CrossRef]
- Kaplan, E.L.; Meier, P. Nonparametric estimation from incomplete observations. J. Am. Stat. Assoc. 1958, 53, 457–481. [Google Scholar] [CrossRef]
- Rasmussen, L.; Pratt, N.; Hansen, M.R.; Hallas, J.; Pottegård, A. Using the “proportion of patients covered” and the Kaplan Meier survival analysis to describe treatment persistence. Pharmacoepidemiol. Drug Saf. 2018, 27, 867–871. [Google Scholar] [CrossRef]
- Cytryńska, M.; Zdybicka-Barabas, A.; Jakubowicz, T. Studies on the role of protein kinase A in humor al imane response of Galleria mellonella larvae. J. Insect Physiol. 2006, 52, 744–753. [Google Scholar] [CrossRef]
- Chalk, R.; Suliaman, W.Y. Antimicrobial peptides from small insects: Methods for insect culture and for the detection, visualization, isolation and sequencing of active hemolymph peptides. In Techniques in Insect Immunology; Wiesner, A., Dunphy, G.B., Marmaras, V.J., Morishima, I., Sugumaran, M., Yamakawa, M., Eds.; SOS Publications: Cambridge, MA, USA, 1998; pp. 109–124. [Google Scholar]
- Cytryńska, M.; Zdybicka-Barabas, A.; Jabłoński, P.; Jakubowicz, T. Detection of antibacterial polypeptide activity in situ after sodium dodecyl sulphate-polyacrylamide gel electrophoresis. Anal. Biochem. 2001, 299, 274–276. [Google Scholar] [CrossRef] [PubMed]
- Hultmark, S.; Engström, Å.; Bennich, H.; Kapur, R.; Boman, H.G. Insect immunity: Isolation and structure of cecropin D and four minor antibacterial components from Cecropia pupae. Eur. J. Biochem. 1982, 127, 207–217. [Google Scholar] [CrossRef]
- Taszłow, P.; Wojda, I. Changes in the hemolymph protein profiles in Galleria mellonella infected with Bacillus thuringiensis involve apolipophorin III. The effect of heat shock. Arch. Insect Biochem. Physiol. 2015, 88, 123–143. [Google Scholar] [CrossRef] [PubMed]
- Wojda, I.; Taszłow, P. Heat shock affects host-pathogen interaction in Galleria mellonella infected with Bacillus thuringiensis. J. Insect Physiol. 2013, 59, 894–905. [Google Scholar] [CrossRef] [PubMed]
- Laemmli, U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 1970, 227, 680–685. [Google Scholar] [CrossRef] [PubMed]
- Blake, M.S.; Johnston, K.H.; Russel-Jones, G.J.; Gotschlich, E.C. A rapid, sensitive method for detection of alkaline phosphatase-conjugated anti-antibody on Western blots. Anal. Biochem. 1984, 136, 175–179. [Google Scholar] [CrossRef]
- Fazekas de St Groth, S.; Webster, R.G.; Datyner, A. Two new staining procedures for quantitative estimation of proteins on electrophoretic strips. Biochim. Biophys. Acta 1963, 71, 377–546. [Google Scholar] [CrossRef]
- Schägger, H. Tricine-SDS-PAGE. Nat. Protoc. 2006, 1, 16–22. [Google Scholar] [CrossRef]
- Bradford, M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Gelman, A. Analysis of variance—Why it is more important than ever. Ann. Statist. 2005, 33, 1–53. [Google Scholar] [CrossRef] [Green Version]

| Protein Band No. | Approximate Molecular Mass (from SDS-PAGE) | Identified N-Terminal Sequence (Bands 1–5) Identification Based on nanoLC-MS/MS (Bands 6–7) | Identification of the Protein in Galleria mellonella | Most Similar Protein from the Other Organism |
|---|---|---|---|---|
| (Protein identity, database accession number, percent of identical amino acid residues with identified N-terminal sequence/the sequence coverage percent *) | ||||
| 1 | 30 kDa | IIGDKDGEAK FGEFPWMVAI | Galleria mellonella phenoloxidase- activating factor 2, NCBI XP_026749280.1, 100% | Zerene cesonia phenoloxidase- activating factor 2-like isoform X1, NCBI XP_038221010.1, 95% |
| 2 | 37kDa | LKDGPCPSYM DTCCLSPDRR | Galleria mellonella unidentified protein, whole genome shotgun sequence, GenBank NHTH01000050.1, 100% | Papilio polytes serine protease 44 isoform X1, NCBI XP_013146563.1, 89% |
| 3 | 55 kDa | ETAAVFPRGI PRTPIIIPRD | Galleria mellonella prophenoloxidase subunit 2, GenBank AAQ75026.1, 100% | Helicoverpa armigera phenoloxidase subunit 2-like, NCBI XP_021182404.1, 80% |
| 4 | 57 kDa | YEVPPAKLEA IWPKGLRVSL | Galleria mellonella beta-1,3-glucan recognition protein, GenBank CAK22401.1, 100% | Papilio xuthus beta-1,3-glucan- binding protein, GenBank KPI97191.1, 95% |
| 5 | 61 kDa | YVVPAAKLEA IYPKGLRVSI | Galleria mellonella unidentified protein, whole genome shotgun sequence, GenBank NHTH01000079.1, 100% | Plodia interpunctella beta-1,3-glucan- binding protein, UniProtKB/Swiss-Prot Q8MU95.1, 95% |
| 6 | 75kDa | (29–40) KLDPSLLNIQTK (41–49) VLSLLENWK (77–87) EVVTEFLSLYK (282–291) LANGLGEIPR | Galleria mellonella Arylphorin, Uniprot ID: Q24995_GALME, Mascot score: 170, sequence coverage 6% | |
| 7 | 250 kDa | (40–50) ANAPQAENLKK (306–323) LGISPEFGLQITNEISEK (341–353) YHLNIYSHPELGK (406–415) FLVDVTGSDK (471–481) FVLQVSPSSFK (482–490) LLLDTPIVK (491–501) VIELEGTAVVK (612–622) QLGEEINSDFK (792–804) ETQNSVVTALQAK (854–861) FDEQAQLR (941–951) SLDPFDEIPAK (1017–1035) NNGQVAVNGASHGYPVEEK (1100–1112) ISTSESEFGNSYR (1162–1177) QACIHAVAGNAENALR (1287–1298) FPYPIVYDTDLK (1331–1340) TIANVIDEIR (1479–1491) QFLLAAANAITQR (1492–1512) ILQESIVEECVCNYANPFVGR | Galleria mellonella Apolipophorin, Uniprot ID: Q68YP1_GALME, Mascot score: 794, sequence coverage 15.2% | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Stączek, S.; Zdybicka-Barabas, A.; Wojda, I.; Wiater, A.; Mak, P.; Suder, P.; Skrzypiec, K.; Cytryńska, M. Fungal α-1,3-Glucan as a New Pathogen-Associated Molecular Pattern in the Insect Model Host Galleria mellonella. Molecules 2021, 26, 5097. https://doi.org/10.3390/molecules26165097
Stączek S, Zdybicka-Barabas A, Wojda I, Wiater A, Mak P, Suder P, Skrzypiec K, Cytryńska M. Fungal α-1,3-Glucan as a New Pathogen-Associated Molecular Pattern in the Insect Model Host Galleria mellonella. Molecules. 2021; 26(16):5097. https://doi.org/10.3390/molecules26165097
Chicago/Turabian StyleStączek, Sylwia, Agnieszka Zdybicka-Barabas, Iwona Wojda, Adrian Wiater, Paweł Mak, Piotr Suder, Krzysztof Skrzypiec, and Małgorzata Cytryńska. 2021. "Fungal α-1,3-Glucan as a New Pathogen-Associated Molecular Pattern in the Insect Model Host Galleria mellonella" Molecules 26, no. 16: 5097. https://doi.org/10.3390/molecules26165097
APA StyleStączek, S., Zdybicka-Barabas, A., Wojda, I., Wiater, A., Mak, P., Suder, P., Skrzypiec, K., & Cytryńska, M. (2021). Fungal α-1,3-Glucan as a New Pathogen-Associated Molecular Pattern in the Insect Model Host Galleria mellonella. Molecules, 26(16), 5097. https://doi.org/10.3390/molecules26165097